Abstract
Par3 is a multiple-PDZ containing scaffold protein that is central to the organization of an evolutionarily conserved cell polarity complex consisting of par3/par6/aPKC. The ability of par3 PDZ domains to target various adhesion molecules and enzymes at the plasma membrane leads to the controlled localization of par6 and aPKC, which has firmly established its role in epithelial cell polarity. Of the numerous PDZ ligands associated with par3, interaction of its third PDZ domain with the class II ligand found within the C-terminal tail of vascular endothelial cadherin (VE-Cad) suggests a role in endothelial cell polarity as well, but the molecular details of the interaction are unknown. Previously solved structures of par3-PDZ3 bound to the class I ligand found within the C-terminal tail of the phosphoinositide phosphatase PTEN revealed two discrete binding sites: a canonical PDZ-ligand interaction site and a distal site involving charge-charge complements. Currently it is unclear if par3-PDZ3 employs both canonical and distal binding modes in its association with VE-Cad or if these modes are unique to the PTEN interaction suggesting a possible mechanism for ligand specificity within the polarity network. The structure of par3-PDZ3 bound to the C-terminal tail of VE-Cad presented in this work shows that both canonical and distal interactions are utilized in binding. Biophysical measurements using fluorescence polarization and 2D NMR implicate the intermolecular charge pairing of aspartic acid 777 (VE-Cad) and arginine 609 (par3-PDZ3) as a crucial modulator of complex formation. Phosphorylation of VE-Cad at serine 776 increases its affinity for par3 demonstrating that post-translational modifications outside of the canonical carboxylate binding site can enhance PDZ-ligand interactions. Comparison of the VE-Cad and PTEN complexes highlights how the unique molecular architecture of par3-PDZ3 can accommodate both canonical and distal interaction modes that allow dual class specificity for these two ligand types.
The formation and disassembly of large protein complexes is of critical importance in the regulation of cellular communication. Elucidating the molecular determinants that govern protein-protein interactions greatly enhances our understanding and ability to manipulate signaling pathways in the treatment of disease. Of the ~150 families of protein interaction domains (1), the PDZ domain is one of the most abundant and best characterized (2). PDZ domains derive their name from the first three proteins in which the domain was discovered (PSD-95, Disc large, and ZO-1) (3). The domain consists of ~90 amino acids that fold into a six stranded β-sheet capped by two α-helices. Typically PDZ domains bind to extreme carboxyl-terminal (C-terminal) peptide ligands found within particular protein targets, but are capable of a diverse range of ligand interactions. For instance, PDZ domains can also bind internal protein sequences (4, 5), as well as phosphoinositides (PIPs) (6). These unique properties allow PDZ domains to participate in a wide range of interactions that often result in localization of specific protein complexes within the cellular membrane (7).
C-terminal peptide binding is the most characterized mode of PDZ domain interaction. In this case peptide binding is facilitated by a structurally conserved groove formed between the β2-strand and the α2-helix of the PDZ domain (8). Canonical peptide binding occurs at the P0 and P−2 positions that form amide hydrogen bonds that anchor into the PDZ binding groove. (P0 denotes the C-terminal residue of the peptide ligand, and P−n denotes the nth amino acid from the C-terminus). As a result C-terminal target peptides are organized into classes depending on the composition of residues at the P0 and P−2 positions. Class I, II, and III peptide ligands are comprised of the motifs -(S/T)-X-ΦCOO-, -Φ-X-ΦCOO-, -G-(E/D)-X-ΦCOO-, respectively, with single letters representing amino acids, X = any amino acid, and Φ = any hydrophobic amino acid (9). Of the numerous peptide ligands that have been identified as PDZ binders, most contain hydrophobic residues at the C-terminal position. However in contrast to the above groupings, class IV ligands are identified by the motif -X-Φ-D/ECOO- that contain a negatively charged side chain at P0. Whereas most PDZ domains are thought to associate with a single class of ligand, examples of PDZ domains with dual class specificity have been reported (10–12).
Although positions P0 and P−2 form strong contacts with the PDZ domain, other residues further upstream can also contribute to ligand binding. An earlier study indicated that residues up to P−8 could affect ligand specificity of discs large PDZ3 (13). Distal binding interactions have also been observed in PDZ domains from the human phosphatase hPTP1E (14) and erbin (15) where respective ligand position P−6 and P−7 can interact with an extended loop between the β2 and β3 strands of the PDZ domain.
