Summary
Nuclear tumor suppressor p53 transactivates proapoptotic genes or antioxidant genes depending on stress severity, while cytoplasmic p53 induces mitochondrial-dependent apoptosis without gene transactivation. Although SIRT1, a p53 deacetylase, inhibits p53-mediated transactivation, how SIRT1 regulates these p53 multifunctions is unclear. Here we show that SIRT1 blocks nuclear translocation of cytoplasmic p53 in response to endogenous reactive oxygen species (ROS) and triggers mitochondrial dependent apoptosis in mouse embryonic stem (mES) cells. ROS generated by antioxidant free culture caused p53 translocation into mitochondria in wild type mES cells, but induced p53 translocation into the nucleus in SIRT1−/− mES cells. Endogenous ROS triggered apoptosis of wild type mES through mitochondrial translocation of p53 and BAX, but inhibited Nanog expression of SIRT1−/− mES, indicating that SIRT1 makes mES cells sensitive to ROS and inhibits p53-mediated suppression of Nanog expression. Our results suggest that endogenous ROS control is important for mES cell maintenance in culture.
Introduction
The tumor suppressor p53 has been studied almost exclusively as a transcription factor that functions by transactivating downstream targets involved in cell cycle arrest, apoptosis and DNA repair (Jin and Levine, 2001). p53 transactivates pro-apoptosis genes such as PUMA and bax and induces apoptosis (Miyashita and Reed, 1995; Nakano and Vousden, 2001). p53 also transcriptionally regulates genes which generate reactive oxygen species (ROS) and contribute to p53 mediated cell death (Macip et al., 2003; Polyak et al., 1997). However, p53 also upregulates genes that remove ROS, including glutathione peroxidase (GPX1) (Hussain et al., 2004; Tan et al., 1999), SESN1, and SESN2 (Budanov et al., 2004), suggesting that, depending on the situation which the cells are under, p53 acts as an apoptosis inducer or a survival factor against ROS insults. This is consistent with a recent report that p53 induces antioxidant enzymes at low H2O2 concentrations, and pro-oxidant genes at high concentrations of H2O2, suggesting that p53 may manifest different effects via the induction of different genes in response to ROS, perhaps dependent on the severity of stress (Sablina et al., 2005). p53 also induces apoptosis in transcription independent manners without gene transactivation. p53 has proapoptotic activity through direct action on mitochondria (Leu et al., 2004; Mihara et al., 2003) or p53 acts as a BH3 protein to induce Bax activation when forced to localize into the cytoplasm by nuclear import inhibitors (Chipuk et al., 2004). Collectively, p53 has multifunctional activities dependent on type and severity of stress.
The capacity of p53 to transactivate genes such as p21cip1/waf1 is inhibited by SIRT1, an enzyme that catalyzes deacetylation of acetyl-lysine residues of proteins such as histone and p53 (Vaziri et al., 2001). SIRT1 inhibits apoptosis induced by exogenous H2O2 (Kume et al., 2006), effects mediated by inhibition of p53 function to activate expression of proapoptotic genes. Since p53 has different roles in response to ROS which depend on the severity of stress, SIRT1 might have different effects on p53 function in response to ROS which might also depend on the severity of stress. Furthermore, it is unclear whether or how SIRT1 affects p53 proapoptotic activity independent of transactivation. There is no information on action of SIRT1 in mES cells, yet these cells express high levels of SIRT1 and p53 (Aladjem et al., 1998; McBurney et al., 2003) and mouse embryonic stem (mES) cells are sensitive to ROS, based on their inability to grow as well without the antioxidant 2-mecaptoethanol (2-ME). Thus, we evaluated SIRT1-mediated regulation of p53 in mES cells. Our experiments revealed an unexpected role of SIRT1 on enhanced responsiveness of mES cells to apoptosis by endogenous ROS.
Results
mES cells generate endogenous ROS in 2-ME free culture by SIRT1-mediated inhibition of the capacity of p53 to enhance expression of antioxidants
mES cells cannot grow in vitro as well without the antioxidant 2-ME, suggesting that mES cells are sensitive to ROS. mES cells highly express p53 and SIRT1 (Aladjem et al., 1998; McBurney et al., 2003). This led us to examine a role for SIRT1 on p53 function in mES cells in response to ROS. The requirement of 2-ME for mES cell growth suggests that mES cells may produce ROS endogenously. To assess this possibility, we examined whether withdrawal of 2-ME from culture medium influenced intracellular ROS levels, as assessed by dichlorofluorescein (DCF) staining and flow cytometry. Withdrawal of 2-ME from wild type cells induced a 2 to 4 fold increase in accumulated intracellular ROS within 24 hr after removal (Figure 1A, B) The increase in endogenous ROS is evident in the whole mES cell population (Figure 1B). To assess the effect of SIRT1 on 2-ME withdrawal-induced increase of intracellular ROS, we assayed intracellular ROS in a SIRT1−/− mES cell line in which the sir2α alleles were sequentially targeted with knockout vectors designed to delete exons 5 and 6 (McBurney et al., 2003). Immunoblot of proteins isolated from wild type and SIRT−/− mES cells confirmed the absence of SIRT1 in SIRT1−/− mES cells (Figure 1A, insert). Although ROS was significantly increased from 12 h after 2-ME withdrwal in SIRT1−/− mES cells, the extend to which 2-ME withdrawal increases ROS was much lower in SIRT1−/− mES cells than in wild type mES cells (Figure 1A, B). To rule out the possibility that SIRT1 mES cells have diverged, we replaced SIRT1 in SIRT1−/− mES cells using SIRT1 expressing lentivirus. Immunoblot of proteins isolated from SIRT1−/− mES cells infected with control or SIRT1 expressing lentivirus confirmed the replacement of SIRT1 in SIRT1−/− mES cells infected with SIRT1 expressing lentivirus (Figure 1B right lower panel, insert). The replacement of SIRT1 renders SIRT1−/− mES cells able to increase intracellular ROS levels. These data indicate that mES cells produce endogenous ROS, and this is associated with SIRT1 expression.
