Abstract
A well-defined N-(2-hydroxypropyl)methacrylamide-s-N-(3-aminopropyl)methacrylamide (HPMA-s-APMA) copolymer, synthesized via reversible addition-fragmentation chain transfer (RAFT) polymerization, was utilized for the rational design of multi-conjugates containing both a gene therapeutic, small interfering RNA (siRNA), and a cancer cell targeting moiety, folate. The copolymer contains a biocompatible poly(HPMA) portion (91 mol%) and a primary amine, APMA, portion (9 mol%). A fraction (20 mol%) of the APMA repeats were converted to activated thiols utilizing the amine- and sulfhydryl-reactive molecule N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP). 5’-Thiolated sense strand RNAs were then coupled to the polymer through a disulfide exchange with pendant pyridyldithio moieties giving an 89 ± 4 % degree of conjugation. The unmodified APMA units (80 mol%) were subsequently coupled to amine reactive folates with 81 ± 1 % efficiency. This yielded a multi-conjugate copolymer with 91 mol% HPMA, 2 mol% RNA and 6 mol% folate. siRNA formation was achieved by annealing anti-sense strands to the conjugated RNA sense strands. Subsequent siRNA cleavage under intracellular conditions demonstrated the potential utility of this carrier in gene delivery. The multi-conjugate copolymer and siRNA release were characterized by UV-vis spectroscopy and polyacrylamide gel electrophoresis.
Keywords: RAFT polymerization, small interfering RNA, copolymers, conjugation, pendant functionality
Introduction
Conjugation of therapeutic agents/targeting moieties to synthetic polymers has been the topic of extensive investigation since the introduction of the “depot” model of pharmacologically active polymers by Ringsdorf in 1975.1,2 This “depot” model has four main components which include the polymer backbone, therapeutic agent, spacer, and targeting moiety. These components must meet several requirements to comprise an effective delivery vehicle. First, the polymer backbone must be water soluble and non-immunogenic. Second, coupling of therapeutic agents or other biological motifs (i.e. targeting moieties and diagnostic agents) should be facile and occur under favorable conditions (i.e. temperature, solvent) in order to maintain the integrity of the biologically relevant species. Third, the spacer or linker used in this reaction can be permanent or cleavable, though a cleavable linker may be preferred when the biological species must be released to become active. Spacers can be cleaved enzymatically2,3 or in response to an environmental stimulus, such as a change in pH (i.e. endosome or lysosome)4,5 or redox potential (i.e. extracellular versus intracellular).6,7 Finally, the introduction of targeting groups allows for the carrier to be directed to specific cellular receptors, thus controlling body distribution, enhancing specific cellular uptake, and potentially decreasing adverse side effects because of specific versus universal cellular delivery.
Although the Ringsdorf model was introduced over 30 years ago, limitations of polymerization techniques restricted progress. These limitations included poor control over molecular weight, molecular weight distributions, placement of terminal or pendant reactive groups, polymer architecture (i.e. blocks, stars), and solubility in biologically relevant media.8–10 Traditional “living” polymerization techniques, such as anionic, cationic or group transfer, can address some of these limitations, such as control over molecular weight and polymer architecture, but have stringent reaction conditions and limited monomer selection that often require protecting group chemistries for functional/reactive monomers.11 Subsequent deprotection steps can lead to incomplete protecting group removal or result in other side reactions that affect the polymer backbone structure or other incorporated comonomers.11 Fortunately, recent developments in polymer chemistry have allowed for significant progress in the design of polymer bioconjugates.12 Within the past decade advances in controlled radical polymerization (CRP) have addressed the above drawbacks by providing facile routes for the synthesis of functional (co)polymers with complex architectures and predetermined structopendant and structoterminal functionality.13–28 Reversible addition-fragmentation chain transfer (RAFT) polymerization is arguably the most versatile of the CRP methods because of its ability to control the polymerization of a wide variety of unprotected functional vinyl-based monomers under mild conditions. For example, various laboratories, including our own, have reported the RAFT polymerization of monomers containing primary amines,21,29–32 activated esters,16,33–37 activated thiols,22,23 as well as other functionalities.38–40 The utility of RAFT polymerization and the facile modification of pendant or end groups is not only attractive for small molecule-polymer (i.e. fluorophores, drugs)15,20–23,41 coupling reactions but also biomacromolecule-polymer (i.e. oligonucleotides, proteins, peptides)14,16,18–20,25 conjugations.
Of particular interest due to its potentially high impact in gene therapy, is the conjugation and delivery of the RNA therapeutic, small interfering RNA (siRNA).42 siRNA is the effector molecule of the RNA interference (RNAi) pathway, which leads to messenger RNA cleavage resulting in suppression of the encoded protein.43 The limited dosage required and lower toxicity of siRNA compared to other oligonucleotide treatments makes it particularly attractive for gene-based therapeutics.44,45 Recently, RAFT polymerization has been exploited as a synthetic tool to rationally design siRNA carriers.19–21,25,27,46 For example, our laboratories,21,46 and Hoffman and coworkers,27 have designed block copolymers via RAFT polymerization that can electrostatically complex siRNA for potential gene delivery applications. In addition, Maynard and coworkers19 as well as Bulmus et al.25 have recently reported the successful end group functionalization of RAFT-synthesized polymers with siRNA. However, RAFT-synthesized copolymers that are well-defined and address the requirements of the “depot” model by incorporating dual functionality for facile and robust conjugation of multiple siRNAs and targeting moieties have yet to be reported.
