Abstract
Activin is required for testis development. Activin signals via the phosphorylation and nuclear accumulation of SMAD2 and SMAD3. We present novel findings of developmentally regulated activin signalling leading to specific transcriptional outcomes in testicular Sertoli cells. In immature, proliferating, Sertoli cells, activin A induces nuclear accumulation of SMAD3, but not SMAD2, although both proteins become phosphorylated. In post-mitotic differentiating cells, both SMAD proteins accumulate in the nucleus. Furthermore, immature Sertoli cells are sensitive to activin dosage; higher concentrations induce maximal SMAD3 nuclear accumulation and a small increase in nuclear SMAD2. Microarray analysis identified distinct transcriptional outcomes correlating with differential SMAD utilization and new activin target genes, including Gja1 and Serpina5, which are essential for Sertoli cell development and male fertility. In transgenic mice with altered activin bioactivity that display fertility phenotypes, Gja1 and Serpina5 are significantly altered. Thus, differential SMAD utilization in response to activin features during Sertoli cell maturation.
Keywords: morphogen gradient, development, TGFβ, Sertoli, testis, Rarres1, TIG-1, PDGF, Gja1, Serpina5
Introduction
During embryogenesis, morphogen gradients formed by transforming growth factor-beta (TGFβ) superfamily ligands contribute to the temporal and spatial regulation of diverse developmental events, including the determination of mesodermal competency by activin A in Xenopus (Dyson and Gurdon, 1998), wing disc formation by Dpp in Drosophila (Bollenbach et al., 2008), digit formation in response to BMP/BMP-like ligands in the chick (Hu et al., 2008; Suzuki et al., 2008) and border formation between the cerebral cortex and the telencephalic dorsal midline by BMP4 during embryonic brain development (Hu et al., 2008). Examination of morphogen gradients and target cell responsiveness using mathematical modelling (Bollenbach et al., 2008) as well as in vitro (Dyson and Gurdon, 1998) and in vivo (Hu et al., 2008; Suzuki et al., 2008) approaches established that cell fate is determined by the availability of ligand and the distance of the target cell from the source of ligand production. It is well established that disruption or dysregulation of TGFβ signalling can alter developmental outcomes and is associated with disease (reviewed in (Chang et al., 2002)).
Whereas the above studies examined cellular responses to a morphogen gradient at a specific developmental timepoint, we sought to examine circumstances where ligand production changes during development. As a model developmental system, we examined activin A signalling in the murine fertility-determining Sertoli cell, the nurse cell to developing sperm. Activin A is required for the proliferation of immature Sertoli cells and for quantitatively normal sperm production in the adult. Production of activin A changes dramatically during testis development, being 15-fold higher in the neonatal testis compared to the adult testis, with a dramatic drop in production occurring around puberty (Barakat et al., 2008; Buzzard et al., 2003; Buzzard et al., 2004). During puberty, the Sertoli cell switches to a post-mitotic phenotype associated with its terminal differentiation, which occurs by 12 days of age. The post-mitotic Sertoli cell is functionally different to the immature Sertoli cell, exhibiting dynamic changes in gene expression required for the support of ongoing spermatogenesis. Thus, the study of Sertoli cell maturation, in an environment of changing activin levels, presents the opportunity to examine the mechanisms by which activin responses are developmentally regulated and to determine the consequences of altered activin signalling on target gene expression.
Sertoli cells and the adjacent peritubular myoid cells, which surround the seminiferous cord, are the predominant sources of activin A in the testis, and both immature and post-mitotic Sertoli cells express activin receptors (de Winter et al., 1992; de Winter et al., 1994; Fragale et al., 2001; Kaipia et al., 1992). A discrete upregulation of type IIA activin receptor subunit (Acvr2) mRNA just prior to Sertoli cell maturation has been observed (Fragale et al., 2001), but how this impacts on the Sertoli cell responsiveness to activin or whether post-mitotic cells can respond to activin has not been directly tested. In vitro, activin A is a well established mitogen for immature Sertoli cells (Boitani et al., 1995; Buzzard et al., 2003; Fragale et al., 2001), whereas in vivo, delayed testis maturation, subfertility or infertility, including Sertoli cell-derived cancer, are consequences of altered activin production (reviewed in (Itman et al., 2006)).
Activin signals via the transcription factors SMAD2 and SMAD3. Upon activin binding to cell surface receptors, SMAD2 and SMAD3 are phosphorylated, complex with SMAD4, accumulate in the nucleus and regulate expression of target genes (Heldin et al., 1997; Massague et al., 2005). Several studies have established that SMAD2 and SMAD3 are not functionally redundant. Strikingly different phenotypes are observed in mice lacking SMAD2, which exhibit embryonic lethality (Waldrip et al., 1998), compared to mice lacking SMAD3, which develop to adulthood (Zhu et al., 1998), and common as well as distinct roles for SMAD2 and SMAD3 have been identified in TGFβ signal transduction (Kretschmer et al., 2003; Piek et al., 2001; Yang et al., 2003) with signal intensity reflected by the concentration of SMAD proteins within the nucleus (Inman et al., 2002). These parameters have not yet been examined in relationship to activin signalling.
While documenting the dynamic patterns of SMAD synthesis and localization during mouse testis development, we discovered that SMAD2 and SMAD3 exhibit different subcellular localization, with SMAD2 apparently excluded from the nuclei of immature, proliferating Sertoli cells from 6 day old mice. This was not observed in post-mitotic Sertoli cells from 15 day old mice. The present study profiles the responsiveness of immature Sertoli cells to activin A and also examines responsiveness to the three TGFβs, which also signal via SMAD2 and SMAD3 (Heldin et al., 1997; Massague et al., 2005). We show that immature Sertoli cells are uniformly and dose-dependently responsive to activin A but not to TGFβ1, TGFβ2 or TGFβ3. We characterize this newly revealed developmentally regulated response to activin, identifying a novel phenomenon in which the regulated nuclear accumulation of SMAD3 and of phosphorylated SMAD2 governs activin responses and developmental outcomes. The physiological relevance of these findings is established by the identification of activin target genes that display different expression levels when activin production is altered in testes of mice with altered activin levels, preceding the onset of fertility phenotypes observed in these mice.
Results
Regulated utilization of SMAD2 and SMAD3 in activin A signal transduction features in Sertoli cell development
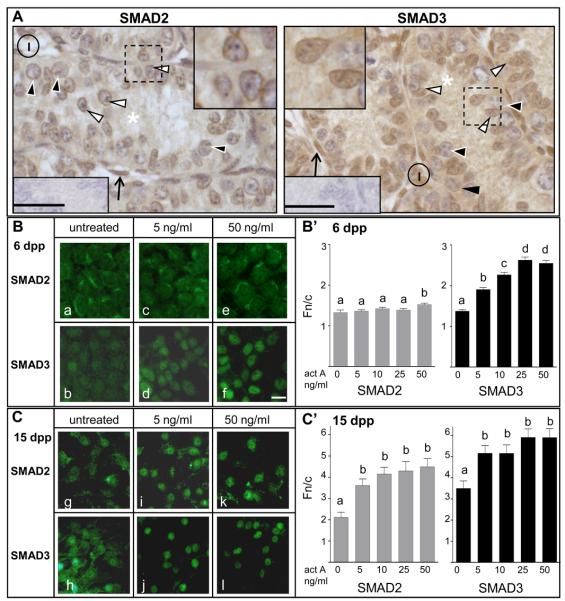
Activin promotes the proliferation of immature rodent Sertoli cells, however a response to activin by post-mitotic Sertoli cells has not been documented (Boitani et al., 1995; Buzzard et al., 2003), nor have the specific transcriptional outcomes been characterized. We began by performing immunohistochemistry to determine the localization of the activin signal transducers SMAD2 and SMAD3, reasoning that a nuclear signal would indicate their active involvement in signal transduction. Each exhibited a different subcellular localization in Sertoli cells of 6 day old mouse testes (Figure 1A). SMAD2 was readily detected within the cytoplasm and displayed distinctive localization at the nuclear membrane with little to no intranuclear staining. In contrast, SMAD3 was also cytoplasmic but was easily detected within the nucleus.
Figure 1. A: SMAD3, but not SMAD2, is detected in Sertoli cell nuclei of 6 dpp mouse testes.
Immunohistochemical localization of SMAD2 and SMAD3 in 6 dpp mouse testes, indicated by brown staining and counterstained with Harris haematoxylin to stain chromatin blue. SMAD2 and SMAD3 were visible within the Sertoli cell cytoplasm (asterisk) and SMAD3 was readily detectable within the nuclei of Sertoli cells (white arrowheads) however little to no SMAD2 was apparent in Sertoli cell nuclei. The dotted black squares in each image are enlarged as insets. Spermatogonia: black arrowhead; Sertoli cell cytoplasm: white asterisk; Sertoli cell nucleus: white arrowhead; interstitial cells: I; peritubular cells: open black arrow. Scale bar represents 50 μm.