Figure 1. 2-ME withdrawal induces intracellular ROS increase by SIRT1-mediated inhibition of the capacity of p53 to enhance expression of antioxidants.
(A) Changes of intracellular ROS levels in wild-type and SIRT1−/− mES cells, 24 h after 2-ME withdrawal. ROS levels are expressed as the mean ± SEM of intensity of cell fluorescence. Intracellular ROS was measured by monitoring conversion of DCFH-DA to DCF using FACscan. Expression levels of SIRT1 in wild type and SIRT1−/− mES cells are shown in the insert. *, p<0.005 versus 0 h after 2-ME withdrawal. (B) Histograms for intracellular ROS level in wild-type, SIRT1−/−, and SIRT1−/− mES cells infected with control or SIRT1 expressing lentivirus at 0 and 24 h after 2-ME withdrawal. Expression levels of SIRT1 in SIRT1−/− mES cells infected with control or SIRT1 expressing lentivirus are shown in the insert of right lower panel. (C) Changes of mRNA expression levels of GPX1, SESN1 and SESN2 in wild type and SIRT1−/− mES cells 24 h after 2-ME withdrawal. Expression levels were detected by quantitative real time PCR. Fold changes were calculated from β-actin normalized Ct (threshold cycle) values; error bars represent SD of triplicate experiments. (D) The effect of tiron, an antioxidant, on 2-ME withdrawal induced expression of GPX1, SESN1 and SESN2. SIRT1−/− mES cells were incubated with or without 2-ME in the presence of the indicated dose of Tiron (07#x2013;2 mM). Twelve hours later, mRNA expression levels of GPX1, SESN1 and SESN2 were measured by quantitative real time PCR. Fold changes were calculated from β-actin normalized Ct values; error bars represent SD of triplicate experiments. (E) The expression levels of p53 in scrambled shRNA and p53 shRNA expressing wild and SIRT1−/− mES cells, as detected by western blot. Wild-type and SIRT−/− mES cells were infected with control or p53 shRNA expressing lentiviral particles and selected with 2 μg/ml puromycin. (F) Changes of intracellular ROS levels in scrambled shRNA and p53 shRNA expressing wild-type and SIRT1−/− mES cells 24 h after 2-ME withdrawal. ROS levels are expressed as mean ± SEM of intensity of cell fluorescence. (G) Changes of mRNA expression of GPX1, SESN1 and SESN2 in scrambled shRNA and p53 shRNA expressing SIRT1−/− mES cells, 24 h after 2-ME withdrawal. Expression levels were detected by quantitative real time PCR. Fold changes were calculated from β-actin normalized Ct values; error bars represent SD of triplicate experiments.
p53 induces antioxidant enzymes such as glutathione peroxidase 1 (GPX 1), Sesn 1 and Sesn 2 in response to low concentrations of H2O2 (Sablina et al., 2005). We therefore evaluated whether the 2-ME withdrawal induced-increase of ROS was caused by SIRT1-mediated inhibition of p53 induction of expression of these enzymes. Expression of these enzymes was analyzed in wild type and SIRT1 −/− mES cells by quantitative real time PCR. Expression of glutathione peroxidase 1 (GPX 1), Sesn 1 and Sesn 2 was increased seven fold, five fold and three fold, respectively, 24 h after 2-ME withdrawal in SIRT1−/−, but not in wild type, mES cells (Figure 1C). To test whether ROS generated by 2-ME stimulates the expression of antioxidant enzymes, we added tiron (4, 5-dihydroxy – 1, 3-benzene disulfonic acid), an antioxidant, instead of 2-ME and analyzed the expression of antioxidant enzymes. The addition of tiron abolished 2-ME withdrawal-induced expression of antioxidant enzymes (Figure 1D). These data suggest that SIRT1 might block expression of antioxidant enzymes by inhibition of p53 activities that are able to induce these enzymes upon exposure to low levels of ROS. To confirm this, we knocked-down p53 using a p53 shRNA lentiviral vector in wild type and SIRT1−/− mES cells (Figure 1E). p53 knock-down in SIRT1−/− mES cells restored the 2-ME withdrawal-induced ROS increase (Figure 1F) and blocked 2-ME withdrawal-induced expression of antioxidant enzymes in SIRT1−/− mES cells (Figure 1G). p53 knock-down in wild type mES cells did not affect the 2-ME withdrawal-induced ROS increase (Figure 1F). This demonstrates that SIRT1 inhibits the capacity of p53 to stimulate expression of antioxidant enzymes in response to endogenous ROS.