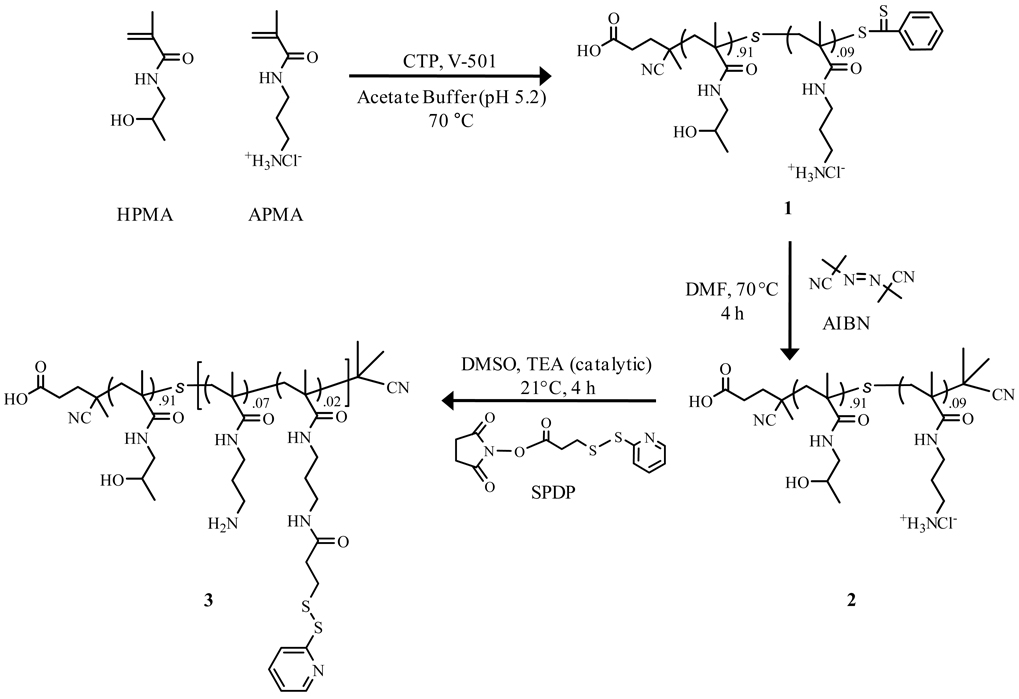
Herein, we report the rational design of a polymeric carrier that encompasses the attributes suggested by Ringsdorf through the incorporation of two reactive functionalities for multi-conjugation of therapeutics (siRNA) and cancer cell targeting moieties (folate) (Scheme 1 and 2). In order to achieve carriers with precise architectures, predetermined structopendant and structoterminal functionalities, and narrow polydispersities, RAFT polymerization was utilized in the synthesis of a N-(2-hydroxypropyl)methacrylamide-s-N-(3-aminopropyl)methacrylamide (HPMA-s-APMA) copolymer. HPMA was chosen as the primary polymer component (90 mol%) due to its non-immunogenic, hydrophilic nature while APMA was statistically incorporated (10 mol%) to provide pendant primary amine functionalities.. A small fraction of the pendant primary amines were subsequently converted to activated thiols, providing a copolymer with two distinct reactive sites. The presence of both activated thiols and primary amines allows for selective and specific conjugation of thiolated-siRNAs and cancer cell targeting moieties (folate), respectively. siRNA release through disulfide cleavage was demonstrated under intracellular conditions while the presence of attached folates would allow for site-directed delivery of the siRNA carrier to cancer cell lines that over-express folate receptors.
Scheme 1.
Reaction pathway for the synthesis of N-(2-hydroxypropyl)methacrylamide-s-N-(3-aminopropyl)methacrylamide (HPMA-s-APMA) copolymer (1), thiocarbonylthio removal and end capped product (2), and copolymer from sequential reaction with N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) (3). Repeat units are designated as mole fractions.
Scheme 2.
Reaction pathway for the synthesis of both RNA (4) and folate (FA) (5) conjugated copolymers and subsequent hybridization (6) with RNA anti-sense strands. Repeat units are designated in mole fractions.
Experimental Section
Materials
All chemicals were purchased from Aldrich (Milwaukee, WI) at the highest available purity and used as received unless otherwise noted. 4,4’-Azobis(4-cyanopentanoic acid) (V-501) and azobisisobutyronitrile (AIBN), gifts from Wako Pure Chemicals Industries, Ltd. (Richmond, VA), were recrystallized twice from methanol. 4-Cyanopentanoic acid dithiobenzoate (CTP) was synthesized according to the literature procedure.47 The RAFT copolymer N-hydroxysuccinimide (NHS) activated folic acid was synthesized as previously reported.21 N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP) was purchased from Thermo Scientific (Rockford, IL) and used as received. Both sense and anti-sense strands of a 59-nucleotide (nt) model small interfering ribonucleic acid (siRNA) against human survivin were chemically synthesized. Unlabeled and 5’-thiolated sense strand, 5’-AGCCCUUUCUCAAGGACCACCGCAUCU-3’, were synthesized by Integrated DNA Technologies. Anti-sense strand, 3’-UUUCGGGAAAGAGUUCCUGGUGGCGUAGAGGA-5’ was synthesized by Biosynthesis Inc.
Synthesis of HPMA-s-APMA Copolymer (1)
HPMA-s-APMA was synthesized as previously reported (Scheme 1).21 Briefly, 10.5 mg (0.0376 mmol) of CTP was weighed and added to a round-bottomed flask equipped with a stir bar. 90 mol% HPMA (3.88 g, 27.0 mmol) and 10 mol% APMA (0.536 g, 3.00 mmol) were then added to the flask and dissolved in 30 mL of acetate buffer (pH 5.2, 0.27 M acetic acid and 0.73 M sodium acetate) bringing the initial monomer concentration ([M]o) to 1 M. After complete dissolution of monomer and CTA, 185 µL (2.10 mg, 7.49 µmol) of a 11.4 mg/mL V-501 stock solution was added to the flask. The round bottomed flask was then septum sealed and purged with nitrogen for 45 min and submerged in a water bath at 70 °C for 3h 30min. HPMA-s-APMA copolymer was prepared with a [M]o/[CTA]o ratio of 800/1, while the [CTA]o/[I]o was kept at 5/1. The copolymer was dialyzed for 3 days (pH 3–4) at 4 °C and dried via lyophilization. HPMA320-s-APMA33 (91 mol% HPMA and 9 mol% APMA) was characterized via aqueous size exclusion chromatography (dn/dc = 0.169 mL g−1)21 and 1H NMR.