B,C: SMAD2 and SMAD3 utilization by Sertoli cells is developmentally regulated. Detection of SMAD2 and SMAD3 by immunofluorescence in 6 and 15 dpp mouse Sertoli cells (Fig 1B and 1C respectively). Cells were cultured for 24 hours in serum-free medium then either left untreated or stimulated with activin A for 45 minutes. In 6 dpp Sertoli cells, SMAD2 sub-cellular localization did not change visibly following activin A treatment, whereas SMAD3 showed increasing nuclear localization with increasing activin concentrations. In 15 dpp Sertoli cells, both SMAD2 and SMAD3 proteins accumulated in the nucleus with 5 ng/ml activin A treatment. Scale bar = 25 μm.
B′, C′: Quantitation of SMAD2 and SMAD3 nuclear accumulation in Sertoli cells Changes in subcellular localization of SMAD2 and SMAD3 in 6 dpp (n=3; Fig 1B′) and 15 dpp (n=2; Fig 1C′) Sertoli cells following activin A treatment (from Figure 1B and 1C respectively) was measured by analysis of confocal images, with an increase in the ratio of nuclear to cytoplasmic fluorescence (Fn/c) indicating accumulation of protein in the nucleus. In 6 dpp Sertoli cells, SMAD3 (black bars) exhibited nuclear accumulation at all concentrations of activin tested, to a point of saturation (50 ng/ml). SMAD2 distribution did not change at lower concentrations of activin A tested (grey bars), yet a small but significant increase in SMAD2 nuclear accumulation was measured when SMAD3 nuclear accumulation was saturated. In 15 dpp Sertoli cells, both SMAD2 and SMAD3 showed similar profiles of nuclear accumulation, with significant and maximal increases in SMAD2 and SMAD3 nuclear accumulation observed with 5 ng/ml activin A. Statistical significance was determined using one-way ANOVA and Tukey's post hoc test (P<0.05). Different letters signify significant differences between samples; error bars indicate SEM. ***P<0.001.
We then compared the in vitro responsiveness to activin of immature (6 dpp) Sertoli cells and post-mitotic Sertoli cells (15 dpp). An established approach was used to enrich Sertoli cells and culture them on laminin for 24 hours in serum-free conditions (Itman and Loveland, 2008), after which they were either left untreated or stimulated with concentrations of activin A ranging from 5 to 50 ng/ml, for 45 minutes. Our aim was to find a concentration that approximated physiological conditions, that of nuclear localization of SMAD3, but not SMAD2, in immature Sertoli cells in vivo (Figure 1A). SMAD proteins were then visualized by immunofluorescence using specific antibodies (see Supplementary Figure S1). In immature (6 dpp) Sertoli cells, SMAD2 and SMAD3 were detected in both the nucleus and cytoplasm in the absence of stimulation (Figure 1B (a,b)). Upon treatment with activin A, SMAD3 exhibited nuclear accumulation, which was enhanced with higher activin doses (Figure 1B (d,f); 5 and 50 ng/ml pictured). However, SMAD2 localization appeared unaltered at all activin concentrations, remaining distributed between the nucleus and cytoplasm (Figure 1B (c,e); 5 and 50 ng/ml). In post-mitotic (15 dpp) Sertoli cells, SMAD2 and SMAD3 were nuclear and cytoplasmic in the absence of stimulation (Figure 1C (g,h)) but both SMAD2 and SMAD3 accumulated in the nucleus following treatment with 5 and 50 ng/ml activin A (SMAD2: Figure 1C (i,k); SMAD3: Figure 1C (j,l)). We continued our study with the consideration that 5 ng/ml activin A (0.1 pmol per 2 cm2 surface area of well) is likely to be physiologically relevant as this induced nuclear accumulation of only SMAD3 in immature Sertoli cells. We also predicted that 50 ng/ml activin A exceeded the physiological concentration in the immature testis.
As the lack of SMAD2 nuclear accumulation in response to activin in immature Sertoli cells was somewhat surprising, this was further examined in 6 dpp spermatogonia (Supplementary data Figure S2A, B). Treatment with 10 ng/ml activin A induced nuclear accumulation of both SMAD2 and SMAD3 in spermatogonia, confirming that the lack of nuclear accumulation of SMAD2 in immature Sertoli cells is a particular feature of these cells.
As SMAD2 and SMAD3 also transduce TGFβ signals, we examined the response of immature Sertoli cells to TGFβ1, TGFβ2 and TGFβ3. This was performed in an analogous fashion to the activin studies, using concentrations of 1 and 5 ng/ml. As depicted in Supplementary Figure S3, no nuclear accumulation of SMAD2 or SMAD3 was observed in response to TGFβ1 or TGFβ3 whereas following treatment with TGFβ2, a small proportion (less than 0.5%) of cells exhibited nuclear accumulation of SMAD3. The bioactivity of each of the three TGFβs was confirmed by treatment of mouse dermal fibroblasts (Verbruggen and Salomon, 1980), in which nuclear localization of SMAD3 was observed for all three growth factors. We therefore focussed on the response of Sertoli cells to activin A for the remainder of the study.
Analysis of confocal images of untreated Sertoli cells and cells treated with 5 to 50 ng/ml activin A was performed to quantify activin A-induced SMAD2 and SMAD3 nuclear accumulation. These Fn/c values confirmed that in immature Sertoli cells, activin A exposure led to significant dose-dependent nuclear accumulation of SMAD3, with maximal nuclear SMAD3 measured in samples treated with 25 and 50 ng/ml (Figure 1B′). SMAD2 nuclear levels did not change with lower activin concentrations but a small, significant, increase in SMAD2 nuclear accumulation was measured when the SMAD3 level in the nucleus was maximal. In post-mitotic Sertoli cells, both SMAD2 and SMAD3 exhibit similar profiles of nuclear accumulation, with maximum nuclear accumulation for both SMAD proteins observed at 5 ng/ml activin A and no graded response observed (Figure 1C′).
To determine whether immature Sertoli cells exhibited temporal responsiveness to activin A, SMAD2 localization was examined by immunofluorescence 10, 45 and 90 minutes after treatment with 10 ng/ml activin A. No change in SMAD2 subcellular distribution was observed (data not shown). These data establish that Sertoli cells exhibit developmental-stage specific SMAD responses to activin A.
SMAD2 and SMAD3 are phosphorylated in response to activin A but nuclear accumulation of SMAD2 at lower activin concentrations is impaired
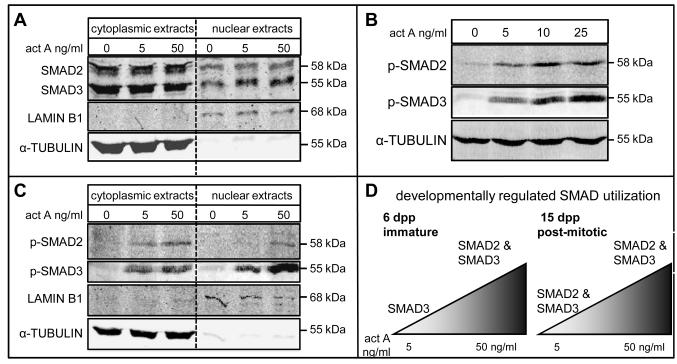
To confirm the immunofluorescence results depicting differential SMAD2 and SMAD3 nuclear accumulation in immature Sertoli cells in response to activin A, Western blot analysis was performed with nuclear and cytoplasmic extracts from untreated Sertoli cells and cells treated with 5 or 50 ng/ml activin A (Figure 2A). In untreated cells, both SMAD2 and SMAD3 were detected in nuclear lysates. Following activin A treatment, no change in SMAD2 protein levels in nuclear lysates was apparent at either 5 or 50 ng/ml, but SMAD3 protein levels in nuclear lysates increased. Antibodies to LAMIN B1 (Goldman et al., 2002) and to α-TUBULIN (Piperno et al., 1987) demonstrated that the nuclear and cytoplasmic fractions were highly purified. These results are consistent with the data presented in Figure 1, confirming that SMAD3 is preferentially used in activin A induced signal transduction by immature Sertoli cells. The absence of a detectable increase in SMAD2 nuclear localization by Western blot in immature cells treated with 50 ng/ml of activin A suggests that this assay is less sensitive than immunofluorescence.
Figure 2. A: Activin A increased the amount of SMAD3, but not SMAD2, in nuclear lysates.