SIRT1 controls intracellular translocation of p53 by deacetylation
Consistent with a previous report that p53 expression is cytoplasmic in ES cells (Aladjem et al., 1998), we also found that most p53 exists in the cytoplasmic fraction (Figure 2A and 2C). Based on inhibition of p53 transactivation by SIRT1, we hypothesized that SIRT1 might block p53 translocation into the nucleus induced by ROS. Wild type or SIRT1−/− mES cells were cultured with or without 100 μM 2-ME for 24 h, cell lysates fractionated into cytoplasmic and nuclear fractions and western blot analysis performed. Of note, endogenous ROS induced by 2-ME withdrawal didn't induce nuclear translocation of p53 whereas SIRTl knockout induced nuclear translocation of p53 when 2-ME was omitted from the medium (Figure 2A left panel and 2B). The replacement of SIRT1 in SIRT1−/− mES cells abolished p53 nuclear translocation induced by 2-ME withdrawal (Figure 2A right panel and 2B). To show more direct evidence for the nuclear translocation, we performed confocal microscopy using FITC:anti-p53 antibody. 2-ME withdrawal induced nuclear translocation of p53 in SIRT1−/− mES cells and the replacement of SIRT1 abolished the nuclear translocation of p53 (Figure 2C). The p53 translocated into the nucleus was acetylated at 379 lysine, a known target for SIRT1(Cheng et al., 2003) (Figure 2D). This indicates that SIRT1 blocks nuclear translocation of p53 induced by oxidative stress via its deacetylation, and this results in inhibition of p53 function as a transcriptional regulator. This is confirmed by the finding that p21 expression was increased only in SIRT1−/− mES cells when 2-ME was omitted from the medium (Figure 2E). However, expression of PUMA, another transcriptional target for p53, was not changed by 2-ME withdrawal in SIRT1−/− mES cells (Figure 2E). To examine the effect of 2-ME withdrawal on the state of p53 phosphorylation, we explored the change of p53 phosphorylation at several sites using site-specific anti-phospho-p53 antibody in wild mES cells cultured with or without 2-ME for 24 h. We observed that serine 392 phosphorylation was increased (Figure 2F), but other phosphorylation on other sites such as serine 15 and 20 didn't change upon 2-ME withdrawal (data not shown). In addition, p53 translocated to mitochondria was highly phosphorylated at serine 392. This suggests that ROS stress generated by 2-ME withdrawal may modify the posttranslational state of p53 and drive p53 to the mitochondria. Taken together, SIRT1 controls subcellular localization of p53; this manifests in different cellular effects in response to ROS.
Figure 2. SIRT1 controls intracellular translocation of p53 by deacetylation.
(A) 2-ME withdrawal-induced nuclear translocation of p53 in SIRT1−/− mES cells. Wild-type, SIRT1−/− and SIRT1−/− mES cells infected with control or SIRT1 expressing lentiviral particles were cultured for 24 h with complete medium in the absence or presence of 2-ME. Cytoplasmic and nuclear fractions were prepared for p53 western blotting. The same blot was probed for β-actin (a control for cytoplasmic fractionation) and PCNA (a control for nuclear fractionation). (B) Ratios of cytosol to nuclear p53 as calculated by the quantification of protein band intensities from triplicate experiments. Error bars represent SD of triplicate experiments. *, p<0.05 versus wild-type mES with 2-ME. (C) Confocal images depicting the nuclear translocation of p53 in SIRT−/− mES removed of 2-ME. Wild-type, SIRT1−/− and SIRT1−/− mES cells infected with control or SIRT1 expressing lentiviral particle were cultured on gelatin coated cover slips for 9 h with complete medium in the absence or presence of 2-ME. The cells were fixed and stained with FITC labeled anti-p53 antibody and 4',6-diamidino-2-phenylindole (DAPI). Images were captured with a confocal microscope. (D) Increase of p53 acetylation in the nuclear fraction of SIRT1−/− mES cells. p53 in cytoplasmic and nuclear fractions of SIRT1−/− mES cells was immunoprecipitated and blotted with anti-p53 antibody and anti-acetyl p53 (lysine 379) antibody. (E) Changes of p21cip1/waf1 and PUMA mRNA expression, 24 h after 2-ME withdrawal, in wild type and SIRT1−/− mES cells. Wild type and SIRT1−/− mES cells were cultured for 24 h with complete medium in the absence or presence of 2-ME. Expression levels were detected by quantitative real time PCR. Fold changes were calculated from β-actin normalized Ct values; error bars represent SD of triplicate experiments. (F) 2-ME withdrawal-induced phosphorylation of p53 at serine 392 in wild-type mES cells. Wild-type mES cells were cultured for 24 h with complete medium in the absence or presence of 2-ME. Whole lysate or mitochondrial fraction in 2 ME withdrawal mES cells was prepared for p53 and phospho-p53 (s392) western blotting.
Endogenous ROS enhances apoptosis of wild-type, but not SIRT1−/−, mES cells
p53 has transcription-independent killing activity through direct action on mitochondria (Mihara et al., 2003). p53 dependent apoptosis through mitochondria is associated with the translocation of p53 and BAX (Mihara et al., 2003). In addition, p53 can be translocated into mitochondria by stimuli such as ROS and hypoxia (Sansome et al., 2001). Thus, we examined whether 2-ME withdrawal induced mitochondrial translocation of p53. Withdrawal of 2-ME resulted in translocation of p53 into mitochondria in wild type, but not in SIRT1−/−, mES cells (Figure 3A). This suggests that 2-ME withdrawal might induce mitochondrial dependent apoptosis. Indeed, 2-ME withdrawal induced apoptosis of wild type, but not of SIRT1−/−, mES cells (Figure 3B). The replacement of SIRT1 rendered SIRT1−/− mES cells sensitive to apoptosis when 2-ME was omitted from the medium (Figure 3B). 2-ME withdrawal enhanced mitochondrial translocation of BAX as well as p53 (Figure 3A) in wild-type cells. In SIRT1−/− mES cells, the mitochondria fraction contained BAX even in the presence of 2-ME. We tested whether 2-ME withdrawal-induced apoptosis is mediated by p53 and BAX through mitochondria. To confirm this, we selected mES cells expressing p53 and BAX shRNA by treatment with antibiotics after infection with each mRNA specific shRNA expressing lentivirus. p53 and BAX shRNA expressing lentivirus respectively suppressed p53 and BAX (Figure 3C). The 2-ME withdrawal induced apoptosis of wild type mES cells was completely blocked by p53 knockdown, but not by BAX knockdown (Figure 3D). These data indicate that SIRT1 allows p53 to accumulate in the cytosol upon ROS exposure, which results in mitochondrial dependent apoptosis.
Figure 3. 2-ME withdrawal induces apoptosis of wild-type, but not SIRT1−/−, mES cells.