Removal of Thiocarbonylthio End Groups from HPMA-s-APMA (2)
Thiocarbonylthio functionalities from compound 1 were removed (Scheme 1) using a previously reported procedure by Perrier and coworkers.48 Briefly, HPMA320-s-APMA33 1 (300 mg, 5.8 µmol) was added to a 10 mL round-bottomed flask and dissolved with 2.9 mL of N,N-dimethylformamide (DMF). Azobisisobutyronitrile (AIBN) (19 mg, 0.116 mmol) was then added to the flask giving an AIBN:copolymer ratio of 20:1. The solution was septum-sealed and purged with nitrogen for 50 min and submersed in a water bath at 70 °C for 4 h. The resulting copolymer was precipitated from DMF in cold anhydrous diethyl ether and washed repeatedly. This step was repeated three times and the copolymer was then dried in vacuo overnight. Quantitative removal of thiocarbonylthio group was determined with UV-vis spectroscopy.
Thiol Activation of HPMA-s-APMA with SPDP (3)
Activated thiols were introduced along the copolymer backbone of compound 2 through the reaction of N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP) with the pendant functional primary amine groups of APMA (Scheme 1). 24 mol% (8 of the available 33) APMA units were targeted for functionalization with SPDP. First, 20 mg (0.39 µmol) of end-capped (no thiocarbonylthio group) copolymer 2 was dissolved in 335 µL of anhydrous DMSO. 48 µL of a 20 mg/mL stock solution of SPDP in anhydrous DMSO (0.97 mg, 3.1 µmol) was then added to the copolymer solution. Triethylamine (TEA) (3.5 µL, 25 µmol) was added to serve as a catalyst and ensure deprotonation of primary amines, thus giving a final concentration of 1 mM and 8 mM for copolymer 2 and SPDP, respectively. The resulting solution was allowed to react for 4 h at room temperature. Following reaction, thiol activated copolymer 3 was precipitated in cold anhydrous diethyl ether and dried overnight in vacuo. The dried copolymer was then dissolved in deionized (DI) water and lyophilized for 24 hours. As determined by UV-vis and 1H NMR spectroscopy, 7 out of 33 APMA groups (20 mol%) were converted to activated thiols (pyridyl disulfides).
Conjugation of HS-RNA Sense Strand to Thiol Activated Copolymer 3 via Disulfide Exchange (4)
Prior to RNA conjugation, the chemically synthesized 5’ thiolated sense strand RNA purchased from Integrated DNA Technologies was treated with dithiothreitol (DTT) to remove the disulfide protecting group, thus giving a RNA with reactive thiol functionality. The protected RNA was first dissolved in nuclease free water to a final concentration of 1 mM. Disulfide cleavage was performed by incubating 50 µL of the RNA with 100 mM DTT and 20 mM sodium phosphate buffer (pH 8.0, total volume 60 µL) for 30 min at room temperature. Following disulfide cleavage, 40 µL of nuclease free water, 20 µL of 3 M sodium acetate buffer (pH 5.2), and 360 µL ethanol were added to the solution to promote RNA precipitation. After cooling the sample for 30 min at −20 °C, the sample was centrifuged for 6 min at 14,000 rpm. After careful removal of the supernatant, the RNA pellet was redissolved in 100 µL of nuclease free water. The solution was then loaded onto a Microcon 10 kDa molecular weight cut-off (MWCO) centrifuge tube and spun at 14,000 rpm for 20 min at room temperature. 100 µL nuclease free water was added to the membrane filter and was subsequently centrifuged for 20 min at 14,000 rpm. Finally, the recovered 5’ thiolated sense strand RNA (HS-RNA) concentration was determined by UV-vis spectroscopy.
Conjugation of HS-RNA to thiol activated copolymer 3 was performed through a disulfide exchange reaction (Scheme 2). In a typical reaction, 29 µL of 700 µM HS-RNA (20 nmol) was added to 57 µL of 0.2 M phosphate buffer (pH 8.0). Second, 14 µL of a 100 µM thiol activated copolymer (3) solution (1.4 nmol polymer, 10 nmol pyridyl disulfide) was then added to the RNA solution bringing the final volume to 100 µL and ratio of pyridyl disulfides to HS-RNA 1:2. This reaction was carried out at room temperature for 3 h. The reaction was repeated four times and a degree of conjugation was determined to be 89 ± 4 % using polyacrylamide gel electrophoresis (PAGE). This corresponds to 6.3 repeats (2 mol%) of RNA along the copolymer backbone.
RNA conjugated copolymer 4 was isolated and purified through PAGE. The reaction mixture was first concentrated to 15 µL using a 30 kDa MWCO Microcon centrifuge tube. Gel-loading buffer containing 15 µL of 0.2% bromophenol blue, 8 M urea, and 1X TBE buffer (Trisborate-EDTA) was added to the reaction tube. The sample was then loaded onto an 8% denaturing polyacrylamide gel and run for 50 min at 15 W. Location of the RNA conjugated copolymer 4 on the gel was determined using UV light and the corresponding band was cut out and crushed inside a 1.5 mL centrifuge tube. Compound 4 was extracted through washing of the crushed gel with nuclease free water (400 µL) and heating at 70 °C for 5 min. This step was repeated in triplicate to increase product recovery. The resulting product was concentrated using a 30 kDa MWCO Microcon centrifuge tube. For UV-vis spectroscopic analysis, 1 µL of the recovered product was diluted in 1 mL of 20 mM phosphate buffer (pH 7.4) and the absorbance at 260 nm was recorded to determine the RNA concentration. The resulting concentration of RNA conjugated copolymer 4 was 23 µM giving a total RNA concentration of 146 µM. The recovery yields via PAGE extraction for the RNA conjugated copolymer 4 and the unreacted HS-RNA were 48 % and ~80 %, respectively.