Western blot analysis of SMAD2 and SMAD3 in nuclear and cytoplasmic extracts of 6 dpp Sertoli cells cultured for 24 hours in serum-free medium and either left untreated or stimulated with activin A at 5 or 50 ng/ml for 45 minutes (n=2). α-TUBULIN and LAMIN B1 signals indicate highly purified cytoplasmic and nuclear lysates, respectively. An increase in SMAD3 protein in the nucleus of cells treated with 5 and 50 ng/ml activin A was evident whereas there was no change in the amount of SMAD2 present in nuclei of treated cells.
B: Both SMAD2 and SMAD3 are phosphorylated upon stimulation of immature Sertoli cells with activin A: Western blot analysis of SMAD2 (n=5) and SMAD3 (n=3) phosphorylation in 6 dpp Sertoli cells cultured for 24 hours in serum-free medium and either left untreated or stimulated with activin A at 5, 10 or 25 ng/ml for 45 minutes. ALPHA-TUBULIN was used as a loading control. Increased phosphorylation of SMAD2 and SMAD3 was readily detected upon treatment of cells with activin A, each exhibiting the same profile of phosphorylation across the range of concentrations tested.
C: SMAD2 is phosphorylated but is not increased in nuclear lysates from Sertoli cells treated with 5 ng/ml activin A: Western blot analysis of phospho-SMAD2 and phospho-SMAD3 in nuclear and cytoplasmic extracts of 6 dpp Sertoli cells cultured for 24 hours in serum-free medium and either left untreated or stimulated with activin A at 5 or 50 ng/ml for 45 minutes (n=4). ALPHA-TUBULIN and LAMIN B1 indicate purity of cytoplasmic and nuclear lysates, respectively. Phosphorylated SMAD2 and phosphorylated SMAD3 were detected in cytoplasmic fractions of cells treated with both 5 and 50 ng/ml activin A. Phosphorylated SMAD2 was absent from nuclear fractions of cells treated with 5 ng/ml activin A but is readily detected in nuclear fractions from cells treated with 50 ng/ml activin A. Intense signals for phosphorylated SMAD3 were detected in nuclear fractions of 5 and 50 ng/ml activin A treated samples.
D: Summary of developmentally regulated SMAD usage in Sertoli cells in response to activin A: Immature Sertoli cells exhibited a dose-dependent response to activin A stimulation in vitro, with nuclear accumulation of SMAD3, but not SMAD2 at lower concentrations of activin. At higher concentrations, both SMAD2 and SMAD3 accumulated in the nucleus. Nuclear accumulation of both SMAD2 and SMAD3 was observed in post-mitotic Sertoli cells at all concentrations of activin tested.
Although SMAD proteins constantly shuttle between the nucleus and cytoplasm, nuclear accumulation of SMADs following ligand stimulation requires the phoshorylation of two C-terminal serine residues by activated receptors complexes (reviewed by (Heldin et al., 1997; Massague et al., 2005)). Western blot analysis performed on whole cell lysates from cells treated with 5, 10 or 25 ng/ml activin A using antibodies to the phosphorylated forms of SMAD2 and SMAD3 confirmed that SMAD2 was phosphorylated at all concentrations of activin A examined, exhibiting a similar profile to phosphorylated SMAD3 (Figure 2B). Hence, the inability of SMAD2 to accumulate in the nucleus following activin A treatment of immature Sertoli cells is not due to a lack of C-terminal phosphorylation.
Having established that, in immature Sertoli cells treated with low levels of activin, SMAD2 and SMAD3 are phosphorylated but only SMAD3 accumulates in the nucleus, the intracellular distributions of phosphorylated SMAD2 and phosphorylated SMAD3 were examined by Western blot of nuclear and cytoplasmic extracts from untreated cells and cells treated with 5 or 50 ng/ml activin A (Figure 2C). Treatment with 5 ng/ml activin A induced a marked increase in phosphorylated SMAD3 in the nuclear and cytoplasmic lysates relative to untreated cells. Phosphorylated SMAD2 was visible in the cytoplasmic fraction however, in the nuclear fraction, a very faint signal for phosphorylated SMAD2 was detected in some samples (data not shown) whereas in other samples, no signal was detected. In cells treated with 50 ng/ml activin A, phosphorylated SMAD2 and SMAD3 were consistently detected in cytoplasmic and in nuclear fractions. These results demonstrate that when immature Sertoli cells are stimulated with activin A, phosphorylated SMAD2 can accumulate in the nucleus, but this capacity is greatly reduced at lower activin concentrations.
In summary, these data have established that immature and post-mitotic Sertoli cells exhibit different SMAD-responses to activin A stimulation and that immature cells are sensitive to activin doses between 5 and 50 ng/ml (Figure 2D).
Identification of candidate genes differentially expressed under conditions correlating with specific SMAD usage
As immature and post-mitotic Sertoli cells exhibit different signal transduction responses to activin A and immature Sertoli cells exhibit sensitivity to activin levels, we set out to identify whether differential SMAD2/3 nuclear accumulation altered transcriptional outcomes downstream of activin signalling. Microarray analyses were conducted under conditions correlating with specific SMAD nuclear accumulation profiles to address several questions.
Cells were cultured as described above and treated with activin A for 6.5 hours. Cells from 6 dpp pups were exposed to 5 or 50 ng/ml activin A, corresponding to differential SMAD nuclear accumulation whereas cells from 15 dpp mice were treated with 5 ng/ml activin A only (Figure 2D). Affymetrix microarray analysis was performed on RNA prepared from two independent culture experiments. A gene was considered to be expressed if the signal value was 50 or greater and to be activin regulated if a minimum two-fold change was detected relative to other treatment groups. Genes identified by microarray analysis as activin regulated are deposited for public viewing, query and download in the gene expression data repository Gene Expression Omnibus (GSE13106).
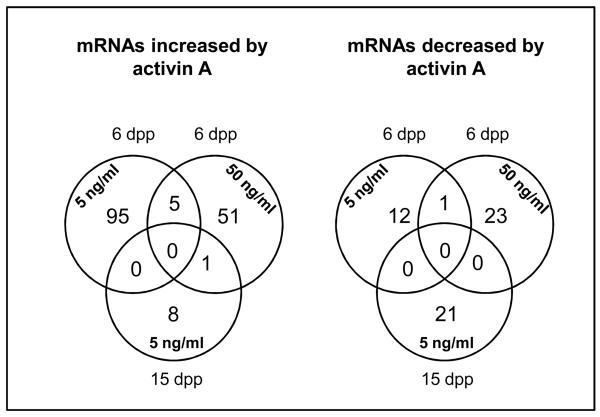
Both upregulated and downregulated genes were detected following exposure of cells to activin. As defined in the Gene Ontology Database (http://www.ebi.ac.uk/GOA/), these relate to a diverse range of biological processes, including DNA and chromatin modification, the ubiquitin cycle, protein transport, RNA processing and ion transport. An important finding is that some of these genes are required for male fertility. Also of note is that most genes exhibiting a two-fold expression change were only found in one treatment group, suggesting that SMAD2 and SMAD3 may have distinct activities in the regulation of activin target gene expression in Sertoli cells (Figure 3). For example, 95 candidate genes were upregulated relative following treatment of 6 dpp cells with 5 ng/ml activin A whereas 51 different candidate genes were upregulated in 6 dpp cells treated with 50 ng/ml activin A. Only five genes were commonly upregulated at both concentrations (Arhgap27, BB022048, Hsd17b1, Mup1 and Spna1). A similar pattern was evident in genes exhibiting downregulation in immature cells, with 12 candidates downregulated following treatment with 5 ng/ml activin A, whereas 23 different candidate genes were downregulated with 50 ng/ml activin A; only Iqseq1 was downregulated at both concentrations. A single gene, Rnf14, was commonly upregulated in 6 dpp treated with 50 ng/ml activin A and 15 dpp cells, whereas an entirely different gene set was downregulated by activin in 15 dpp cells; none of these overlapped with those identified in 6 dpp cells. A selection of genes spanning a range of biological processes that were identified by microarray is listed in Table 1 alongside their fold-change relative to untreated cells.
Figure 3.
Candidate genes identified by microarray exhibiting a minimum two-fold change in expression in response to activin A were grouped according to the conditions under which they were regulated. Most genes were upregulated or downregulated under specific conditions consistent with the hypothesis that SMAD2 and SMAD3 have distinct activities in the regulation of gene expression in Sertoli cells at different developmental stages.
Table 1. mRNAs altered in response to activin A.