(A) Western blot analysis of p53 and BAX from mictochondrial fractions, 12 h after 2-ME withdrawal, in wild-type and SIRT1−/− mES cells. Mitochondrial fractions were prepared for p53 and BAX western blotting as described in “Experimental Procedures”. The results of HSP-70 analysis are shown as a marker for mitochondrial fractions. The results of β-actin and PCNA analysis are shown to compare the extend to which the cysolic and nuclear fractions are contaminated. (B) Apoptosis of wild type, SIRT1−/− mES cells and SIRT1−/− mES cells infected with control or SIRT1 expressing lentiviral particles 60 h after 2-ME withdrawal as detected by FACS after Annexin V staining. Error bars represent SD of triplicate experiments. (C) Expression levels of p53 and BAX in scrambled shRNA, p53 shRNA or BAX shRNA expressing wild type mES cells, as detected by western blot analysis. (D) Apoptosis of scrambled shRNA, p53 shRNA or BAX shRNA expressing wild type mES cells 60 h after 2-ME withdrawal as detected by FACS after Annexin V staining. Error bars represent SD of triplicate experiments. Results shown in A–D are for 1 of 3 reproducible experiments with R1 mES cells.
Endogenous ROS induces inhibition ofNanog expression in mES cells
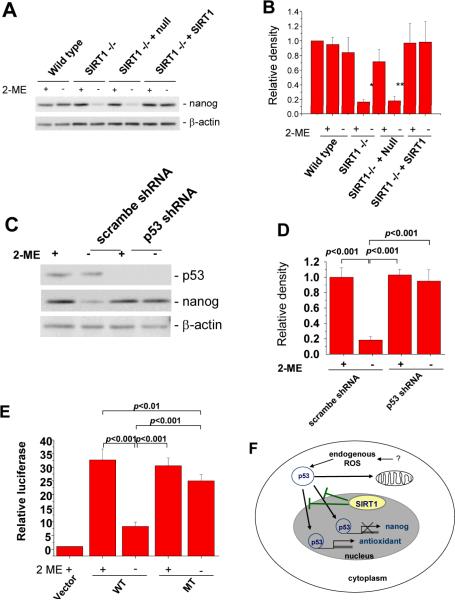
p53 suppresses expression of Nanog in ES cells, which is required to maintain cells in an undifferentiated status (Lin et al., 2005). Since 2-ME withdrawal caused p53 translocation into the nucleus in SIRT1−/− mES cells (Figure 3A), it was possible that the translocated p53 induced by 2-ME withdrawal also bound to the Nanog promoter and inhibited expression of Nanog. Indeed, 2-ME withdrawal suppressed Nanog expression in SIRT1−/−, but not in wild type, mES cells (Figure 4A and 4B). The replacement of SIRT1 in SIRT1−/− mES cells abolished 2-ME withdrawal-induced suppression of Nanog (Figure 4A and 4B). 2-ME withdrawal-induced suppression of Nanog was blocked by knock-down of p53 in SIRT1−/− mES cells (Figure 4C and 4D). Nanog promoter activity was decreased in SIRT1−/− mES cells without 2-ME, but mutation of p53 binding sites in the promoter region restored Nanog promoter activity (Figure 4E), indicating that Nanog suppression in SIRT1−/−mES cells is mediated by the inhibitory action of p53 on Nanog transcription. These observations suggest that SIRT1 maintains proliferation/self-renewal under a ROS stress by inhibition of p53-mediated suppression of Nanog expression. The combined data support a model whereby SIRT1 allows mES cells to proliferate/self-renew in the context of endogenous ROS stress by sacrificing mES cells exposed to ROS. A schematic representation for this model is shown in Figure 4F.
Figure 4. 2-ME withdrawal induces inhibition of Nanog expression.
(A) Nanog expression levels in wild type, SIRT1−/− mES cells and SIRT1−/− mES cells infected with control or SIRT1 expressing lentiviral particles upon 2-ME withdrawal, as detected by western blot analysis. Wild-type, SIRT1−/− mES cells and SIRT1−/− mES cells infected with control or SIRT1 expressing lentiviral particles were cultured for 24 h with complete medium in the absence or presence of 2-ME. (B) Relative density of nanog as calculated by β-actin normalized protein band intensities from triplicate experiments. Error bars represent SD of triplicate experiments. *, p<0.01 versus SIRT1−/− mES with 2-ME; **, p<0.05 versus SIRT1−/− + null mES with 2-ME. (C) p53 knockdown blocks 2-ME withdrawal-induced suppression of Nanog expression in SIRT1−/− mES cells. Scrambled shRNA and p53 shRNA expressing SIRT1−/− mES cells were cultured for 24 h with complete medium in the absence or presence of 2-ME. Nanog and p53 levels were detected by western blot. (D) Relative expression of nanog as calculated by β-actin normalized protein band intensities from triplicate experiments. Error bars represent SD of triplicate experiments. (E) 2-ME removal-induced inhibition of Nanog promoter activity and restoration of the activity by mutation of the p53 binding sites in SIRT1−/− mES cells. SIRT1−/− mES cells were transfected with wild type Nanog luciferase and mutant luciferase constructs, cultured for 24 h and then further incubated for 24 h in the absence or presence of 2-ME. Relative luciferase activities of wild type (WT) and mutant (MT) promoters for p53 binding sites are shown. Error bars represent SD of triplicate experiments. (F) Proposed model of SIRT1 role in p53 dependent apoptosis and Nanog expression. SIRT1 blocks nuclear translocation of p53 induced by ROS and triggers mitochondrial dependent apoptosis. SIRT1 also blocks inhibition of the p53 role in suppressing Nanog expression.