NHS-Folate Conjugation to RNA Conjugated Copolymer 4 (5)
Following the isolation of RNA conjugated copolymer 4, the remaining APMA pendant functionalities (APMA functionalities not reacted with SPDP) were labeled with NHS-activated folate (Scheme 2). First, the nuclease free water was removed in vacuo from the copolymer 4 solution using a Speed Vac for 3h. Following complete water removal, copolymer 4 was dissolved in 10 µL of anhydrous DMSO followed by the addition of 4 µL of 80 mM (0.32 µmols) NHS activated folate. 1 µL of TEA was added as a catalyst bringing the total reaction volume to 15 µL giving a copolymer 4 concentration of 46.0 µM, RNA concentration of 290 µM and NHS activated folate concentration of 20 mM (1:6.3:435). This reaction was allowed to proceed at room temperature for 2 h. The reaction mixture was then diluted to 500 µL using 100 mM phosphate buffer (pH 8.0). Excess folic acid was removed using a 30 kDa MWCO Microcon centrifuge tube. The product (5) was washed four times with 200 µL of 40 mM phosphate buffer (pH 8.0) and once with 100 µL nuclease free water. RNA/folate conjugated copolymer 5 was concentrated to a final volume of 50 µL. For UV-vis spectroscopic analysis 2 µL of the recovered product was diluted in 0.5 mL with 20 mM phosphate buffer (pH 7.4) and the absorbance was recorded. It was determined through UV-vis spectroscopy that 81 ± 2 % of the remaining APMA units coupled with folate. This results in a multi-conjugated copolymer that is 91 mol% HPMA, 2 mol % RNA and 6 mol % folate. The remaining 1 mol % would be comprised of unmodified APMA units.
Anti-sense RNA Strand Hybridization (6)
Following the synthesis of multi-conjugate copolymer 5, the hybridization between the copolymer conjugated RNA sense strand and the RNA anti-sense strand was performed (Scheme 2). Typical hybridization conditions are as follows: 15 µL of 11.9 µM RNA/folate conjugated copolymer 5, 75 µM RNA (1.13 nmols of RNA), was added to 11.3 µL of a 100 µM (1.13 nmols) solution of anti-survivin RNA anti-sense strand. The resulting solution was added to 15 µL of 0.39 M NaCl/60 mM phosphate buffer (pH 7.0) and diluted to 45 µL with nuclease free water giving a final RNA concentration of 25 µM and salt concentration of 0.15 M. The two RNA strands were annealed at 72 °C for 2 min and allowed to cool back down to room temperature resulting in the siRNA conjugated copolymer 6. Quantitative hybridization to the resulting anti-survivin siRNA was confirmed by PAGE.
Copolymer Characterization
HPMA320-s-APMA33 copolymer 1, and copolymer 2 and 3 were characterized by aqueous size exclusion chromatography (ASEC) utilizing an eluent of 1 wt% acetic acid/0.10 M Na2SO4 (aq) at a flow rate of 0.25 mL/min at 25 °C, Eprogen, Inc. CATSEC columns (100, 300, and 1000 Å), a Polymer Labs LC1200 UV/Vis detector, a Wyatt Optilab DSP interferometric refractometer (λ = 690 nm), and a Wyatt DAWN-DSP multiangle laser light scattering (MALLS) detector (λ = 633 nm). Removal of the thiocarbonylthio functionality was confirmed by monitoring the UV signal at 310 nm before and after removal.
The thiol activation of copolymer 2 with SPDP was verified by 1H NMR and UV-vis spectroscopy. 1H NMR was performed with a Varian MercuryPLUS 300 MHz spectrometer in DMSO-d6 utilizing delay times of 2s. The amount of SPDP reacted with the APMA functionalities of copolymer 2 was estimated to be 7 out of 33 (20 mol %) of the APMA units by integration of the methyne-proton resonance of HPMA at 3.7 ppm and the proton resonance of the SPDP pyridyl ring at 8.4 ppm. These integration values were estimated employing a Lorentzian/Gaussian line fit using MestReC NMR data processing software version 4.7. UV-vis spectroscopy was carried out using a Varian Cary 50 Bio spectrophotometer for copolymer 1, 2 and 3 to demonstrate thiocarbonylthio removal and the presence of pyridyl disulfide functionalities. All UV-vis measurements were repeated in triplicate.
Characterization of Copolymer 4 and Copolymer 5 Multi-Conjugates
Conjugation of RNA sense strand to copolymer 3 and subsequent folate conjugation was verified by PAGE and/or UV-vis spectroscopy. All gel electrophoresis was carried out using 8% polyacrylamide denaturing gel using 1X TBE buffer/Urea as running buffer. All gels were stained with ethidium bromide (EtBr) for band visualization and imaged by a BIO-RAD Molecular Imager FX using Quantity One software package version 4.2.2. For analysis of RNA conjugated copolymer 4, 2 µL of the reaction solution was diluted to 16 µL with gel-loading buffer containing 0.2% bromophenol blue, 8 M urea, and 1X TBE buffer. 2 µL (approx. 50 pmol RNA) was then loaded onto the gel and run for 10 min at 15 W. The degree of conjugation of HS-RNA was determined using BIO-RAD Molecular Analyst software package version 1.5 through the comparison of the product band (RNA conjugated copolymer 4) to that of free/unreacted HS-RNA using Equation 1 and was found to be 89 ± 4 %.
| (1) |
The 2 in Equation 1 is due to the fact that two times excess HS-RNA was employed.