Partition defective protein-3 (par3) is a large scaffold protein containing three PDZ domains (PDZ1-3) that interact with multiple target proteins. It was first demonstrated that par3-PDZ1 associated with C-terminal ligands found within junction adhesion molecules (JAM’s), which led to speculation about its role in epithelial and endothelial cell polarity (17). Subsequent studies revealed that PDZ1 also binds the C-terminal tail of nectin (18), another adhesion molecule, as well as phospholipase C-β (19). Permissive binding has been demonstrated for par3-PDZ2, which has been shown to associate with certain PIPs within lipid membranes (20). This interaction involves residues from the α2-β6/β1-β2 loops, and appears to be critical for epithelial cell polarity. Investigations of par3-PDZ3 have shown affinities toward C-terminal ligands found in JAM-C (21), vascular endothelial cadherin (VE-Cad) (22), and the PIP phosphotase PTEN (16). Given that par3 interacts with both PIPs and PTEN, it is now believed that par3 plays a major role in the integration of PIP signaling events during epithelial cell polarization (16).
Structural investigation of the par3-PDZ3/PTEN complex has revealed canonical PDZ interactions involving P0 and P−2, as well as distal interactions that reach the P−10 register of the target peptide (16). These distal contacts result, in part, from distinctive structural aspects of the PDZ domain. Relative to other PDZ domains par3-PDZ3 possesses a slightly longer α2-helix, resulting in an elongated ligand binding groove. This feature creates additional surface area facilitating complementary side chain interactions involving the acidic residues D393, E394, and D395 (i.e. P−8, P−9, and P−10) of PTEN, and basic residues (K606, R609, K611, and K622) within the extended β2 and β3 regions of par3-PDZ3 (16). Interestingly, PTEN is recognized as a class I C-terminal ligand containing -T-K-Vcoo-, while both JAM-C and VE-Cad contain putative class II ligands -F-V-Icoo- and L-I-Icoo- at their respective c-termini. Given the dual specificity of par3-PDZ3 for C-terminal ligands we investigated the degree to which the reported non-canonical distal interactions contribute to its affinity for different binding partners.
Here we report the NMR structure of par3-PDZ3 in complex with the C-terminal tail of VE-Cad. Canonical interactions in the carboxylate binding pocket are augmented by an association between distal parts of the peptide and the extended β2 region of the PDZ domain. Using fluorescence polarization to measure peptide binding affinity and 2D NMR to monitor specific interactions, we show that D777 (P−7) of VE-Cad and R609 of par3-PDZ3 make significant contributions to binding. Interestingly, phosphorylation of VE-Cad at P−8 (S776) also enhances PDZ3 binding.
Experimental Procedures
Protein expression, Purification and Mutagenesis
The amino acid sequence for par3-PDZ3 (residues 581 to 689) was PCR amplified from M. musculus par3 and cloned into a modified pQE30 vector (Qiagen) containing a cleavable N-terminal His8-tag. Protein was expressed in SG13009 E. coli cells and purified by Ni2+ affinity chromatography. The resulting protein was cleaved with TEV protease to remove the His8 affinity tag and further purified by cation exchange chromatography. Unlabeled protein was prepared from growth in Luria-Bertani media. Uniformly labeled 15N or 15N/13C-labeled proteins used in NMR experiments were prepared from growth in M9 minimal media incorporating [U-13C]-glucose (Cambridge Isotopes Laboratories, CIL) and/or [U-15N]-ammonium chloride (CIL) as the sole carbon or nitrogen sources. A cDNA fragment encoding the C-terminal 16 residues of M. musculus VE-Cad was also cloned into a modified pQE30 vector containing a cleavable N-terminal His8-GB1-tag. Uniformly labeled 15N/13C-VE-Cad peptide was expressed in SG13009 E. coli cells from growth in M9 minimal medium incorporating [U-13C]-glucose (CIL) and [U-15N]-ammonium chloride (CIL) as the sole carbon and nitrogen sources, and purified by Ni2+ affinity chromatography. After removal of the His8-GB1-tag, the peptide was further purified by reverse-phase HPLC using a 10–70% acetonitrile gradient, followed by lyophilization. Identity and molecular mass of 15N/13C-labeled peptide were verified by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and NMR (Supplementary Material, Figure S1). To create the alanine substituted par3-PDZ3 R609A protein site directed mutagenesis was performed using pairs of complementary primers and the QuickChange kit (Stratagene) following the manufacturer’s protocol.
Peptide synthesis
The Protein Nucleic Acid Facility (PNA) at the Medical College of Wisconsin synthesized all unlabeled and rhodamine labeled VE-Cad peptides with rhodamine coupled to the N-terminus using standard Fmoc chemistry.