Since SIRT1 is considered a survival factor for ROS challenge (Kume et al., 2006), this might seem contrary to our data. To address this, we analyzed effects of exogenous H2O2 on apoptosis in wild type and SIRT1−/− mES cells. Low concentrations of H2O2 (0.2 mM to 0.4 mM) induced more apoptosis in wild type than in SIRT1−/−, mES cells (Figure S1A) and induced higher expression of antioxidant enzymes in SIRT1−/−, than in wild type, mES cells (Figure S1B), which is consistent with the above noted 2-ME withdrawal-induced apoptosis and expression of antioxidant enzymes. In contrast, higher concentrations of H2O2 (1 mM) induced more apoptosis in SIRT1−/−, than in wild type, mES cells (Figure S1A) and induced higher expression of BAX and PUMA in SIRT1−/−, than wild type, mES cells (Figure S1B), which is consistent with previous reports showing the survival effect of SIRT1 upon ROS challenge (Kume et al., 2006). p53 has opposite functions in response to ROS depending on ROS concentration: p53 acts as a survival factor by inducing antioxdants at low ROS and as an apoptosis inducer at high ROS (Sablina et al., 2005). SIRT1 appears to inhibit these opposite functions of p53, which results in opposite effects of SIRT1 on ROS-induced apoptosis. The survival effect of SIRT1 upon ROS challenge has been viewed at relatively high concentrations (0.5 mM to 1 mM H2O2) of ROS in which cells couldn't be placed physiologically. Intracellular H2O2 concentrations are within several μM even in stimulated cells (Arai et al., 2000; Werner, 2003). Our findings suggest that SIRT1 acts as an apoptosis inducer, instead of a survival factor, at physiological levels of ROS. This suggestion is consistent with reports that resveratrol, a known SIRT1 activator, induces apoptosis of cancer cells (Huang et al., 1999; She et al., 2001).
Differentiation of SIRT1−/− mES cells cultured in absence of 2-ME
We further explored the state of SIRT1−/− mES cells that are cultured in the condition of 2 ME withdrawal. When the cells were passaged one time in the absence of 2ME, alkaline phosphatase, SSEA-1 and Oct-4 levels of the cells were highly decreased compared to wild mES and SIRT1−/− mES cells cultured in the presence of 2-ME (Figure 5), indicating differentiation of SIRT1−/− mES cells upon 2-ME withdrawal. In addition, the morphology of the mES cell colonies was changed (Figure 5). These data indicate that SIRT1 regulates mES cell differentiation by controlling p53 activity that negatively modulates Nanog expression.
Figure 5. Differentiation of SIRT1−/− mES cells cultured in absence of 2-ME.
SIRT1−/− mES cells were passaged one time in absence of 2-ME. Alkaline phosphatase was stained as described in Materials and Methods. SSEA-1 and Oct-4 were measured by flow cytometry.
Discussion
p53 is stabilized and activated only in cells stressed with either DNA damage, abnormal proliferation signals, hypoxia, or ROS(Pluquet and Hainaut, 2001). However, mES cells highly express p53, even in the absence of stress (Aladjem et al., 1998). Moreover, p53 in mES cells is nonfunctional in the transactivation of genes such as p21cip1/waf1 (Aladjem et al., 1998). Although p53 in mES cells has no transcriptional activity for apoptosis-related genes, p53 is still able to induce apoptosis in a transcription-independent manner through mitochondria in mES cells exposed to ROS (Figure 3). The transcription-independent killing action of p53 appears to be preferable to p53-mediated transcription-dependent apoptosis in rapidly growing ES cells, since the latter is more rapid (Erster et al., 2004). Damaged ES cells may be removed by apoptosis instead of being repaired. mES may be ready to apoptose in response to endogenous ROS, since mES cells have high levels of p53 which are required for mitochondrial dependent apoptosis. The generation of endogenous ROS appears to be intrinsic and independent of cell growth rate since wild type and SIRT1−/− mES cells grow at similar rates (Figure S2), although the mechanism of ROS generation is not yet clear. It is possible that ROS generation in wild-type mES cells is associated with exposure of HIF-1α expressing mES cells to normoxia because hypoxic culture of wild-type mES cells did not induce endogenous ROS accumulation upon 2-ME withdrawal, and the growth rate in normoxic culture with 2-ME was similar to that in hypoxic culture without 2-ME (Figure S3). Hypoxic culture of human ES cells maintains a more undifferentiated state than during normoxic culture (Ezashi et al., 2005). This phenomenon might be due to the prevention of endogenous ROS generation.
p53 posttranslational modifications regulate various function of p53 (Lavin and Gueven, 2006). Numerous serine residues in both the N- and C- terminal regions of p53 are phosphorylated by various stimuli (Lavin and Gueven, 2006). The N-terminal phosphorylations stabilize p53 by inhibiting its interaction with its negative regulator Mdm2 (Canman et al., 1998; Chehab et al., 2000; Khosravi et al., 1999), while the C-terminal phosphorylatons enhance its transcriptional activity by inducing a conformational change (Hupp et al., 1992; Wang and Prives, 1995). Similarly, other posttranslational modifications such as ubiquitination, acetylation, and sumoylation also affect its proteolytic turnover and transcriptional activities (Brooks and Gu, 2006; Rodriguez et al., 1999). In this study we found that p53 posttranslational modification also regulates its transcription-independent activity, mitochondria dependent apoptosis, in 2-ME omitted culture of mES cells. In this condition, p53 phsophorylation seems to be a trigger for subcellular localization and its acetylation seems to act as a handle to determine its direction, nucleous or mitochondria.
p53 shows two different transactivation abilities, cell-cycle arrest- and apoptosis-inducing transactivation, by interacting with other proteins such as ASPP family members and hematopoietic zinc finger (Hzf) (Braithwaite et al., 2006; Das et al., 2007). ASPP1 and ASPP2 interact with p53 and specially enhance p53-induced apoptosis but not cell-cycle arrest, while iASPP binds p53 and inhibits p53-mediated apoptosis (Bergamaschi et al., 2006). Recently, Hzf has been found to preferentially transactivate p53 target genes involved in cell cycle arrest over its target genes involved in apoptosis (Das et al., 2007). It seems that p53 stimulated by 2-ME withdrawal in SIRT1−/− mES cells has only cell-cycle arrest inducing, but not apoptosis-inducing, transactivation abilility, which is based on our finding that 2-ME withdrawal in SIRT1−/− mES cells induced p21, but not PUMA, expression (Figure 2E). This suggest that 2-ME withdrawal in SIRT1−/− mES cells may induce the interaction of p53 with cell cycle arrest-related proteins such as iASPP and Hzf, potential effects which require study further.