For gel analysis of purified copolymer 4, RNA/folate conjugated copolymer 5, and hybridization (siRNA/folate conjugated copolymer 6), all sample solutions were diluted to approximately 25 µM RNA concentration and 1 µL of the resulting solution was diluted to 2 µL with gel-loading buffer containing 0.2% bromophenol blue, 8 M urea, and 1X TBE buffer. The samples were loaded immediately and allowed to run for 12 min at 15 W, with the exception of the hybridization reaction which was run for 30 min at the same wattage. For samples involving DTT reduction 1 µL of the above sample solutions were diluted with 1 µL of 0.2 M DTT/80 mM phosphate buffer (pH 8.0) and allowed to incubate at 37 °C for 30 minutes. Following reduction the resulting samples were diluted with 2 µL of gel-loading buffer containing 0.2% bromophenol blue, 8 M urea, and 1X TBE buffer. The samples were loaded immediately and allowed to run under the same conditions as above.
UV-vis spectroscopy was carried out using a Varian Cary 50 Bio spectrophotometer for RNA conjugated copolymer 4 and RNA/folate conjugated copolymer 5. An extinction coefficient (ε) of 250,000 M−1 cm−1 for polynucleotides at 260 nm was used to determine the RNA concentration for the RNA conjugated copolymer 4. Conjugation of NHS activated folate to RNA conjugated copolymer 4 was determined by comparing the absorbance at 260 nm (maxima for RNA) to 281 nm (maxima for folate). Extinction coefficients for free folic acid in 40 mM phosphate buffer (pH 8.0) at 260 and 281 nm are 16,600 M−1 cm−1 and 30,300 M−1 cm−1, respectively. Extinction coefficient for RNA at 281 nm is 107,500 M−1 cm−1. Using the following two equations, the concentration of RNA and folate linked to the copolymer was determined:
| (2) |
| (3) |
where A is the absorbance at either 260 or 281 nm, C is the concentration of either RNA of folate (FA) and ε is the extinction coefficient for RNA or FA at either 260 or 281 nm. All UV-vis measurements were repeated in triplicate. It is important to note that the absorbance due to the copolymer at the measured concentrations is negligible at both 260 and 281 nm.
siRNA Release via Glutathione
siRNA release from siRNA/folate conjugated copolymer 6 was carried out in the presence of glutathione to simulate intracellular conditions. First, 4 µL of a 4 µM siRNA/folate conjugated copolymer 6 solution (25 µM anti-survivin siRNA) was added to 4 µL of a 10 mM glutathione / 40 mM phosphate buffer (pH 7.4) solution bringing the final concentration of glutathione to 5 mM and siRNA to 12.5 µM. The resulting solution was incubated at 37 °C and 1 µL aliquots were taken at increasing time intervals and subsequently diluted with 2 µL of gel-loading buffer containing 0.2% bromophenol blue, 8 M urea, and 1X TBE buffer. The resulting samples were loaded and run on an 8% polyacrylamide denaturing gel using 1X TBE buffer/Urea as the running buffer for 20 min at 15 W. The percent release of anti-survivin siRNA was determined using BIO-RAD Molecular Analyst software package version 1.5 where the intensity of the released siRNA band was directly compared to the remaining conjugate band.
Results and Discussion
Synthesis of Thiol Activated HPMA-s-APMA (3)
The polymeric platform utilized for multi-conjugation reactions, was designed to contain 90 mol % hydrophilic HPMA repeats and 10 mol % primary amine pendant APMA units distributed statistically along the polymer backbone. The statistical copolymer, shown as compound 1 in Scheme 1, was prepared utilizing aqueous RAFT polymerization with 4-cyanopentanoic dithiobenzoate (CTP) and 4,4’-azobis(4-cyanopentanoic acid) (V-501) as the CTA and initiator, respectively (Scheme 1). As determined by ASEC-MALLS the resulting copolymer had a number average molecular weight (Mn) of 51,800 g/mol and a polydispersity index (PDI) of 1.13. The copolymer composition, HPMA320-s-APMA33, was determined by 1H NMR (data not shown). Low molecular weight tailing is apparent in the ASEC chromatogram (Figure 1 a) which is typically observed in RAFT polymerizations and has been attributed to premature termination and/or loss of the CTA thiocarbonylthio moiety.9,46 Nevertheless, the chromatogram is unimodal and narrow, representative of a controlled polymerization.
Figure 1.
Aqueous SEC-MALLS traces of (a) N-(2-hydroxypropyl)methacrylamide-s-N-(3-aminopropyl)methacrylamide (HPMA-s-APMA) (1), (b) HPMA-s-APMA after thiocarbonylthio removal (2) and (c) thiol activated copolymer 3 after reaction with N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP). Analysis was conducted at pH 3.0.
The ω-thiocarbonylthio functionality was then removed to avoid aminolysis by the pendant primary amine functionalities of APMA (Scheme 1). This removal was carried out via addition of excess AIBN according to a literature procedure developed by Perrier and coworkers48 yielding copolymer 2 illustrated in Scheme 1. The removal of the thiocarbonylthio functionality was confirmed to be quantitative by comparing the absorbance at 310 nm before and after removal (Figure 2). Further evidence is provided by monitoring the thiocarbonylthio absorbance at 310 nm with a UV-vis detector in-line with ASEC instrumentation (Supporting Information, Figure S1). The ASEC chromatogram (RI response) for end-capped HPMA320-s-APMA33 (2) is unimodal with no detectable byproducts (Figure 1 b). The Mn and PDI obtained from ASEC-MALLS (Mn = 51,600 g/mol, PDI = 1.10) are similar to those of copolymer 1 prior to end group removal. Although, other methods for the removal of thiocarbonylthio groups are available8,49,50 the utilized method proved to be satisfactory.
Figure 2.
UV-vis spectra of (—) N-(2-hydroxypropyl)methacrylamide-s-N-(3-aminopropyl)methacrylamide (HPMA-s-APMA) (1), (…) HPMA-s-APMA after thiocarbonylthio removal (2) and (----) thiol activated copolymer 3 after reaction with N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) in 20 mM phosphate buffer (pH 7.4).