NMR Spectroscopy
NMR experiments were performed on a Bruker DRX 600 equipped with a 1H, 15N, 13C cyroprobe. NMR titration experiments were performed with 100μM 15N-labeled par3-PDZ3 in 20mM sodium phosphate, 50 mM NaCl, 3mM dithiothreitol (DTT), pH 6 at 298K. Complete resonance assignment were achieved for 1mM 15N/13C-labeled par3-PDZ3 saturated with (2mM) unlabeled VE-Cad peptide (MLAELYGSDPQEELIIcoo-) using the following experiments: 2D 15N-1H HSQC, 3D HNCA, 3D HNCO, 3D HNCACB, 3D HNCACO, 3D HN(CO)CA, 3D C(CO)NH, 3D HBHACONH, 3D HC(CO)NH, 3D HCCH-TOCSY. Complete resonance assignments were achieved for 1mM 15N/13C-labeled VE-Cad peptide (MLAELYGSDPQEELIIcoo-) saturated with 1mM par3-PDZ3 in 20mM sodium phosphate, 50mM NaCl, 3mM dithiothreitol, pH 6 at 298K with the following experiments: 2D 15N-1H HSQC, 3D HNCA, 3D HNCO, 3D HNCACB, 3D C(CO)NH, 3D HC(CO)NH, 3D HCCH-TOCSY. All NMR data were processed with NMRPipe (23) and XEASY (24) was used for resonance assignment and analysis of NOE spectra.
Structure Calculation and Analysis
NOE distance restraints were obtained from 3D 15N-edited NOESY-HSQC, 13C(aliphatic)-edited NOESY-HSQC, 13C(aromatic)-edited NOESY-HSQC, and 13C-12C-filtered NOESY-HSQC (25) spectra (τmix = 80 ms). Backbone dihedral angle restraints (φ and ψ angles) were derived from 1Hα, 13Cα, 13Cβ, 13C′, 15N chemical shift data using TALOS (26). Structures were generated in an automated manner using the NOEASSIGN module of the torsion angle dynamics program CYANA 2.1 (27). This procedure resulted in an ensemble of high precision structures that required minimal manual refinement. The 20 CYANA conformers with lowest target function were further refined by a molecular dynamics protocol in explicit solvent (28) using XPLOR-NIH (29).
Fluorescence Polarization Assay
The fluorescence polarization (FP) assays were performed on a PTI (Photon Technologies International) fluorimeter equipped with automated polarizers at 298K. Synthetic VE-Cad peptides were rhodamine labeled at the N-terminus. Titrations were conducted by monitoring fluorescence polarization as a function of increasing amounts of par3-PDZ3 added to 100nM of rhodamine-VE-Cad peptide in 50mM HEPES, 100mM NaCl, 1mM DTT, and 1mM EDTA at pH 7.4, using excitation and emission wavelengths of 562nm and 582nm, respectively. The dissociation constants (Kd) were determined by fitting titration curves using the program profit 6.1 to the equation:
Where FPobs is the observed FP value, FPo is the value of free peptide, FPmax is maximum value of completely bound peptide, and x is par3-PDZ3 protein concentration.
Results and Discussion
Identification of the VE-Cadherin binding site on par3-PDZ3
The association of VE-Cad with par3 in endothelial cells is mediated by PDZ3 of par3 and the VE-Cad C-terminus (22). To detail the structural basis for this important polarity complex we identified the VE-Cad binding site of par3-PDZ3 using 1H-15N HSQC NMR spectroscopy. In these experiments 15N-labeled par3-PDZ3 was titrated with a synthetic peptide comprising the last 16 amino acids of VE-Cad shown in Figure 1A. Chemical shift perturbations caused by the addition of VE-Cad were mapped onto the 3D structure of the ligand free form of par3-PDZ3 (PDB code: 2K1Z (16)) shown in Figure 1B. Consistent with C-terminal peptide binding, residues within the β2-α2-binding groove (e.g. G600, L601, G602, V605) displayed the most prominent chemical shift perturbations upon VE-Cad addition. We also observed marked peptide-induced chemical shift perturbations for a cluster of basic residues including K606, R609, K611, and K622, all of which extend beyond the β2-α2-binding groove. We speculated that conserved acidic amino acids near the VE-Cad C-terminus (Figure 1A) might interact with the basic patch of par3-PDZ3.
Figure 1.
A) Sequence alignment of the 16 terminal amino acids of VE-Cadherin. Position indicates the register of amino acids relative to the c-terminal denoted as 0. The mouse (M. musculus) sequence shown corresponds to the exact peptide used in NMR titration studies, with numbers above corresponding to amino acid location in primary sequence. Conserved acidic residues are shown in red, and potential phosphorylation sites are depicted in green. B) Chemical shift perturbations induced by VE-Cad peptide binding mapped onto the structure of ligand free par3-PDZ3. Differences in amide chemical shifts were defined as: Δδppm = [(5*ΔδHN)2 + (ΔδN)2]1/2, where ΔδHN and ΔδN represent chemical shift differences of the free and bound forms of par3-PDZ3. Oval highlight represents location of the binding groove between the β2-strand and the α2-helix, site of canonical c-terminal ligand binding. Positions of basic patch residues (K606, R609, K611, K622) located in the β2 and β3-strands are shown to distinguish areas of distal ligand binding.