A selection of candidate activin-regulated genes identified by Affymetrix microarray in immature (6 dpp) and post-mitotic (15 dpp) murine Sertoli cells, expressed as fold change relative to untreated cells. Putative biological functions are indicated. ND = not detected.
| mRNAs upregulated in response to activin A (fold change relative to untreated cells) | |||||
|---|---|---|---|---|---|
| Accession number |
Symbol | Function | 6 dpp | 15 dpp | |
| 5 ng/ml | 50 ng/ml |
5 ng/ ml |
|||
| BF322785 | Mup1 | chemical signalling | 4.2 | 2.3 | ND |
| NM_010475 | Hsd17b1 | steroidogenesis | 2 | 4.3 | ND |
| NM_008291 | Hsd17b3 | steroidogenesis | 1.85 | 2.3 | ND |
| BE446879 | Rbm5 | RNA binding | 1.8 | 0.8 | 0.9 |
| NM_011980 | Zfp14 | zinc finger protein | 2.4 | 1.1 | 1.3 |
| NM_007706 | Socs2 | suppressor of cytokine signalling | 1.4 | 2.6 | 1.2 |
| NM_010950 | Numb-like | Numb is an inhibitor of notch signalling | 0 | 0.8 | 2 |
| NM_008785 | Serpina5 | serine protease inhibitor, male fertility | 1.4 | 2.5 | 1.7 |
| NM_021344 | Tescalcin | Ca2+ binding protein | 1.6 | 2.6 | 1.1 |
| NM_031380 | Fstl3 | repressor of activin signalling | 1.2 | 2.2 | 1 |
| BC023427 | Pdgfb | growth factor | 1.4 | 2 | 1 |
| AV343573 | Cbln4 | member of TNFα superfamily | 1.5 | 2 | ND |
| AF249668 | Rnf14 | ring finger protein | ND | 2.4 | 2.4 |
| BC013493 | Slc13a2 | solute carrier | ND | 9 | 0.8 |
| AK019971 | Prrx2 | homeobox protein | 1.5 | 2.2 | ND |
| mRNAs downregulated in response to activin A (fold change relative to untreated cells) | |||||
| Accession number |
Symbol | Function | 6 dpp | 15 dpp | |
| 5 ng/ml | 50 ng/ml |
5 ng/ ml |
|||
| NM_007522 | Bad | pro-apoptotic | 0.4 | 0.7 | 0 |
| BB035017 | Rarres1 | tumour suppressor | 0.7 | 0.4 | 0.8 |
| NM_007825 | Cyp7b1 | cytochrome P450 family | 0.8 | 0.5 | ND |
| AF300870 | Csen | calsenilin binding protein | 0.6 | 0.2 | ND |
| BC026153 | Epha7 | Eph receptor a7 | 0.7 | 0.4 | ND |
| AI451018 | Plxnb3 | axon guidance receptor | 0.5 | 0.6 | ND |
Activin-regulated gene expression is developmentally regulated
Several genes were selected for further analysis by quantitative PCR. These include Pdgfb, a known SMAD-target gene (Bruna et al., 2007), as well as genes not previously identified as being activin regulated, including Tescalcin, Cryl1, Rarres1, Gja1 and Serpina5. Testicular expression of Tescalcin, which encodes a calcium binding protein, and of Pdgfb has previously been described (Gnessi et al., 1995; Loveland et al., 1995; Perera et al., 2001); the findings presented here are the first identification of Cryl1 and Rarres1 transcripts in the testis. The requirement for Gja1 and Serpina5 expression for normal Sertoli cell development and male fertility has already been established; mice with Sertoli cell-specific ablation of Gja1 (Brehm et al 2007, Sridharan et al 2007) or which do not express Serpina5 (Anway et al 2005) are infertile.
Total RNA isolated from two additional independent cultures of 6 and 15 dpp cells was treated in a manner identical to that described for microarray samples to measure expression levels of selected activin target genes by Real Time PCR. Data normalization with either 18S or beta-actin generated similar results, and these experimental outcomes were consistent with our findings that 6 dpp Sertoli cells exhibit dose-dependent responsiveness to activin A.
Treatment of immature Sertoli cells with 5 ng/ml activin A elicited no significant increase in Gja1, Pdgfb, Serpina5 or Tesc mRNA levels relative to untreated cells (Figure 4A). In contrast, exposure to 50 ng/ml activin A significantly elevated the transcriptional response of immature Sertoli cells for each of these genes (Gja1, 1.5-fold relative to untreated cells; Pdgfb, 2.8-fold; Serpina5, 2.1-fold; Tesc, 3.8-fold). Notably, treatment of 15 dpp cells with just 5 ng/ml activin significantly increased the expression of Pdgfb (2.2-fold relative to untreated 15 dpp cells), Serpina5 (1.9-fold) and Tesc (2.1-fold); Gja1 expression increased 1.9-fold but this was not significant.
Figure 4. Quantitative analysis of several candidate activin A target genes in 6 dpp and 15 dpp Sertoli cells.
Real Time PCR was used to validate the activin-regulation of several candidate activin target genes. 6 dpp Sertoli cells were treated for 6.5 hours with 5 (grey bars) or 50 ng/ml (black bars; bars span lowest and highest values measured) activin A and 15 dpp Sertoli cells were treated with 5 ng/ml activin A (white bars). Two independent cultures were analyzed and gene expression was compared to that measured in untreated cells (white bars), which was set at 1. Normalization to beta-actin or 18S generated equivalent results. Copy number of target genes was determined by the comparative threshold cycle method (ΔΔCT) using the Pfaffl method (Pfaffl, 2001). Fold-change in expression relative to untreated cells is depicted under bars (* P<0.05, ** P<0.01, ***P<0.001).
A: mRNAs increased by activin A. Gja1, Pdgfb, Serpina5 and Tesc were all significantly upregulated in 6 dpp cells treated with 50 ng/ml activin A, but not in cells treated with 5 ng/ml. Pdgfb, Serpina5 and Tesc were significantly upregulated in 15 dpp cells following treatment with just 5 ng/ml; Gja1 exhibited a 1.9-fold increase relative to untreated cells but this did not reach significance.
B: mRNAs decreased by activin A. Dhcr24, Cryl1 and Rarres1 were all significantly downregulated in 6 dpp cells treated with 5 ng/ml activin A, with greater downregulation observed upon treatment with 50 ng/ml. In 15 dpp cells, no change in expression of Dhcr24 was observed in response to activin whereas Cryl1 and Rarres1 were both significantly downregulated.
Dose-dependent downregulation of mRNA levels was also measured (Figure 4B). In immature cells, significant downregulation of Dhcr24 was observed at 5 and 50 ng/ml activin A, to 84% and 80% of levels measured in untreated cells, respectively. Interestingly, Dhcr24 expression in 15 dpp cells was not altered, indicating developmental stage-specific responsiveness of some loci to transcriptional machinery. In 6 dpp cells, Cryl1 was not significantly altered by 5 ng/ml activin A but was downregulated to 0.75 of untreated levels by 50 ng/ml. Rarres1 was downregulated by 5 and 50 ng/ml activin A, to 0.5 and 0.25 of the levels measured in untreated cells, respectively. Downregulation of Cryl1 (to 0.7 of untreated levels) and Rarres1 (to 0.5 of untreated levels) was also observed in 15 dpp cells.
A clear outcome from these experiments is the observation that treatment of immature Sertoli cells with 50 ng/ml activin A induces a transcriptional response similar to that induced in post-mitotic cells at a 10-fold lower activin concentration. In immature cells, this is associated with maximal SMAD3 and uncharacteristic SMAD2 nuclear accumulation correlating with the observation in post-mitotic cells of nuclear accumulation of both SMAD2 and SMAD3. These findings suggest that regulated SMAD utilization in activin signal transduction is a feature that is required for appropriate responses by immature Sertoli cells to activin A.
Altered in vivo activin levels affect activin target gene expression in mouse testes
To examine the potential for activin to regulate these target genes in vivo, we examined whether their expression was altered in testes from mice with reduced activin bioactivity (InhbaBK/BK) and in mice with excessive activin signalling (Inha−/−). Both mouse strains display testicular phenotypes; adult InhbaBK/BK mice have smaller testes and reduced sperm output (Brown et al., 2000) and Inha−/− mice develop Sertoli cell-derived stromal tumours (Matzuk et al., 1992). Reduced Sertoli cell number in InhbaBK/+ and InhbaBK/BK mice relative to wildtype littermates is evident at 7 dpp, although they are not significantly different to each other whereas the number of Sertoli cells in testes from 7 dpp Inha−/− mice is not different to wildtype (our unpublished results).