Many studies have been performed in the context of modulation of cell behavior by SIRT1 through p53 deacetylation. Some suggest that SIRT1 might affect cellular response by modulating the acetylated status of p53 (Cheng et al., 2003; Langley et al., 2002; Luo et al., 2001; Vaziri et al., 2001). Other reports suggest that p53-dependent functions, such as apoptosis and transcription, are independent of SIRT1 (Kamel et al., 2006; Solomon et al., 2006). SIRT1−/− mES cells did not show any marked difference in expression of mES specific markers and growth rate (data not shown), indicating that SIRT1 probably plays no role under normal conditions. However, when mES cells are exposed to ROS, SIRT1 mediates mitochondrial dependent apoptosis through mitochondrial translocation of p53. Our data provide evidence for involvement of SIRT1 in modulation of cellular behavior through p53 deacetylation under special conditions, such as ROS stress. p53 knockdown and consequent loss of a p53 role in expression of antioxidants results in excessive oxidation of DNA, increased mutation rate and karyotype instability, effects which are prevented by the antioxidant N-acetylcysteine (Sablina et al., 2005). mES cells maintained with LIF express high levels of SIRT1, which inhibits p53-mediated induction of antioxidant enzymes. Suppression of p53 function by SIRT1 might make mES cells sensitive to ROS. It is possible that chromosomal abnormalities generated in ES cells might be due to intrinsic ROS generation in these cells. Our results suggest that the control of endogenously generated ROS is important for maintenance of healthy pluripotent mES cells in vitro.
Experimental Procedures
mES cell culture and LIF deprivation
mES cell line R1 (Nagy et al., 1993), and E14 line(Sawai et al., 1991), and SIRT1−/− mES derived from R1 (McBurney et al., 2003), were maintained on irradiated feeder layers in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal calf serum (FCS), glutamine, non-essential amino acids, antibiotics, 100 μM 2-ME and recombinant LIF. 1 × 106 mES cells were seeded onto gelatinized 10-cm plates in normal ES cell medium 12 h before initiation of experiments.
Annexin V flow cytometry assay
Annexin V binding was performed following the manufacturer's direction with a small modification in the procedure (Pharmingen, San Diego, CA, USA). Briefly, Annexin V (FITC labeled, 5 μL), propidium iodide (10 μL), and the binding buffer (400 μL) were added to a final volume of 500 μL, then analyzed by a FACScan flow cytometer (Becton Dickinson, Sunnyvale, CA) in 1 h.
Promoter constructs and luciferase assays for Nanog promoter activity
We cloned a 1-kb genomic DNA encompassing the promoter region of Nanog into a promoterless luciferase construct, pGL3Basic vector (Promega). The C/G to A/T mutations at the consensus p53-binding sites as described elsewhere (Bordone and Guarente, 2005) were introduced by site-directed mutagenesis. mES cells were plated at a density of 1 × 106 cells per 150 mm dish. Wild type and mutant promoter luciferase constructs (5 μg of each) were transfected into mES with 2 μg of RSV β-gal luciferase reporter plasmid by Lipofectamine according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Twenty fours after transfection, the luciferase activities were assayed according to the manufacturer's instructions (Promega) and luminescence was determined using a Lumat LB9507 luminometer (Berthold Systems, Pittsburgh, PA, USA). Luciferase activities were normalized against the β-galactosidase activity.
Western blot analysis
Protein extracts from 2 × 106 mES cells were separated by SDS–PAGE on a 12% polyacrylamide gel and transferred to PVDF membrane. The membrane was blocked with 2% bovine serum albumin and probed with antibodies against p53 (Clone No;FL-393 or Clone No;Pab246), SIRT1, HIF-1α, β-actin, PCNA, BAX (Santa Cruz Biotechnology, Santa Cruz, CA), mouse Nanog, HSP-70 (R&D, Minneapolis, MN) or acetyl p53 (Lys 379) (Cell Signaling Technology, Denvers, MA). The membrane was then incubated with a horseradish-peroxidase-conjugated secondary antibody and developed with enhanced chemilluminescence PLUS reagent from Amersham (Piscataway, NJ). To determine that the amount of protein in each lane was comparable, the membrane was cut at about the expected molecular weight of target proteins and β-actin, and the cut membranes were probed with antibodies against target proteins or a rabbit polyclonal antibody against β-actin, respectively.
Detection of intracellular ROS
ROS levels were determined by incubating cells with 10 μg/ml dichlorfluorescein diacetate (DCF-DA, Sigma-Aldrich) for 20 min at 37°C. We washed the cells twice in phosphate buffered saline (PBS), trysinized them and measured fluorescence with a FACScan flow-cytometer (Becton-Dickenson, San Diego, CA; exitation at 488 nm, emission at 515–545 nm). Data was analyzed with CELLQuest software Becton-Dickenson or WinMDI version 2.5 and mean fluorescence intensity was used to quantify responses.