Structure of par3-PDZ3 in complex with VE-Cad C-terminus
To define the nature of the par3-PDZ3/VE-Cad interaction, the structure of par3-PDZ3 in complex with VE-Cad peptide was solved by NMR. Initially 15N/13C-labeled par3-PDZ3 was titrated with unlabeled VE-Cad (Figure 1A) until saturation was reached. Standard heteronuclear NOESY spectra (i.e. 15N-edited and 13C-edited) were processed and analyzed for use in automated NOE assignment. Intermolecuar NOEs between the labeled PDZ domain and the unlabeled peptide were also obtained from 13C-12C filtered NOESY spectra. Conversely, a complex of 15N/13C-labeled VE-Cad peptide with unlabeled par3-PDZ3 was used for acquisition of a complete set of heteronuclear and 13C-12C filtered spectra. After final analysis 1868 NOE distance restraints were used in calculation of an ensemble of structures representing the par3-PDZ3/VE-Cad complex.
The final ensemble of par3-PDZ3 in complex with VE-Cad peptide is displayed in Figure 2A, with structural statistics shown in Table 1. Comparison of our PDZ domain complex to the reported ligand free form (PDB code: 2K1Z) revealed a Cα-backbone alignment RMSD of 1.59Å. The geometries of the two PDZ domains are in excellent agreement, with the only difference seen in the orientation of the α2 helix resulting from an outward rotation caused by ligand entry into the binding groove. This same helical rotation has been reported in the par3-PDZ3/PTEN complex (16). Consistent with C-terminal peptide binding, P0 of VE-Cad (I784) packs into a cavity created by residues L601 and V603 of the conserved ‘GLGF’ motif between the β1-β2 strands, and M666 in the α2-helix shown in Figure 2B and C. Additional hydrophobic side chain contacts are observed between P−1 (I783) and I625, as well as the P−2 (L782) and M659 located in the α2-helix (Figure 2C). The VE-Cad peptide contains four acidic residues at P−3 (E781), P−4 (E780), P−7 (D777) and P−12 (E772) that could potentially serve as interaction partners with the distal basic patch unique to par3-PDZ3. NOEs between P−3 (E781) and K606 confirmed the pairing of these residues, and the side chain orientation of K622 places the positively charged head group in close proximity to the glutamate side chain of P−3 (E781), well within hydrogen bonding distance. This configuration suggested that movements of amide cross-peaks K606 and K622 observed in 1H-15N HSQC titration experiments are likely caused by interactions with E781 of VE-Cad. NOE contacts between P−4 (E780), and N655 located at the N-terminal end of the α2-helix, were observed resulting in the glutamate side chain pointing away from the β2-strand. Interestingly several structures in the ensemble place the acidic side chain of P−7 (D777) in close proximity to the guanidinium group of R609 (Figure 2C), though no intermolecular NOEs were detected for this pair of residues. In contrast, P−12 (E772) is disordered and does not appear to contact par3-PDZ3. Weak NOEs from both the P−9 (G775) and P−10 (Y774) residue to K611 within the β2-β3 turn of par3-PDZ3 define the most distal interaction with the extended binding groove (Figure 2C). A schematic diagram representing all amino acid residues involved in intermolecular binding observed within the par3-PDZ3/VE-Cad complex are shown in supplementary material (Figure S2).
Figure 2.
A) Solution structure of par3-PDZ3 in complex with VE-Cad peptide. Overlay of 20 conformers with VE-Cad peptide shown in red, unstructured n and c-termini have been deleted for clarity. B) Ribbon diagram of representative conformer showing areas of secondary structure. Cyan box highlights region of canonical interaction, green box highlights region of distal interactions. C) Zoomed in view of the canonical (upper) and distal (lower) binding sites, dashed lines between molecules represent areas of observed NOEs described in text. Bold numbering indicates amino acids within par3-PDZ3, Pn represents amino acid positions within peptide.
Table 1.
Structural statistics for the 20 par3-PDZ3/VE-Cad conformers.
| Experimental constraints | PDB code: 2KOH | |
|---|---|---|
| Distance constraints | ||
| Long | 735 | |
| Medium [1<(i−j)≤5] | 254 | |
| Sequential [ (i−j)=1] | 383 | |
| Intra-residue [i=j] | 434 | |
| Inter-residue | 62 | |
| Total | 1868 | |
| Dihedral angle constraints (φ and ψ) | 114 | |
| Average atomic R.M.S.D. to the mean structure (Å) | ||
| Residues 8–16, 22–30,38–103 | ||
| Backbone ( Cα, C′, N) | 0.44 ± 0.07 | |
| Heavy atoms | 0.93 ± 0.09 | |
| Deviations from idealized covalent geometry | ||
| Bond lengths | RMSD (Å) | 0.019 |
| Torsion angle violations | RMSD (°) | 1.4 |
| WHATCHECK quality indicators | ||
| Z-score | −1.30 ± 0.20 | |
| RMS Z-score | ||
| Bond lengths | 0.88 ± 0.02 | |
| Bond angles | 0.71 ± 0.02 | |
| Bumps | 0 ± 0 | |
| Lennard-Jones energya (kJ mol−1) | −2,408 ± 51 | |
| Constraint violations | ||
| NOE distance | Number > 0.5 Å | 0 ± 0 |
| NOE distance | RMSD (Å) | 0.023 ± 0.001 |
| Torsion angle violations | Number > 5 ° | 0 ± 0 |
| Torsion angle violations | RMSD (°) | 0.674 ± 0.097 |
| Ramachandran statistics (% of all residues) | ||
| Most favored | 81.6 ± 3.2 | |
| Additionally allowed | 16.5 ± 3.0 | |
| Generously allowed | 1.1 ± 1.0 | |
| Disallowed | 0.7 ± 0.8 | |
Nonbonded energy was calculated in XPLOR-NIH.