Following thiocarbonylthio removal, copolymer 2 was reacted with the amine-and sulfhydryl-reactive molecule N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) (Scheme 1) to yield the thiol activated copolymer 3. By modifying a portion of pendant amines with SPDP, a copolymer containing both pyridyl disulfide sites and primary amine sites was prepared. The introduction of these two types of functionality allows copolymer 3 to be modified with both thiol and amine reactive compounds, which is beneficial for conjugation of multiple chemically-distinct therapeutics and targeting moieties.
Stoichiometries were used such that only 24 mol % of the available APMA repeat units (8 of 33) would be converted to activated thiols when reacted with SPDP. The successful reaction of SPDP with copolymer 2 was confirmed by both UV-vis (Figure 2) and 1H NMR spectroscopy (Figure 3). Based on an extinction coefficient of 5100 M−1 cm−1 for pyridyl disulfide at 280 nm,51 the number of pyridyl disulfide groups reacted with copolymer 2 was determined to be 7.1, indicating that 20 mol % of the available APMA units were reacted with SPDP. The significant increase in absorbance at 280 nm is shown in Figure 2. Additionally, 1H NMR spectroscopy (Figure 3) was utilized to confirm the presence of pyridyl disulfide functionalities in copolymer 3 as evidenced by the appearance of aromatic protons at 8.4 ppm and 7.6–7.8 ppm. The amount of conjugated SPDP was estimated by comparing the pyridyl resonance at 8.4 ppm to that of the methyne-proton of HPMA at 3.7 ppm. This analysis indicates that 20 mol % of the APMA units were modified (6.9 pyridyl disulfide groups), thus corroborating the results obtained by UV-vis spectroscopy. For comparison, the 1H NMR spectrum for copolymer 2, along with integrations, can be found in the Supporting Information (Figure S2). In addition, a 2,4,6-trinitrobenzene sulfonic acid (TNBS) primary amine assay was performed on copolymer 2 to determine the number of APMA units and corroborate the result determined by the combination of 1H NMR and SEC (33 APMA units). As expected, upon measuring the absorbance at 420 nm due to reacted TNBS, 32.9 ± 0.6 APMA units were found (Supporting Information, Figure S3).
Figure 3.
1H NMR spectra (d6-DMSO) showing the successful thiol activation of copolymer 2 with N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP).
In addition, ASEC-MALLS was utilized to demonstrate that thiol activated copolymer 3 remained narrowly dispersed. As shown in Figure 2 c, the chromatogram remained unimodal and narrowly dispersed with a PDI of 1.18 suggesting minimal side reactions. Further characterization was provided by monitoring the pyridyl disulfide absorbance at 280 nm with a UV-vis detector in-line with our ASEC instrumentation (Supporting Information, Figure S1). The Mn of copolymer 3 was estimated to be 53,000 g/mol based on the results of 1H NMR and UV-vis spectroscopy that 20 mol% of the APMA units were modified by SPDP. ASEC-MALLS, UV-vis, and 1H NMR spectroscopy data corroborate the successful synthesis of the target, dually reactive (amines and activated thiols) copolymer (3) allowing for subsequent conjugation of both RNA and folate.
siRNA and Folate Conjugation to Thiol Activated Copolymer 3
Following the successful preparation of copolymer 3, a 5’-thiolated RNA sense strand (HS-RNA) was then introduced via a disulfide exchange reaction between pendant pyridyl disulfides and HS-RNA to yield RNA conjugated copolymer 4 as shown in Scheme 2. The disulfide exchange reaction was successfully carried out at pH 8.0 with a molar ratio of pendant pyridyl disulfide groups to HS-RNA of 1:2. Successful HS-RNA conjugation was evidenced using polyacrylamide gel electrophoresis (PAGE) (Figure 4 A and B) and UV-vis spectroscopy (Figure 5).
Figure 4.
Polyacrylamide gel electrophoresis (PAGE) of (A) control reactions, (B) RNA conjugated copolymer 4 and RNA/folate conjugated copolymer 5 and (C) hybridization of RNA/folate copolymer conjugate 5 with RNA anti-sense strand to yield small interfering RNA (siRNA)/folate conjugate 6. A) Lane 1 – copolymer 2 with 5’-thiolated RNA sense strand (HS-RNA), Lane 2 – thiol activated copolymer 3 with RNA sense strand, Lane 3 – thiol activated copolymer 3 reacted with HS-RNA and Lane 4 – components of lane 3 treated with 0.1 M dithiothreitol (DTT). B) Lane 1 – HS-RNA, Lane 2 – thiol activated copolymer 3 reacted with HS-RNA, Lane 3 – purified RNA conjugated copolymer 4, Lane 4 – folate conjugated to copolymer 2 (contains no RNA), Lane 5 – Purified RNA/folate conjugated copolymer 5, Lane 6 – lane 5 components treated with 0.1 M DTT. C) Lane 1 – RNA anti-sense strand, Lane 2 – RNA/folate conjugated copolymer 5 hybridized with RNA anti-sense strand yielding siRNA/folate conjugate 6, Lane 3 – lane 2 components treated with 0.1 M DTT, Lane 4 – double stranded anti-survivin siRNA. All images were stained with ethidium bromide (EtBr) and the run direction is from the top to bottom of images. Intensity differences between lane 1 and 4 in C) is due to intercalation efficiency of EtBr for double stranded RNA versus single stranded, not concentration.
Figure 5.
UV-vis spectra normalized by their extinction coefficients at 260 nm for (—) free RNA sense strand, (---) RNA conjugated copolymer 4 and (…) RNA/folate conjugated copolymer 5 carried out in 20 mM phosphate buffer (pH 7.4).
Linkage of HS-RNA to copolymer 3 is shown by PAGE through the appearance of a high molecular weight band (Figure 4). Using Equation 1 from the Experimental Section, the degree of conjugation of HS-RNA was determined by comparing the intensity of the unreacted HS-RNA and RNA dimer band to that of the conjugate band. From four replicates and standard gel imaging, the degree of conjugation was found to be 89 ± 4 %. This implies that 6.3 of the 7 pendant pyridyl disulfides participated in the disulfide exchange reaction. This coupling efficiency yields an RNA conjugated copolymer (4) which contains 2 mol% pendant RNA groups, 91 mol% HPMA and 7 mol% APMA (Scheme 2).