Real Time PCR confirmed that Gja1 and Serpina5 mRNA levels are unchanged in Inhba+/+ and InhbaBK/+ testes but are significantly lower in InhbaBK/BK testes (Table 3), with Gja1 transcripts present at less than half and Serpina5 approximately 40% of the levels measured in testes from wildtype and heterozygous littermates. In testes from Inha−/− pups, Gja1, Serpina5 and Tesc mRNA levels were all significantly greater (2-, 1.3- and 2.5-fold respectively) than in wildtype littermates. These data establish that activin-regulation of Gja1 and Serpina5 occurs in vivo, demonstrating the impact of modulated activin production on transcription in immature Sertoli cells and its important physiological outcomes.
Table 3. Expression of activin target genes is altered in testes from mice with altered activin levels.
Real Time PCR was used to measure differences in the expression of Gja1 and Serpina5 in testes of 7 dpp InhbaBK/BK pups (n=6) and their wildtype littermates (n=6) and Inha−/− pups (n=6) and their wildtype littermates (n=6). Gja1 and Serpina5 expression was significantly reduced in testes from InhbaBK/BK pups whereas significantly increased expression of Gja1, Serpina5 and Tescalcin was measured in testes from 7 dpp Inha−/− pups.
| Altered expression of mRNAs in testes of Inha−/− mice | |
|---|---|
| Gja1 | 2.03-fold increase (P<0.001) |
| Serpina5 | 1.31-fold increase (P<0.01) |
| Tesc | 2.48-fold increase (P<0.01) |
| Altered expression of mRNAs in testes of InhbaBK/BK mice | |
| Gja1 | 0.51 of wildtype levels (P<0.001) |
| Serpina5 | 0.41 of wildtype levels (P<0.01) |
Discussion
Activin A signal transduction is regulated at the level of SMAD nuclear accumulation
The testis is a powerful model for the study of activin signalling in development as activin is required for normal testis development and its production is highly regulated. We have determined that immature Sertoli cells are particularly sensitive to extracellular cues and are vulnerable to subtle changes in activin levels that can trigger them to deviate from their normal developmental pathway. By effectively mimicking a morphogen gradient in vitro, we have shown that Sertoli cell responses to activin A are different at different developmental stages. Furthermore, immature Sertoli cells are sensitive to activin dosage, apparent in the increased phosphorylation of both SMAD2 and SMAD3 that is observed with increasing activin concentration. It has previously been shown that receptor occupancy within a morphogen gradient is a key determinant in specifying the outcome of activin signalling (Dyson and Gurdon, 1998) and that sensitivity to activin concentration leads to a dose-dependent increase in nuclear accumulation of exogenous Smad2 (Saka et al., 2007). Our data add to these findings by illustrating that activin can also induce dose-dependent nuclear accumulation of SMAD3 and that the phosphorylated forms of SMAD2 and SMAD3 differ in their capacity to accumulate in the nucleus.
Similar to the temporally regulated nuclear accumulation of SMAD2 described here during Sertoli cell maturation, developmentally regulated SMAD2 nuclear accumulation in response to activin has previously been described in Xenopus (Grimm and Gurdon, 2002; Saka et al., 2007). During Xenopus embryogenesis, nuclear accumulation of GFP-tagged SMAD2 corresponds to the time of mesoderm induction. However, prior to and following this period of mesodermal competence, activin fails to induce GFP-SMAD2 nuclear accumulation or to induce the expression of mesodermal markers. Phosphorylation within the linker region of SMAD2 was established as the determining factor blocking SMAD2 nuclear accumulation, with this event required for the termination of mesodermal induction. Thus, blocking SMAD2 nuclear accumulation is a developmental switch that restricts the size of the cell population directed into the mesodermal lineage and permits progressive development of the embryo (Grimm and Gurdon, 2002). In an analogous manner, our finding that activin-induced nuclear accumulation of SMAD2 in Sertoli cells is restricted to post-mitotic cells may reflect specific requirements for SMAD2-mediated transcription in post-mitotic, but not immature, Sertoli cells.
Our current understandings of SMAD2 and SMAD3 target gene regulation is based on studies in cell lines using overexpression or knockdown technologies (Kim et al., 2005; Kretschmer et al., 2003; Labbe et al., 1998; Uemura et al., 2005; Yang et al., 2003), Smad2- or Smad3-deficient fibroblasts (Piek et al., 2001) or from phenotypic analysis of knockout mice (Waldrip et al., 1998; Zhu et al., 1998). We undertook to characterize activin signal transduction by measuring the activation and utilization of endogenous SMAD proteins and by distinguishing between SMAD2 and SMAD3. The novel findings we present are therefore particularly insightful and highlight a level of complexity in activin signal transduction that has not previously been described. These data suggest that, in cells that express both SMAD2 and SMAD3, intrinsic mechanisms exist to modulate the participation of phosphorylated SMAD2 versus phosphorylated SMAD3 in regulating gene expression. Ongoing studies in our laboratory are addressing the mechanisms underpinning this differential utilization and the selective roles that SMAD2 and SMAD3 play in the regulation of activin target genes which we expect will considerably expand our understanding of TGFβ superfamily signalling.
Regulated activin responsiveness is required for Sertoli cell transition from an immature to terminally differentiated state
This study identified numerous activin target genes previously known to be expressed in the testis, including Gja1 and Serpina5. Gja1 encodes the gap junction protein connexin43 required for Sertoli cell maturation; Sertoli cell-specific knockout mice are infertile (Sridharan et al., 2007). Serpina5 encodes a serine protease inhibitor, also required by post-mitotic Sertoli cells; male mice that do not express Serpina5 are infertile (mPCI knockout mice) (Uhrin et al., 2000). The select upregulation of these genes in post-mitotic cells supports the hypothesis that temporal control of these genes confers developmentally regulated gene expression. These genes, and others identified by microarray herein, also represent a valuable selection of novel markers that can distinguish immature from post-mitotic Sertoli cells, the utilization of which will advance investigations into testis development and testicular pathologies where altered Sertoli cell maturation is suspected.
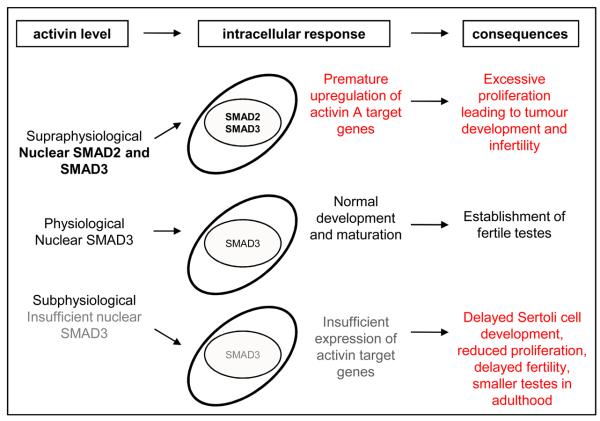
The hypothesis that regulated responsiveness to activin is a feature of normal Sertoli cell maturation and that altered responses by immature Sertoli cells would influence developmental outcomes in vivo was confirmed using two mouse models of altered activin synthesis (Figure 5). In a chronic model of altered activin signalling, we predict that the delayed testis development in InhbaBK/BK mice is, at least in part, due to impaired Sertoli cell development arising from insufficient production of Gja1 and Serpina5. In contrast, elevated activin exposure led to excessive nuclear accumulation of SMAD3 and premature nuclear accumulation of SMAD2 in immature Sertoli cells, associated with increased transcription of activin target genes. Importantly, altered expression of these genes preceded the onset of the phenotypes associated with Sertoli cell malfunction in mouse models: the delayed fertility observed in InhbaBK/BK mice and the Sertoli cell-derived tumours in Inha−/− mice (Sridharan et al., 2007; Uhrin et al., 2000). Collectively, these data indicate that maximal nuclear accumulation of SMAD3 and premature nuclear accumulation of SMAD2 in immature Sertoli cells alters gene expression profiles resulting in detrimental consequences on their development. The demonstration that Smad3 deletion relieves the Sertoli cell tumour-forming phenotype of the inhibin alpha knockout mouse (Li et al., 2007; Looyenga and Hammer, 2007) reinforces the concept that regulation of SMAD3 nuclear accumulation at this developmental stage is of particular importance.
Figure 5. Tightly regulated activin production and Sertoli cell responsiveness to activin are required for normal testis develoment.
Immature Sertoli cells preferentially transduce activin A signals via SMAD3. Insufficient exposure of Sertoli cells to activin during testis development delays the onset of fertility and reduces the expression of activin target genes, associated in vitro with insufficient SMAD3 nuclear accumulation. Excessive exposure to activin A, associated with nuclear accumulation of both SMAD2 and SMAD3, results in premature expression of activin target genes preceding the development of Sertoli cell-derived stromal tumours and infertility.