VE-Cadherin composition has an effect on par3-PDZ3 binding
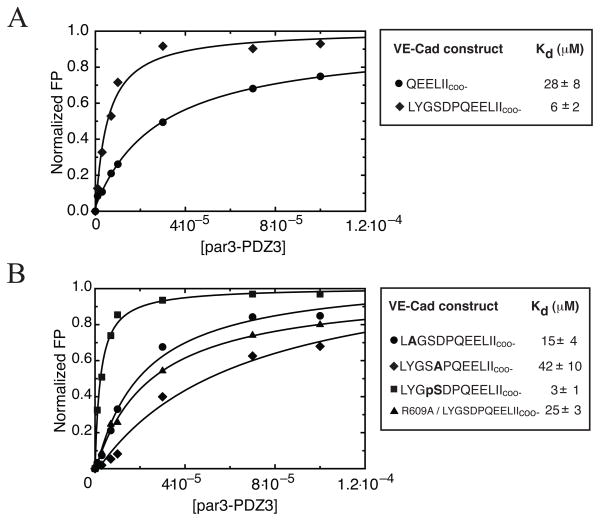
Our structural analysis indicated that in addition to the canonical P0 and P−2 binding sites, amino acids as distant from the VE-Cad C-terminus as P−10 contributed to par3-PDZ3 recognition. Based on the contacts between P−10 (Y774) and K611, we speculated that P−7 (D777) of VE-Cad might interact with R609 of par3. To probe specific contributions of distal contacts to the par3-PDZ3/VE-Cad interaction, we determined binding constants (Kd) for several different VE-Cad constructs using fluorescence polarization. If the P−10 and P−7 positions enhance the interaction with par3-PDZ3, there should be a measurable difference in binding affinities between C-terminal constructs lacking these residues. Rhodamine-labeled peptides containing either the C-terminal 6 or 12 residues of VE-Cad were used to measure par3-PDZ3 binding constants by FP (Figure 3A). VE-Cad (773–784) bound PDZ3 ~5-fold more tightly than VE-Cad (779–784), indicating that residues in the P−11 to P−6 register contribute ~1 kcal/mol to the free energy of binding. Guided by the par3-PDZ3/VE-Cad structure we measured binding affinities for VE-Cad (773–784) peptides containing Y774A or D777A mutations (Figure 3B). While the Y774A substitution has a modest effect on Kd, the D777A mutation at P−7 shifts the Kd from 6 ± 2μM to 42 ± 10μM, comparable to the 6-residue peptide (Kd 28 ± 8μM). This suggests that the P−7 residue of VE-Cad (D777) is the major contributor to distal binding.
Figure 3.
A) Determination of affinity for different length VE-Cad peptides binding to par3-PDZ3 using a fluorescence polarization assay. All peptides contained n-terminally linked rhodamine label. Comparison of binding curves of short (●) and long (◆) constructs of VE-Cad revealed a ~5-fold increase in binding affinity for the longer VE-Cad segment. B) Comparison of binding curves generated for YtoA (●) and DtoA (◆) VE-Cad mutants revealed that P−7(D) contributes to VE-Cad interaction with par3-PDZ3 as indicated by the decrease in binding affinity. Mutation R609A of par3-PDZ3 (▲) shows decrease in binding affinity when titrated against long VE-Cad construct. Further modification of VE-Cad by phosphorylation of P−8(S) (■) revealed that additional negative charge introduced to distal amino acids can increase binding relative to the long construct by ~2-fold. Bold letters signify location of alanine substitutions or serine phosphorylation (pS). Error in Kd is estimated from fitting.
Mutation of R609 of par3-PDZ3 reduces VE-Cad binding affinity
To further test for the presence of distal interaction between R609 and D777 in the par3-PDZ3/VE-Cad complex, we constructed a par3-PDZ3 protein containing a R609A mutation. We speculated that R609A should disrupt charge pairing with D777 in wild-type VE-Cad, resulting in a measurable decrease in binding affinity. The R609A mutant revealed a VE-Cad binding Kd of 25 ± 3μM (Figure 3B), reinforcing our hypothesis that a D777-R609 intermolecular salt-bridge is the major contributor to distal binding in the par3-PDZ3/VE-Cad complex.