Real time and RT-PCR analyses
Real time PCR or RT-PCR primer sets for GPX-1, SESN1, SESN2, p21cip1/waf1, Nanog, β-actin, BAX and PUMA were purchased from Superarray (Frederick, MD). Total RNA was isolated from mES cells with RNAeasy RNA Cleanup (Qiagen, Valencia, CA). RNA was treated with RNase-free DNAse for 20 min (Ambion, Austin, TX) at room temperature before reverse transcription with Superscript II RT (Invitrogen, Carlsbad, CA). Real-time PCR was performed on an ABI 7500 machine with SyBr Green PCR Master Mix (Qiagen, Valencia, CA). PCR conditions consisted of a 10 min hot start at 95 °C, followed by 40 cycles of 30 s at 95 °C and 1 min at 61 °C. The average threshold cycle (Ct) for each gene was determined from triplicate reactions and the levels of gene expression relative to β-actin were determined. RT-PCR was performed with One step RT-PCR system (Invitrogen, Carlsbad, CA).
lentivirus expression of shRNA
Oligonucleotides (5'-AAGTCTGTTATGTGCACGTACTTCAAGAGAGTACGTGCA CATAACAGACTTTTTTTT-3' for mouse p53, 5'AACAGATCATGAAGAC AGGGGTTCAAGAGACCCCTGTCTTCATGATCTGTTTTTTTT-3' for mouse BAX and 5'-GATGCATACCAGTGGCTATTTTTCATGAAAATAGCCACTGGTATGCAT CTTTTTT-3' for a scrambled sequence) containing the sense, loop, antisense sequence, and a polythymidine tract were annealed and ligated into into pLK0.1 downstream of the U6 promoter. To produce virus containing the shRNA-generating cassette, 293FT cells were transfected with 9 μg of Virapower™ packaging mix 4 and 3μg of pLK0.1 with Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA). Target cells were infected in the presence of Polybrene (8 μg/ml) and selected with puromycin at 3.0 μg/ml for 6 days.-
Replacement of SIRT1 in SIRT1−/− mES cells
For expression of SIRT1 in SIRT1−/− mES cells, a lentiviral vector expressing mouse SIRT1 was generated (Lenti-SIRT1). The mouse SIRT1 cDNA was isolated by PCR using a high fidelity polymerase (Invitrogen, Carlsbad, CA) and cloned into the AgeI and XhoI sites of pcDNA-cs-cgw lentiviral vector to generate Lenti-SIRT1. To produce virus containing the SIRT1-expressing cassette, 293FT cells were transfected with 9 μg of Virapower™ packaging mix 4 and 3μg of Lenti-SIRT1 with Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA). Target cells were infected in the presence of Polybrene (8 μg/ml) and selected with zeocin at 25 μg/ml for 6 days.
Subcellular fractionations
Nuclear and cytoplasmic fractions from mES cells were prepared using NucBusterTM protein extraction kit according to the manufacturer's instructions (Novagen, Sandiego, CA, USA). Mitochondria of mES cells were isolated by standard differential centrifugation. Mitochondrial pellet was resuspended in buffer containing 0.3 M mannitol and 5 mM MOPS (pH 7.2), overlayed onto a 30/70% percoll gradient and centrifuged for 40 min. at 20,000 rpm. Purified mitochondria were collected and solubilized in PBS containing 2% CHAPS.
Confocal microscopy
Wild type or SIRT1−/− mES cells were grown on gelatin-coated glass coverslips with or without 2-ME. Six hour later the cells were washed 3x with PBS, fixed for 20 min in 3% paraformaldehyde. The cells were then washed 3x with PBS and incubated with 50 mM NH4Cl for 10 min to quench crosslinking reaction. The cells were further washed 3x with PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min and washed 3x with PBS. The cells were then treated with blocking buffer (10% fetal bovine serum in PBS) for 30 min at room temperature and then incubated with FITC:monoclonal anti-mouse p53 antibody (Clone No:Pab246, Gene Tex, San Antonio, TX) for 2 h, followed by washing in PBS. Staining of nuclei was accomplished by incubation with DAPI (5 μg/ml) for 10 min. The slides were then washed extensively in PBS and mounted using vectashield medium (Vector Laboratories, Burlingame, CA). Specimens were viewed using an Olympus FluoView™ laser scanning confocal microscope.
Flow cytometry analysis of SSEA-1 and Oct-4
An aliquot of 1 × 106 cells was washed in ice cold PBS containing 2% FCS (PBS-2%FCS) serum and incubated with anti-mouse CD16/CD32 receptor monoclonal antibody at 1 μg/100 μl (Pharmigen; San Diego, CA) to block nonspecific binding of immunoglobulins to the mouse FcIII/II receptors. Cells to be stained for Oct-4 were fixed with 100 μl of IC Fixation buffer (Ebioscience; San Diego, CA) for 30 minutes at room temperature, washed 1x with PBS-2%FCS, permeabilized with Permeabilization Buffer (Ebioscience; San Diego, CA) and incubated with a 1:100 dilution rabbit-mouse Oct3/4 polyclonal antibody (Chemicon; Temecula, CA) for 30 minutes at room temperature. Cells were washed 2x with 1 ml of PBS-2%FCS, followed by staining with 1:100 dilution of FITC:goat anti-mouse IgG antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Finally, the cells were washed 2x with PBS-2%FCS, and resuspended in PBS-2%FCS for analysis on a FACScan flow cytometer (Becton Dickinson, Sunnyvale, CA). Cells analyzed for SSEA-1 were incubated first with a 1:10 dilution of monoclonal anti-SSEA-1 (Chemicon; Temecula, CA) for 40 minutes at 4°C, washed 2x with ice-cold PBS-2%FCS and incubated with a 1:500 dilution of FITC:goat anti-mouse IgM antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Alkaline phosphatase staining
mES cells were cultured for 3 days to obtain spheroidal colonies. Cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 5 minutes. After washing, the cells were stained with an Alkaline Phosphatase Staining Kit (Chemicon, Temecula, CA) at room temperature for 15 minutes in the dark. Cells were again washed with PBS, and colonies were visualized using an Olympus Microscope.
Statistical analysis
Statistical significance was determined using a two tailed Student's t test and one way Anova test followed by Scheffe's post hoc test.
Supplementary Material
(a) Apoptosis of wild type and SIRT1−/− mES cells, as detected by FACS after Annexin-V staining 12 h after treatment of cells with various doses of H2O2 (0 to 1 mM). Error bars represent standard errors. (b) Changes of mRNA expression levels of GPX1, SESN1, SESN2, BAX and PUMA in wild-type and SIRT1−/− mES cells 12 h after treatment of cells with various doses of H2O2 (0 to 1 mM). Expression levels were detected by RT-PCR. Results shown are for 1 of 3 (a) or of 2 (b) reproducible experiments with R1 mES cells.