Activin as a coordinator of other biological processes
The approach undertaken here to identify activin target genes has generated considerable information about the potential influence of activin on other biological processes. For example, substantial dose-dependent downregulation of Rarres1 (also called TIG-1, tazarotene-induced gene 1) was observed. Rarres1 was previously identified as a tumour-suppressor that is upregulated by retinoic acid signalling (Nagpal et al., 1996), and loss of Rarres1 expression is a seen in a broad spectrum of cancers (Youssef et al., 2004) including colorectal (Wu et al., 2006), esophageal (Mizuiri et al., 2005), breast (Wilson et al., 2006) and prostate cancer, in which ectopic expression significantly reduces invasiveness and tumorigenicity (Jing et al., 2002). In these cases, downregulation of retinoic acid signalling was thought to be the cause of reduced Rarres1 expression, however this should be revisited in the light of a recent report of elevated serum activin A levels in patients with prostate or breast cancer (Incorvaia et al., 2007).
The calcium-binding protein tescalcin, negative signalling regulators Fstl3, Soc2 and Numb-like, the growth factor Pdgfb and two genes encoding steroidogenic enzymes (Hsd17b1, Hsd17b3) were upregulated by activin A. Each of these genes influence developmental and disease processes. Tescalcin directs megakaryocytic differentiation by controlling expression of the transcription factors Fli-1, Ets-1 and Ets-2 (Levay and Slepak, 2005); Fstl3, Socs2 and Numb-like downregulate the response of cells to cytokines whereas PDGFB is a potent mitogen and angiogenic factor (Betscholtz et al., 2001). The potential for altered steroidogenic capacity in response to activin suggests that modulation of activin responsiveness could influence androgen production, of relevance not only to male fertility but also to other androgen-regulated processes such as prostate development and disease (Taplin, 2008).
To conclude, we have identified a novel developmental regulation of activin signalling in which the nuclear accumulation of SMAD2 and SMAD3 is modulated, presumably to direct appropriate activin signalling outcomes during development. The physiological relevance of these findings is evident in the identification of activin target genes required for normal Sertoli cell function and male fertility. Insufficient or excessive in vivo exposure of immature Sertoli cells to activin leads to altered activin target gene expression, preceding the onset of fertility phenotypes observed in mice with altered activin levels. These data highlight that activin production and target cell responsiveness must be tightly regulated to drive specific transcriptional outcomes appropriate to developmental stage.
Experimental Procedures
Experimental animals and tissues
Wildtype mice (CBAxC57Bl6 F1) were housed at the Monash University Central Animal Services, InhbaBK/BK mice at the Monash Medical Centre Animal Facility, and Inha−/− at the Baylor College of Medicine, Houston, Texas (as described in the NIH Guide for the Care and Use of Laboratory Animals). All investigations conformed to the NHMRC/CSIRO/AAC Code of Practice for the Care and Use of Animals for Experimental Purposes and were approved by the Monash University Standing Committee on Ethics in Animal Experimentation.
Testes for RNA extraction were decapsulated, snap frozen and stored at −80°C until use. Intact tissue for immunohistochemistry was fixed for five hours in Bouin's fixative immediately after collection, dehydrated through a graded ethanol series, paraffin embedded and placed as 4 μm sections onto Superfrost Plus II slides (Lomb Scientific, Sydney Australia).
Cell culture
Enrichment and culture of Sertoli cells was performed as described (Itman and Loveland, 2008). After 24 hours, floating spermatogonia were aspirated and medium replaced. Medium was changed again after one hour and cells were exposed to medium with or without activin A, TGFβ1, TGFβ2 or TGFβ3 for 45 minutes (R&D Systems, Minneapolis, MN). Following activin stimulation, cells used for immunofluorescence were fixed in a solution of 2% paraformaldehyde (BDH Laboratory Supplies, Poole, UK) in PBS for 10 minutes, washed twice in PBS and stored in PBS at 4°C. For protein analysis of whole cell lysates, cells were scraped and collected by centrifugation at 13,000 rpm at room temperature for 1 minute; cell pellets were snap frozen on dry ice and stored at −80°C. Nuclear and cytoplasmic lysates were prepared as per (Rosner et al., 2007). For microarray and Real Time PCR analysis, cells were exposed to activin A for 6.5 hours.
Western Blot
Cell lysate preparation and Western blots were performed as described (Itman and Loveland, 2008). Anti-SMAD2 and anti-SMAD3 (Zymed, San Francisco, CA) were used at 0.25 μg/ml; anti-phospho-SMAD2 and anti-phospho-SMAD3 (Cell Signalling Technologies, Beverly, MA), anti-LAMINB1 (Zymed), anti-β-TUBULIN and anti-α-TUBULIN (Sigma-Aldrich, St. Louis, MO) antibodies were used at 1:1000 dilution. Bound primary antibodies were detected using goat-anti-rabbit Alexa Fluor 680 (Invitrogen, Carlsbad CA) or goat-anti-mouse IR-800 (Rockland, PA, USA) and measured with the Li-Cor Odyssey System. Measurement of SMAD2 phosphorylation was performed five times and SMAD3 three times. In nuclear and cytoplasmic lysates, total SMAD2 and SMAD3 were measured twice and phosphorylated SMAD2 and SMAD3 four times.
Immunohistochemistry and immunofluorescence
Immunohistochemistry (Meehan et al., 2000) and immunofluorescence were performed as previously described (Itman and Loveland, 2008) using anti-SMAD2 and anti-SMAD3 antibodies (Zymed; 2μg/ml and 0.5μg/ml respectively). Verification of SMAD2 and SMAD3 antibody specificity is presented in Figure S1. Bound primary antibody was detected using biotinylated anti-rabbit antibody (Chemicon, Temecula, CA) diluted 1:500 in 0.5% BSA/PBS and streptavidin-FITC (Chemicon) or streptavidin-TR (Calbiochem, La Jolla, CA) diluted 1:500 in 0.5% BSA/PBS. Culture and immunofluorescence experiments for activin A titrations were performed three times for 6 dpp (days post partum), twice for 15 dpp with a minimum of 200 cells measured per treatment. Images were captured using a Fluorview FV300 Olympus Scientific confocal laser scanning microscope and analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). The level of fluorescence in the nucleus (Fn) and in the cytoplasm (Fc) were each measured and presented as a ratio (Fn/Fc [Fn/c]); an increase in Fn/c indicates nuclear accumulation (Argentaro et al., 2003).
Statistical Analysis
Statistical analysis was performed with the statistical software GraphPad Prism 5 by one-way ANOVA and Tukey post hoc test. Differences were considered significant when P < 0.05, with data expressed as mean +/− SEM.
Mouse genotyping
Inha−/− mice were genotyped as described (Li et al., 2007). InhbaBK/BK mice were genotyped using the following primers: BK1 forward primer (CTGTTGAGTGGAAGGAGAG, designed from NM_008380 [Inhba promoter]), BK2 reverse primer (CGATGAGCCGAAAGTCGATG, designed from NM_008381 [Inhbb mature domain]) and BK3 reverse primer (GAGATGGGAAGAAGAAGA, designed from NM_008380 [Inhba mature domain]). Combined within one reaction, these primers generate a 750 bp wildtype product (BK1 and BK3) and a 500 bp mutant product (BK1 and BK2).
RNA isolation and cDNA synthesis
RNA was prepared using TRIzol (Invitrogen) and DNAse1 (Ambion, TX, USA) and 500 ng was reverse transcribed using 100 U Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's guidelines using random hexamers (2.5 μM final concentration).
Microarray Processing
Microarray analysis was performed on RNA prepared from two independent culture experiments. Biotinylated cRNA target was created from 10 micrograms of total RNA generated by reverse transcription using an oligo(dT) primer with a T7 promoter followed by in vitro transcription using the MEGAScript kit (Ambion) with biotinylated cytosine and uridine triphosphate. Labeled cRNA was fragmented, hybridized to Mouse Genome 430 2.0 arrays (Affymetrix, Santa Clara, CA) and stained in accordance with the manufacturer's standard protocol. Washes and post hybridization processing were performed using the Fluidics Station 450 (Affymetrix) as per manufacturers' instructions and the GeneChip arrays were scanned using a laser confocal slide scanner (GeneChip Scanner 3000, Affymetrix). Array quality was assessed and images were quantified using GeneChip Operating Software (GCOS) v1.2 (Affymetrix). Data were viewed and analyzed using GeneSpring software with expression determined as a minimum signal of 50 and a two-fold change considered significant.