Possible role of serine phosphorylation in the VE-Cad C-terminus
Having recognized the importance of side chain mediated contacts governing the par3-PDZ3/VE-Cad complex, we next determined if post translational modifications could further increase binding affinity by introducing additional complementary charges. VE-Cad contains two potential phosphorylation sites at P−10(Y774) and P−7(S776) that could interact with basic residues of par3-PDZ3 and S776 is predicted to be a target for casein kinase2 (30). Both phosphopeptides were synthesized and titrated against 15N-labeled par3-PDZ3. Screening by 1H-15N HSQC revealed binding affinities of ~2μm and ~6μm for the pS776 and pY774 containing VE-Cad peptides, respectively. Since wild-type VE-Cad peptide (773–784) binds par3-PDZ3 with a binding affinity of 6μM, these results suggested that phosphorylation of S776 but not Y774, enhances par3-PDZ3 binding, and we confirmed the increased binding affinity by FP (Figure 3B). This suggests that specific serine phosphorylation within the VE-Cad C-terminus creates an additional electrostatic interaction with basic residues of par3-PDZ3.
D777A VE-Cadherin mutant has an effect on distal par3-PDZ3 interactions
If a stable D777-R609 salt bridge is critical in forming the par3-PDZ3/VE-Cad complex it seems NOEs would be detected. However, if the side chains of D777 and R609 point toward each other, the shortest interproton distance (Hε of R609 to the Hβ protons of D777) would be ~6Å. Hence, the lack of observable NOEs between R609 and D777 is not inconsistent with a side chain mediated electrostatic interaction. To confirm the function of D777 in side chain mediated binding, the alanine substituted peptide was titrated against 15N-labeled par3-PDZ3 to detect site-specific perturbations to backbone amide resonances. We hypothesized that if the D777A-VE-Cad mutant disrupted side chain mediated binding, differences in backbone amide resonances along the β2-strand of par3-PDZ3 should be evident when compared to a wild-type VE-Cad titration spectrum. Comparisons of the 1H-15N HSQC spectra of 15N-labeled par3-PDZ3 saturated with either D777A-VE-Cad (red) or wild-type VE-Cad (gray) show that while most cross-peaks are in identical positions (Δδppm < 0.09), several residues (e.g. K606, G607, N608, R609) display differences in resonance position (Figure 4). Identical experiments performed with Y774A-VE-Cad peptide showed no differences attributable to the substitution (data not shown). These results confirm that D777 is the major contributor of distal VE-Cad contacts to par3-PDZ3, and likely serves as an anchor to upstream regions of the peptide that allows further interaction along the extended β2-strand of par-PDZ3.
Figure 4.
A) Highlighted regions of 15N-1H HSQC spectra of par3-PDZ3 saturated with VE-Cad peptide (MLAELYGSDPQEELIIcoo-) shown in gray, overlaid with spectra saturated with mutant VE-Cad peptide (MLAELYGS(A)PQEELIIcoo-) shown in red. B) Chemical shift difference in par3-PDZ3 calculated from Δδppm = [(5*ΔδHN)2 + (ΔδN)2]1/2, where ΔδHN and ΔδN represent chemical shift differences between the two saturated forms of par3-PDZ3. All residues with Δδppm > 0.09 are shown. C) Par3-PDZ3 chemical shift map resulting from D777A binding relative to wt-VE-Cad binding. Highlighted residues indicate changes in the electronic environment for a region along the β2-strand consistent with a disruption of distal contact.
Par3-PDZ3 complexes reveal the source of dual ligand class specificity
Par3-PDZ3 employs two distinct binding modes in the interaction with the class II ligand found within the C-terminal tail of VE-Cad, similar to interactions reported for the class I C-terminal PTEN ligand (16). Both ligands exploit the hydrophobic cavity formed by residues L601, V603 and M666 on par3-PDZ3, and both ligands use additional distal contacts to further stabilize binding (Figure 5). However there are subtle differences that may explain the molecular basis of dual specificity that par3-PDZ3 displays towards these two ligand classes. One key factor is the absence of a conserved histidine within the α2-helix of par3-PDZ3 that is associated with the recognition of class I PDZ ligands. Typically this histidine residue forms a hydrogen bond with the hydroxyl group of the serine or theronine at P−2 of class I ligands (31). Par3-PDZ3 contains a methonine (M659) at the this position, which is more suitable for hydrophobic contact expected for class II ligand binding (31). Interestingly, PTEN requires both canonical and distal interactions in order to form a complex with par3-PDZ3 (16). Consistent with this claim, our laboratory failed to detect any interaction between par3-PDZ3 and a rhodamine labeled PTEN peptide QHSQITKVcoo-. Par3-PDZ3 is reported to bind a longer PTEN peptide DEDQHSQITKVcoo- with a Kd of 19μm (16), which corresponds to a free energy of ~ –6kcal/mol. Assuming conservatively that 3 kcal/mol of stabilization is derived from distal hydrogen bonding (1 kcal/mol for each hydrogen bond from the DED motif, figure 5B), the remaining energy would be extracted from classical PDZ binding of the C-terminal tail. However, this translates into a C-terminal binding affinity of only ~6mM, which may explain the lack of detectable interaction observed for the short PTEN peptide. In contrast, a short C-terminal VE-Cad peptide QEELIIcoo- binds par3-PDZ3 with an affinity of 28 μM (Figure 3A). This is likely due to favorable hydrophobic contact (figure 2C), and electrostatic contributions betwen P−3 (E781) and K622 (Figure 5). Considering just canonical C-terminal interactions, VE-Cad QEELIIcoo- binds par3-PDZ3 ~200 times more strongly than PTEN QHSQITKVcoo-. As a result, PTEN binding depends on compensatory distal contacts to overcome extremely weak canonical contacts, while VE-Cad binding exploits a complementary interaction surface with distal contacts augmenting PDZ binding. While both ligands employ similar binding modes, there is a difference in the balance between canonical and distal contributions to the free energy of binding. Thus the unique molecular architecture of par3-PDZ3 allows dual class specificity by providing an additional interaction surface outside the canonical binding pocket that can be optimized to overcome deficiencies resulting from non-complementary ligand class interactions.