Wild type and SIRT1−/− mES cells were cultured for the indicated times in mES culture medium and the cell number counted. Results shown are for 1 of 3 reproducible experiments with R1 mES cells.
Wild type mES cells (2.5 × 104 cells) were plated on gelatin coated dish, cultured for 40 h in the presence of various doses of 2-ME, and cell numbers counted. *, p<0.05 versus hypoxia without 2-ME. Results shown for R1 mES cells are representative of 2 additional experiment with E14 mES cells.
Acknowledgements
We are grateful to Michael W. McBurney (Ottawa Health Research Institute, Ottawa, Canada) for providing SIRT1−/− mES cells, to Kenneth Cornetta (Indiana University Vector Facility, Indiana, USA) for pcDNA-cs-cgw lentiviral vector, and to Young-June Kim for reading the manuscript.
References
- Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 1998;8:145–55. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- Arai M, Yoguchi A, Takizawa T, Yokoyama T, Kanda T, Kurabayashi M, Nagai R. Mechanism of doxorubicin-induced inhibition of sarcoplasmic reticulum Ca2+-ATPase gene transcription. Circ. Res. 2000;86:8–14. doi: 10.1161/01.res.86.1.8. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, Crook T, Lu X. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat. Genet. 2006;38:1133–41. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell. Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Braithwaite AW, Del Sal G, Lu X. Some p53-binding proteins that can function as arbiters of life and death. Cell Death Differ. 2006;13:984–93. doi: 10.1038/sj.cdd.4401924. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–9. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 2000;14:278–88. [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Nat'l Acad. Sci. U. S. A. 2003;100:10794–9. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–4. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–37. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol. Cell. Biol. 2004;24:6728–41. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Nat'l Acad. Sci. U. S. A. 2005;102:4783–8. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Ma WY, Goranson A, Dong Z. Resveratrol suppresses cell transformation and induces apoptosis through a p53-dependent pathway. Carcinogenesis. 1999;20:237–42. doi: 10.1093/carcin/20.2.237. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–86. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Amstad P, He P, Robles A, Lupold S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, Okamura S, Hofseth LJ, Moake M, Nagashima M, Forrester KS, Harris CC. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 2004;64:2350–6. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- Jin S, Levine AJ. The p53 functional circuit. J. Cell Sci. 2001;114:4139–40. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- Kamel C, Abrol M, Jardine K, He X, McBurney MW. SirT1 fails to affect p53-mediated biological functions. Aging Cell. 2006;5:81–8. doi: 10.1111/j.1474-9726.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc. Nat'l Acad. Sci. U. S. A. 1999;96:14973–7. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isono M, Isshiki K, Uzu T, Kashiwagi A, Koya D. Silent information regulator 2 (SIRT1) attenuates oxidative stress-induced mesangial cell apoptosis via p53 deacetylation. Free Radic. Biol. Med. 2006;40:2175–82. doi: 10.1016/j.freeradbiomed.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. Embo J. 2002;21:2383–96. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–50. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat. Cell Biol. 2004;6:443–50. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005;7:165–71. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Macip S, Igarashi M, Berggren P, Yu J, Lee SW, Aaronson SA. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol. Cell Biol. 2003;23:8576–85. doi: 10.1128/MCB.23.23.8576-8585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Bieman M, Th'ng J, Lemieux M. The absence of SIR2α protein has no effect on global gene silencing in mouse embryonic stem cells. Mol. Cancer Res. 2003;1:402–9. [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–90. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–9. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Nat'l Acad. Sci. U. S. A. 1993;90:8424–8. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 2001;7:683–94. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- Pluquet O, Hainaut P. Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett. 2001;174:1–15. doi: 10.1016/s0304-3835(01)00698-x. [DOI] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–5. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. Embo J. 1999;18:6455–61. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–13. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488:110–5. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- Sawai S, Shimono A, Hanaoka K, Kondoh H. Embryonic lethality resulting from disruption of both N-myc alleles in mouse zygotes. New Biol. 1991;3:861–9. [PubMed] [Google Scholar]
- She QB, Bode AM, Ma WY, Chen NY, Dong Z. Resveratrol-induced activation of p53 and apoptosis is mediated by extracellular-signal-regulated protein kinases and p38 kinase. Cancer Res. 2001;61:1604–10. [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Li S, Swaroop M, Guan K, Oberley LW, Sun Y. Transcriptional activation of the human glutathione peroxidase promoter by p53. J. Biol. Chem. 1999;274:12061–6. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- Werner E. Determination of cellular H2O2 production. Sci STKE. 2003;2003:PL3. doi: 10.1126/stke.2003.168.pl3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Apoptosis of wild type and SIRT1−/− mES cells, as detected by FACS after Annexin-V staining 12 h after treatment of cells with various doses of H2O2 (0 to 1 mM). Error bars represent standard errors. (b) Changes of mRNA expression levels of GPX1, SESN1, SESN2, BAX and PUMA in wild-type and SIRT1−/− mES cells 12 h after treatment of cells with various doses of H2O2 (0 to 1 mM). Expression levels were detected by RT-PCR. Results shown are for 1 of 3 (a) or of 2 (b) reproducible experiments with R1 mES cells.
Wild type and SIRT1−/− mES cells were cultured for the indicated times in mES culture medium and the cell number counted. Results shown are for 1 of 3 reproducible experiments with R1 mES cells.
Wild type mES cells (2.5 × 104 cells) were plated on gelatin coated dish, cultured for 40 h in the presence of various doses of 2-ME, and cell numbers counted. *, p<0.05 versus hypoxia without 2-ME. Results shown for R1 mES cells are representative of 2 additional experiment with E14 mES cells.