Real Time PCR
Quantitative real-time PCR analysis was performed using the Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems) in a Master mix consisting of 125 nM each of dACT, dCCt, dTCT, dGCT (Bioline Alexandria, NSW, Australia), 2.5 mM MgCl2 (Sigma-Aldrich), 20 μg/ml 6-ROX (Invitrogen), 100 nM primers, 1 × AmpliTaq Gold buffer and 10 U/ml AmpliTaq Gold DNA polymerase (Applied Biosystems) in 8% DMSO (Sigma-Aldrich). Primer sequences are listed in Table 2. Amplification parameters were 95°C for 10 mins, 40 cycles of 95°C (15s), 62°C (15s) and 72°C (30s) using cDNAs diluted 1:50. Standard curves were generated using 6 dpp mouse testis cDNA diluted at 1:20/60/180/540/1620. Amplification was normalized to β-actin and 18S for each sample except for Inha−/− tissues, which were only normalized to β-actin. Data were analyzed using SDS software version 2.3 (Applied Biosystems) and copy number of target genes was determined by the comparative threshold cycle method (ΔΔCT) using the Pfaffl method (Pfaffl, 2001). Melting curve data acquisition to confirm product purity was conducted from 60°C to 95°C and specificity was confirmed by sequencing as described (Itman and Loveland, 2008). Real time PCR on cultured wildtype cells was performed on two independently prepared samples, separate to those used for microarray analysis. Real Time PCR on transgenic Inha and InhbaBK/BK mice was performed using testes from six wildtype, five heterozygous and six homozygous mutant pups.
Table 2.
Primer sequences for Real Time PCR amplification of candidate activin regulated genes identified by Affymetrix microarray.
| Gene name | Accession number |
Primer sequences (5′ to 3′) | Product size (bp) |
|
|---|---|---|---|---|
| Cryl1 | C85932 | forward reverse |
AGGAGTGTGTTCCAGAGAACC TGGTGGATTGACAGGATGAGC |
176 |
| Dhcr24 | BG295389 | forward reverse |
CATCTTCCGCTACCTCTTCG CTCTGCTTCATCTCCCTTGG |
250 |
| Gja1 | BB043407 | forward reverse |
TCTGTGCCCACACTCCTGTA TTGCCGTGTTCTTCAATCCCA |
176 |
| PDGFB | BC023427 | forward reverse |
GATCTCTCGGAACCTCATCG GGCTTCTTTCGCACAATCTC |
174 |
| Rarres1 | BB035017 | forward reverse |
CTGCGCTGCACTTCTTCAAC TGCTAAATACCAAGTCCACTTCG |
126 |
| Serpina5 | NM_008785 | forward reverse |
ATCCAGGACGTCTTCACCAC CCGAGCAGATCTGAATGTGA |
162 |
| Tescalcin | NM_021344 | forward reverse |
ACAATGTCCCTGACCTGGAG GCTTCTCCTTTCGTGACAGC |
201 |
| 18S | X00686.1 | forward reverse |
GTAACCCGTTGAACCCCATT CCATCCAATCGGTAGTAGCG |
151 |
| β-actin | NM_007393 | forward reverse |
AGGCTGTGCTGTCCCTGTAT AAGGAAGGCTGGAAAAGAGC |
388 |
Supplementary Material
Acknowledgements
We are grateful to Professor Peter Koopman for valuable comments on the manuscript, to Associate Professor Sarah Robertson for the gift of TGFβ proteins, Derek Pouchnik for microarray processing and Richard Lizhong Yang for microarray data analysis.
Grant Support: Support from the NHMRC of Australia (#384108 and #334011 to KL, #331005, #384105 to DAJ), the Australian Research Council (#348239 to KL & DAJ), Monash University (Postgraduate Scholarship and Bridging Postdoctoral Fellowship to CI), the NIH (4R37 HD10808-30/2R01 HD035494-09 to MG, HD01156/HD27823 to CB, CA60651 to MM), the March of Dimes Birth Defects Foundation (5-FY01-482 to CB) and the Robert Wood Johnson Foundation (CB) is hereby acknowledged.
References
- Argentaro A, Sim H, Kelly S, Preiss S, Clayton A, Jans DA, Harley VR. A SOX9 defect of calmodulin-dependent nuclear import in campomelic dysplasia/autosomal sex reversal. Journal of Biological Chemistry. 2003;278:33839–33847. doi: 10.1074/jbc.M302078200. [DOI] [PubMed] [Google Scholar]
- Barakat B, O'Connor AE, Gold E, de Kretser DM, Loveland KL. Inhibin, activin, follistatin and follicle stimulating hormone serum levels and testicular production are highly modulated during the first spermatogenic wave in mice. Reproduction. 2008;136:345–359. doi: 10.1530/REP-08-0140. [DOI] [PubMed] [Google Scholar]
- Betscholtz C, Karlsson L, Lindahl P. Developmental roles of platelet-derived growth factors. BioEssays. 2001;23:494–507. doi: 10.1002/bies.1069. [DOI] [PubMed] [Google Scholar]
- Boitani C, Stefanini M, Fragale A, Morena AR. Activin stimulates Sertoli cell proliferation in a defined period of rat testis development. Endocrinology. 1995;136:4538–5444. doi: 10.1210/endo.136.12.7588293. [DOI] [PubMed] [Google Scholar]
- Bollenbach T, Pantazis P, Kicheva A, Bokel C, Gonzalez-Gaitan M, F J. Precision of the Dpp gradient. Development. 2008;135:1137–1146. doi: 10.1242/dev.012062. [DOI] [PubMed] [Google Scholar]
- Brown CW, Houston-Hawkins DE, Woodruff TK, Matzuk MM. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nature Genetics. 2000;25:453–457. doi: 10.1038/78161. [DOI] [PubMed] [Google Scholar]
- Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, Seoane J. High TGF-beta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Buzzard JJ, Farnworth PG, de Kretser DM, E OCA, Wreford NG, Morrison JR. Proliferative phase Sertoli cells display a developmentally regulated response to activin in vitro. Endocrinology. 2003;144:474–483. doi: 10.1210/en.2002-220595. [DOI] [PubMed] [Google Scholar]
- Buzzard JJ, Loveland KL, O'Bryan MK, O'Connor AE, Bakker M, Hayashi T, Wreford NG, Morrison JR, de Kretser DM. Changes in circulating and testicular levels of inhibin A and B and activin A during postnatal development in the rat. Endocrinology. 2004;145:3532–3541. doi: 10.1210/en.2003-1036. [DOI] [PubMed] [Google Scholar]
- Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian Transforming growth factor-beta superfamily. Endocrine Reviews. 2002;23:L787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- de Winter JP, Themmen APN, Hoogerbrugge JW, Klaij IA, Grootegoed JA, de Jong FH. Activin receptor mRNA expression in rat testicular cell types. Molecular and Cellular Endocrinology. 1992;83:R1–R8. doi: 10.1016/0303-7207(92)90201-g. [DOI] [PubMed] [Google Scholar]
- de Winter JP, Vanderstichele HMJ, Verhoeven G, Timmerman MA, Wesseling JG, de Jong FH. Peritubular myoid cells from immature rat testes secrete activin A and express activin receptor type II in vitro. Endocrinology. 1994;135:759–767. doi: 10.1210/endo.135.2.8033824. [DOI] [PubMed] [Google Scholar]
- Dyson S, Gurdon JB. The Interpretation of Position in a Morphogen Gradient as Revealed by Occupancy of Activin Receptors. Cell. 1998;93:557–568. doi: 10.1016/s0092-8674(00)81185-x. [DOI] [PubMed] [Google Scholar]
- Fragale A, Puglisi R, Morena AR, Stafanini M, Boitani C. Age-dependent activin receptor expression pinpoints activin A as a physiological regulator of rat Sertoli cell proliferation. Molecular Human Reproduction. 2001;7:1107–1114. doi: 10.1093/molehr/7.12.1107. [DOI] [PubMed] [Google Scholar]
- Gnessi L, Emidi A, Jannini EA, Carosa E, Maroder M, Arizzi M, Ulisse S, Spera G. Testicualr development involves the spatiotemporal control of PDGFs and PDGF receptors gene expression and action. Journal of Cell Biology. 1995;131:1105–1121. doi: 10.1083/jcb.131.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DR, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Nuclear lamins: building blocks of nuclear architecture. Genes and Development. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Grimm OH, Gurdon JB. Nuclear exclusion of Smad2 is a mechanism leading to loss of competence. Nature Cell Biology. 2002;4:519–522. doi: 10.1038/ncb812. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hu JS, Doan LT, Currle DS, Paff M, Rheem JY, Schreyer R, Robert B, Monuki ES. Border formation in a Bmp gradient reduced to single dissociated cells. Proceedings of the National Academy of Sciences. 2008;105:3398–3403. doi: 10.1073/pnas.0709100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incorvaia L, Badalamenti G, Rini G, Arcara C, Fricano S, Sferazza C, di Trapani D, Gebbia N, Leto G. MMP-2, MMP-4 and activin A blood levels in patients with breast cancer or prostate cancer metastatic to the bone. Anticancer Research. 2007;27:1519–1525. [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS. Nucleocytoplasmic shuttling of Smads 2, 3 and 4 permits sensing of TGF-beta receptor activity. Molecular Cell. 2002;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Itman C, Loveland KL. SMAD expression in the testis: an insight into BMP regulation of spermatogenesis. Developmental dynamics. 2008;237:97–111. doi: 10.1002/dvdy.21401. [DOI] [PubMed] [Google Scholar]