Figure 5.
A) Structure of par3-PDZ3 in complex with VE-Cad c-terminus. VE-Cad is shown in red with Pn denoting amino acid positions within the peptide. Numbered amino acids correspond to side chain locations within par3-PDZ3 involved in peptide binding. Dashed lines show nitrogen to oxygen distances less then 3 angstroms that indicate areas of probable electrostatic contact. B) Structure of par3-PDZ3 in complex with PTEN c-terminus (16). PTEN is shown in blue with Pn denoting amino acid position within the peptide. Numbered amino acids correspond to side chain locations within par3-PDZ3 involved in peptide binding. Dashed lines show nitrogen to oxygen distances less then 3 angstroms that indicate areas of probable electrostatic contact.
Summary
We characterized the canonical PDZ binding expected for a class II C-terminal ligand involving the P0 and P−2 register of VE-Cad. Our NMR data also revealed evidence for distal contacts outside the canonical binding mode involving upstream residues P−10 and P−7 of VE-Cad and a patch of basic residues in par3-PDZ3. We inferred from structural results that the acidic side chain of D777 in VE-Cad pairs with R609 in β2-strand of par3-PDZ3 to enhance binding through favorable electrostatic interaction. This distal pairing was further confirmed by fluorescence polarization, which demonstrated a decrease in binding affinity upon mutation of either residue. This work provides a detailed view of par3-VE-cad binding and clarifies the role distal binding modes play in ligand specificity associated with PDZ domain based protein-protein interactions.
Supplementary Material
Acknowledgments
We thank Professor Ken Prehoda at the University of Oregon for graciously donating murine par3 DNA. RCT thanks Davin Jensen for technical support.
This work was supported by NIH Protein Structure Initiative Grant U54 GM074901 (J.L. Markley, PI, G.N. Phillips, and B.G. Fox, Co-Investigators) and NIH instrumentation grant S10 RR024665.
Abbreviations
- FP
fluorescence polarization
- Par3
partition defective protein 3
- PDZ
PSD-95/Disc large/ZO-1
- VE-Cad
vascular endothelial cadherin
Footnotes
Structure of PDZ3/VE-Cad complex has been deposited in the RCSB under PDB ID 2KOH. All NMR data has been deposited in the BMRB under accession number 16507.
Supporting information available includes MALDI-MS and NMR verification of double-labeled VE-Cad peptide, and a diagram depicting all intermolecular NOE contacts. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Robert C. Tyler, Email: rtyler2@mcw.edu.
Francis C. Peterson, Email: fpeterso@mcw.edu.
Brian F. Volkman, Email: bvolkman@mcw.edu.