Control reactions with copolymer 2 and 3 and HS-RNA were carried out to verify that the HS-RNA was indeed attached via a disulfide linkage and not merely associated with the polymer through non-covalent interactions (Figure 4 A). Lane 1 in Figure 4 A shows the incubated product of copolymer 2 with HS-RNA while lane 2 shows the results from incubation of thiol activated copolymer 3 with un-thiolated RNA. These control reactions were carried out under identical conditions of the RNA conjugation reaction. No appreciable conjugate formation is evidenced in lanes 1 & 2 (Figure 4 A) when compared to lane 3 which shows specific conjugation between thiol activated copolymer 3 and HS-RNA. Weak electrostatic associations between the copolymer and RNA may be attributed to the statistical nature of the copolymer and to the large excess of HS-RNA compared to primary amine pendant groups (nitrogen/phosphate = 0.07). Since HS-RNA is used, a dimer band is visible above the unreacted HS-RNA which is attributed to disulfide formation between two HS-RNA strands. RNA attachment to the copolymer backbone through a disulfide linkage was established by showing RNA cleavage under reducing conditions (0.1 M DTT for 30 min) which led to the disappearance of the conjugate band (Figure 4 A lane 4) as well as the HS-RNA dimer band.
The current method provides a facile and robust route for the coupling of multiple RNAs to RAFT-synthesized copolymers utilizing low molar ratios and relatively fast reaction times under mild conditions. The introduction of multiple thiol reactive sites renders attachment of modified RNAs/oligonucleotides to high molecular weight polymers straightforward with the potential to couple multiple gene targets to the same carrier. Due to low molar ratios, purification of the RNA copolymer conjugate 4 is facile and allows for isolation and subsequent modifications with targeting moieties. Therefore, this pathway is advantageous for producing well-defined targeted gene delivery vehicles as compared to previously reported methods for reacting modified oligonucleotides to the end groups of RAFT generated polymers.19,20,25 Those methods required 20 to 50 times excess of polymer relative to siRNA/oligonucleotide and long reaction times (18–20 h) to achieve high degrees of end-group conjugation. Furthermore, these higher molar ratios may further complicate purification steps if conjugation to high molecular weight polymers (> 10,000 g/mol) is desired due to difficulty in separating two high molecular weight species (i.e. the polymer and the polymer-oligonucleotide conjugate) by dialysis or membrane filtration separation methods. However, separation of excess polymer or RNA when employing end group modification methods is possible via HPLC. Additionally, molecular weights below 10,000 g/mol were used to achieve high coupling efficiency whereas molecular weights above this led to low or incomplete coupling efficiency even when excess polymer was present. A possible explanation for this low degree of conjugation to higher molecular weight polymers is that the RAFT polymers utilized contained only one reactive site at either the α or ω terminus (i.e. monofunctional). This could lead to poor collision frequency or proper orientation between two reactive functionalities, thus further complicating an already difficult reaction between two macromolecular species. Another possible reason is incomplete chain end functionalization during platform synthesis, thus resulting in a lower degree of oligonucleotide conjugation.25
In contrast to previously reported methods which required 20 to 50 times excess of thiol-oligonucleotide to active site to obtain high yields, a 2 times excess of HS-RNA in our approach resulted in 90 % conjugation efficiency to a copolymer with Mn > 50,000 g/mol. Since two times excess HS-RNA is used relative to reactive copolymer sites, the removal of unreacted HS-RNA from the RNA conjugated copolymer 4 is straightforward. This removal is evidenced by comparing lanes 2 and 3 in Figure 4 B showing the conjugate band visible only in lane 3. As a side note, the unreacted HS-RNA in these reactions can be recovered through extraction from the PAGE gel and reused for future studies, which is beneficial given the high cost of modified oligonucleotides. A similar recovery method could be employed for end group modified RNA-RAFT copolymer conjugates. Following purification and recovery of RNA conjugated copolymer 4, the concentration of RNA was determined by UV-vis spectroscopy. As shown in Figure 5, the free RNA spectrum is analogous to that of RNA conjugated copolymer 4 thus supporting the PAGE results. The slightly higher absorbance present in the RNA conjugated copolymer 4 spectrum below 250 nm is due to the absorbance of the copolymer repeating units.
Following isolation and subsequent analysis of RNA conjugated copolymer 4, the remaining APMA groups (26 repeats, 7 mol%) were reacted with excess NHS-activated folate (NHS-FA) for 2 h as illustrated in Scheme 2. Any excess or unreacted folate was removed by membrane filtration. After purification and recovery, RNA/folate conjugated copolymer 5 was analyzed by UV-vis spectroscopy. As shown in Figure 5 (dotted line) the absorbance increase is due to conjugated folate. The shoulder peaks that appear near 350 and 310 nm are also due to the presence of folate. Given the higher extinction coefficient for folate at 281 nm (30300 M−1 cm−1) versus 350 nm (8000 M−1 cm−1)21 and the low concentration of the measured sample, the former extinction coefficient was used for a more accurate analysis. Using Equations 2 and 3 (Experimental Section), the relative concentrations of both RNA and folate were calculated. The number of conjugated folates was determined to be approximately 21 out of 26 available sites, yielding a coupling efficiency of 81 ± 1 %. Similarly, using the extinction coefficient for folate at 350 nm, it was estimated that 19 sites contained folate, which is in close agreement to the results obtained from Equations 2 and 3. The resulting copolymer consists of 91 mol% HPMA, 2 mol% RNA, and 6 mol% folate. The remaining 1 mol% can be assigned to unmodified APMA units.