- Itman C, Mendis S, Barakat B, Loveland KL. All in the family: Transforming Growth Factor Beta Family Action in Testis Development. Reproduction. 2006;132:233–246. doi: 10.1530/rep.1.01075. [DOI] [PubMed] [Google Scholar]
- Jing C, El-Ghany MA, Beesley C, Foster CS, Rudland PS, Smigh P, kKe Y. Tazarotene-induced gene 1 (TIG1) expression in prostate carcinomas and its relationship to tumorigenicity. Journal of the National Cancer Institute. 2002;94:482–490. doi: 10.1093/jnci/94.7.482. [DOI] [PubMed] [Google Scholar]
- Kaipia A, Penttila T-L, Shimasaki S, Ling N, Parvinen M, Toppari J. Expression of Inhibin beta-A and beta-B, follistatin and activin A receptor messenger ribonucleic acids in the rat seminiferous epithelium. Endocrinology. 1992;131:2703–2710. doi: 10.1210/endo.131.6.1332846. [DOI] [PubMed] [Google Scholar]
- Kim SG, Kim HA, Jong HS, Park JH, Kim NK, Hong SH, Kim TY, Bant YJ. The Endogenous Ratio of Smad2 and Smad3 Influences the Cytostatic Function of Smad3. Molecular Biology of the Cell. 2005;15:4672–4683. doi: 10.1091/mbc.E05-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer A, Moepert K, Dames S, Sternberger M, Kaufmann J, Klippel A. Differential regulation of TGF-beta signalling through Smad2, Smad3 and Smad4. Oncogene. 2003;22:6748–6763. doi: 10.1038/sj.onc.1206791. [DOI] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 Positively and Negatively Regulate TGF-beta-Dependent Transcription throughthe Forkhead DNA-Binding Protein FAST2. Molecular Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Levay K, Slepak VZ. Tescalcin is an essential factor in megakaryocytic differentiation associated with Ets family gene expression. Journal of Clinical Investigation. 2005;117:2672–2683. doi: 10.1172/JCI27465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Graff JM, O'Connor AE, Loveland KL, Matzuk MM. SMAD3 regulates gonadal tumorigenesis. Molecular Endocrinology. 2007;21:2472–2486. doi: 10.1210/me.2007-0147. [DOI] [PubMed] [Google Scholar]
- Looyenga BD, Hammer GD. Genetic removal of Smad3 from inhibin-null mice attenuates tumor progression by uncoupling extracellular mitogenic signals from the cell cycle machinery. Moelcular Endocrinology. 2007;21:2440–2457. doi: 10.1210/me.2006-0402. [DOI] [PubMed] [Google Scholar]
- Loveland KL, Zlatic K, Stein-Oakley A, Risbridger G, de Kretser DM. Platelet-derived growth factor ligand and receptor subunit mRNA in the Sertoli cell and Leydig cells of the rat testis. Molecular and Cellular Biology. 1995;108:155–159. doi: 10.1016/0303-7207(94)03471-5. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes and Development. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- Meehan T, Schlatt S, O'Bryan M, de Kretser DM, Loveland KL. Regulation of germ cell and Sertoli cell development by activin, follistatin and FSH. Developmental Biology. 2000;220:225–237. doi: 10.1006/dbio.2000.9625. [DOI] [PubMed] [Google Scholar]
- Mizuiri H, Yoshida K, Toge T, Oue N, Aungg PP, Noguchi T, Yasui W. DNA methylation of genes linked to retinoid signalling in squamous cell carcinoma of the esophatus: DNA methylation of CRBP1 and TIG1 is associated with tumor stage. Cancer Science. 2005;96:571–577. doi: 10.1111/j.1349-7006.2005.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal S, Patel S, Asano AT, Johnson AT, Duvic M, Chandraratna RA. Tazarotene-induced gene 1 (TIG1), a novel retinoic acid receptor-responsive gene in skin. Journal of Investigative Dermatology. 1996;106:269–274. doi: 10.1111/1523-1747.ep12340668. [DOI] [PubMed] [Google Scholar]
- Perera EM, Martin H, Seeherunvong T, Kos L, Hughes IA, Hawkins JR, Berkovitz GD. Tescalcin, a novel gene endoding a putative EF-Hand Ca2+ binding protein, Col9a3 and Renin are expressed in the mouse testis during the early stages of gonadal differentiation. Endocrinology. 2001;142:455–463. doi: 10.1210/endo.142.1.7882. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek E, Ju WJ, Heyer J, Escalante-Alcalde D, Stewart CL, Weinstein M, Deng C, Kucherlapati R, Bottinger EP, Roberts AB. Functional characterization of transforming growth factor beta signalling in Smad2- and Smad3- deficient fibroblasts. Journal of Biological Chemistry. 2001;276:19945–19953. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. Journal of Cell Biology. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M, Freilinger A, Hengstschlager M. Akt regulates nuclear/cytoplasmic localization of tuberin. Oncogene. 2007;24:521–531. doi: 10.1038/sj.onc.1209812. [DOI] [PubMed] [Google Scholar]
- Saka Y, Hagemann AI, Piepenburg O, Smith JC. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development. 2007;134:4209–4218. doi: 10.1242/dev.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan S, Simon L, Meling DD, Cyr DG, Gutstein DE, Fishman GI, Guillou F, Cooke P. Proliferation of adult Sertoli cells following conditional knockout of the gap junction progein GJA1 (Connexin43) in mice. Biology of Reproduction. 2007;76:804–812. doi: 10.1095/biolreprod.106.059212. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Hasso SM, F FJ. Unique SMAD1/5/8 activity at the phalanx-forming region determines digit identity. Proceedings of the National Academy of Sciences. 2008;105:4185–4190. doi: 10.1073/pnas.0707899105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taplin M. Androgen receptor: role and novel therapeutic prospects in prostate cancer. Expert Review of Anti-Cancer Therapy. 2008;8:1495–1508. doi: 10.1586/14737140.8.9.1495. [DOI] [PubMed] [Google Scholar]
- Uemura M, Swenson ES, Gaca MDA, Giordano FJ, Reiss M, Wells RG. Smad2 and Smad3 play different roles in rat hepatic stellate cell function and alpha-smooth muscle actin organization. Molecular and Cellular Biology. 2005;16:4214–4224. doi: 10.1091/mbc.E05-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrin P, Dewerchin M, Hilpert M, Chrenek P, Schofer C, Zechmeister-Machhart M, Kronke G, Vales A, Carmeliet P, Binder BR, Geiger M. Disruption of the protein C inhibitor gene results in impaired spermatogenesis and male infertility. The Journal of Clinical Investigation. 2000;106:1531–1539. doi: 10.1172/JCI10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen LA, Salomon DS. Glucocorticoid receptors and inhibition of neonatal mouse dermal fibroblast growth in primary culture. Archives of Dermatological Research. 1980;269:111–126. doi: 10.1007/BF00406531. [DOI] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ. Smad2 signaling in the extraembrynoic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Sims AH, Howell A, Miller CJ, Clarke RB. Effects of oestrogen on gene expression in epithelium and stroma of normal human breast tissue. Endocrine-related Cancer. 2006;13 doi: 10.1677/erc.1.01165. [DOI] [PubMed] [Google Scholar]
- Wu CC, Shyu RY, Chou JM, Jao SW, Chao PC, Kang JC, Wu ST, Huang SL, Jiang SY. RARRES1 expression is significantly related to tumour differentiation and staging in colorectal adenocarcinoma. European Journal of Cancer. 2006;42:557–565. doi: 10.1016/j.ejca.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Yang YC, Piek E, Zavadil J, Liang D, Xie D, Heyer J, Pavlidis P, Kucherlapati R, Roberts AB, Bottinger EP. Hierarchical model of gene regulation by transforming growth factor beta. Proceedings of the National Academy of Sciences. 2003;100:10269–10274. doi: 10.1073/pnas.1834070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef EM, Chen XQ, Higuchi E, Kondy Y, Garcia-Manero G, Lotan R, Issa JP. Hypermethylation and silencing of the putative tumor suppressor Tazarotene-induced gene 1 in human cancers. Cancer Research. 2004;64:2411–2417. doi: 10.1158/0008-5472.can-03-0164. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.