References
- 1.Ceol A, Chatr-aryamontri A, Santonico E, Sacco R, Castagnoli L, Cesareni G. DOMINO: a database of domain-peptide interactions. Nucleic Acids Res. 2007;35:D557–560. doi: 10.1093/nar/gkl961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castagnoli L, Costantini A, Dall’Armi C, Gonfloni S, Montecchi-Palazzi L, Panni S, Paoluzi S, Santonico E, Cesareni G. Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett. 2004;567:74–79. doi: 10.1016/j.febslet.2004.03.116. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy MB. Origin of PDZ (DHR, GLGF) domains. Trends Biochem Sci. 1995;20:350. doi: 10.1016/s0968-0004(00)89074-x. [DOI] [PubMed] [Google Scholar]
- 4.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. [PubMed] [Google Scholar]
- 5.Penkert RR, DiVittorio HM, Prehoda KE. Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nat Struct Mol Biol. 2004;11:1122–1127. doi: 10.1038/nsmb839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J. PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell. 2002;9:1215–1225. doi: 10.1016/s1097-2765(02)00549-x. [DOI] [PubMed] [Google Scholar]
- 7.Jelen F, Oleksy A, Smietana K, Otlewski J. PDZ domains - common players in the cell signaling. Acta Biochim Pol. 2003;50:985–1017. [PubMed] [Google Scholar]
- 8.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 9.Vaccaro P, Dente L. PDZ domains: troubles in classification. FEBS Lett. 2002;512:345–349. doi: 10.1016/s0014-5793(02)02220-2. [DOI] [PubMed] [Google Scholar]
- 10.Borg JP, Marchetto S, Le Bivic A, Ollendorff V, Jaulin-Bastard F, Saito H, Fournier E, Adelaide J, Margolis B, Birnbaum D. ERBIN: a basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat Cell Biol. 2000;2:407–414. doi: 10.1038/35017038. [DOI] [PubMed] [Google Scholar]
- 11.Kurschner C, Mermelstein PG, Holden WT, Surmeier DJ. CIPP, a novel multivalent PDZ domain protein, selectively interacts with Kir4.0 family members, NMDA receptor subunits, neurexins, and neuroligins. Mol Cell Neurosci. 1998;11:161–172. doi: 10.1006/mcne.1998.0679. [DOI] [PubMed] [Google Scholar]
- 12.Kang BS, Cooper DR, Devedjiev Y, Derewenda U, Derewenda ZS. Molecular roots of degenerate specificity in syntenin’s PDZ2 domain: reassessment of the PDZ recognition paradigm. Structure. 2003;11:845–853. doi: 10.1016/s0969-2126(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 13.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 14.Kozlov G, Banville D, Gehring K, Ekiel I. Solution structure of the PDZ2 domain from cytosolic human phosphatase hPTP1E complexed with a peptide reveals contribution of the beta2-beta3 loop to PDZ domain-ligand interactions. J Mol Biol. 2002;320:813–820. doi: 10.1016/s0022-2836(02)00544-2. [DOI] [PubMed] [Google Scholar]
- 15.Birrane G, Chung J, Ladias JA. Novel mode of ligand recognition by the Erbin PDZ domain. J Biol Chem. 2003;278:1399–1402. doi: 10.1074/jbc.C200571200. [DOI] [PubMed] [Google Scholar]
- 16.Feng W, Wu H, Chan LN, Zhang M. Par-3-mediated junctional localization of the lipid phosphatase PTEN is required for cell polarity establishment. J Biol Chem. 2008 doi: 10.1074/jbc.M802482200. [DOI] [PubMed] [Google Scholar]
- 17.Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Meyer Zu Brickwedde MK, Ohno S, Vestweber D. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) Embo J. 2001;20:3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takekuni K, Ikeda W, Fujito T, Morimoto K, Takeuchi M, Monden M, Takai Y. Direct binding of cell polarity protein PAR-3 to cell-cell adhesion molecule nectin at neuroepithelial cells of developing mouse. J Biol Chem. 2003;278:5497–5500. doi: 10.1074/jbc.C200707200. [DOI] [PubMed] [Google Scholar]
- 19.Cai Y, Stafford LJ, Bryan BA, Mitchell D, Liu M. G-protein-activated phospholipase C-beta, new partners for cell polarity proteins Par3 and Par6. Oncogene. 2005;24:4293–4300. doi: 10.1038/sj.onc.1208593. [DOI] [PubMed] [Google Scholar]
- 20.Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Ebnet K, Aurrand-Lions M, Kuhn A, Kiefer F, Butz S, Zander K, Meyer zu Brickwedde MK, Suzuki A, Imhof BA, Vestweber D. The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: a possible role for JAMs in endothelial cell polarity. J Cell Sci. 2003;116:3879–3891. doi: 10.1242/jcs.00704. [DOI] [PubMed] [Google Scholar]
- 22.Iden S, Rehder D, August B, Suzuki A, Wolburg-Buchholz K, Wolburg H, Ohno S, Behrens J, Vestweber D, Ebnet K. A distinct PAR complex associates physically with VE-cadherin in vertebrate endothelial cells. EMBO Rep. 2006;7:1239–1246. doi: 10.1038/sj.embor.7400819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Bartels CX, Billeter T-H, Güntert M, Wüthrich PK. The program XEASY for Computer-Supported NMR Spectral Analysis of Biological Molecules. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 25.Stuart AC, Borzilleri KA, Withka JM, Palmer AG., 3rd Compensating for Variations in 1H–13C Scalar Coupling Constants in Isotope-Filtered NMR experiments. J Am Chem Soc. 1999;121:5346–5347. [Google Scholar]
- 26.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 27.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 28.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 29.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 30.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 31.Bezprozvanny I, Maximov A. Classification of PDZ domains. FEBS Lett. 2001;509:457–462. doi: 10.1016/s0014-5793(01)03214-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.