RNA/folate conjugated copolymer 5 was also analyzed by PAGE (Figure 4 B lane 5) to verify that the coupled RNA was unaffected by the folate reaction. As a control, lane 4 of Figure 4 B contains folate conjugated to copolymer 2 (no RNA) to show that the copolymer or folate cannot be stained by EtBr and that neither fluoresce under standard gel imagining techniques. Importantly, lane 6 in Figure 4 B indicates that the disulfide-linked RNA can still be released in the presence of 0.1 M DTT. Additionally, our studies revealed that the order of reaction is important (data not shown). Reaction of NHS-FA followed by HS-RNA led to substantially lower degrees of RNA conjugation (30–40%) as compared to the synthetic pathway described above. Decreased yields are attributed to pendant folates sterically blocking activated thiol sites or secondary interactions between multiple folates, lowering the number of accessible activated thiol sites for reaction with HS-RNA. Given the lower degree of HS-RNA conjugation, reactions by this alternative route were not further explored.
In the last step, illustrated in Scheme 2, the conjugated RNA strand was hybridized with its complementary anti-sense strand to form double stranded siRNA. The siRNA model chosen for this work was an anti-survivin siRNA which our laboratories have used previously due to its potential in cancer treatment.21 PAGE was employed in order to confirm hybridization as shown in Figure 4 C. Lane 1 indicates only the anti-sense strand while lane 2 shows the hybridized product. As seen in lane 2 of Figure 4 C, no band is present for the anti-sense strand indicating quantitative hybridization. The release of the siRNA from the copolymer backbone under reducing conditions (0.1 M DTT) is visualized in lane 3 of Figure 4 C and for comparison lane 4 shows unmodified anti-survivin siRNA. The slightly slower migration of the released siRNA in lane 3 as compared to the reference unmodified siRNA in lane 4, Figure 4 C, is attributed to high salt concentration (150 mM) used during the hybridization step. The unmodified siRNA used as a reference in lane 4 contains no salt. A longer running time was used (30 versus 10 min) for the gel in Figure 4C compared to that in Figure 4A to compensate for the faster migration of double stranded siRNA (59 nt) versus single stranded RNA with a similar number of nucleotides (e.g. the RNA dimer (54 nt) in Figure 4A). This difference arises from the RNA chain confirmation in solution (i.e. random coil versus double helix). Although it was demonstrated that the siRNA could be released in the presence of 0.1 M DTT, subsequent studies were conducted to verify siRNA release under conditions similar to those in the cytoplasm of the cell.
siRNA Release Under Intracellular Conditions
Effective gene delivery requires an intracellular release mechanism of the therapeutic agent from the carrier. The reducing environment in the cytoplasm of cells attributed to the presence of glutathione render disulfide linkages attractive for intracellular therapeutic release. In fact, several research groups are using disulfides as a means to conjugate siRNA, thus exploiting intracellular reducing conditions.19,20,23,25,52–55 Typical glutathione concentrations in the intracellular compartment of mammalian cells range from 1–10 mM.6,7 Therefore, we sought to demonstrate that the attached oligonucleotide can be released not just under the artificial conditions (0.1 M DTT) demonstrated above, but also under more typical cytoplasmic conditions. The synthesized siRNA/folate copolymer conjugate 6 was incubated at 37 °C in the presence of 5 mM glutathione (pH 7.4). The profile of siRNA released versus time and the PAGE image can be seen in Figures 6 A & B. Approximately 60 % release was achieved in 4 hours with an estimated half-life of 200 min. These experiments demonstrate that the siRNA/folate copolymer conjugate 6 can indeed release siRNA under these conditions.
Figure 6.
Release of siRNA from siRNA/folate copolymer conjugate 6 incubated in the presence of 5 mM glutathione (pH 7.4). A) Percent release of siRNA versus time and B) polyacrylamide gel electrophoresis (PAGE) of siRNA/folate copolymer conjugate 6 incubated with glutathione at increasing time intervals (left to right).
Conclusions
A unique synthetic pathway allowing the conjugation of gene therapeutics and targeting moieties to a well-defined polymeric platform has been reported. The synthesized copolymer contains biocompatible segments provided by poly(HPMA) and both primary amine and activated thiol functionalities for post-polymerization modification. In this case, the introduction of multiple reactive sites allows attachment of multiple siRNAs and multiple folates for potential targeted delivery to cancerous cells that over-express folate receptors. The degree of siRNA conjugation was determined to be 89 ± 4 % (6 siRNAs) and the coupling efficiency of folate was 80 ± 1 % (21 folates). The final molar composition of the siRNA/folate copolymer conjugate 6 was 91 mol% HPMA, 2 mol% siRNA, and 6 mol% folate. To date, the conjugation of siRNA, specifically to copolymers synthesized via CRP methods, has been limited to the end-groups of low molecular weight polymers.19,20,25 The current method addresses limitations of previous methods by allowing 1) reaction of multiple siRNAs with lower molar ratios, 2) coupling of targeting moieties, 3) conjugation to high molecular weight polymers, 4) facile purification, and 5) short reaction times while still taking advantage of RAFT polymerization. In addition, by linking siRNAs via disulfide bonds, release of siRNA from the polymer was achieved under simulated intracellular conditions (5 mM glutathione) demonstrating the potential utility of these carriers for future delivery to cancer cells. Although attachment of siRNA and folate were demonstrated, multiple types of bio-agents with either thiol or amine reactive functionalities might also be envisioned for various delivery and/or biodiagnostics purposes. We feel that this strategy, providing two reactive side-chain functionalities, will be useful in the preparation of other well-defined bioconjugate systems. We are currently optimizing this system for the in vitro treatment of cancer cells which over-express folate receptors.
Supplementary Material
Acknowledgements
The authors would like to thank MRSEC (DMR-0213883), NIH (CA120566) and the Robert M. Hearin Foundation for financial support. We also thank Dr. Vijay Rangachari for use of his UV-vis spectrophotometer.
Footnotes
Paper number 144 in a series entitled “Water Soluble Polymers”
Supporting Information Available: SEC chromatograms monitoring the UV-vis absorbance at 310 nm and 280 nm, 1H NMR spectrum of copolymer 2, and results from the TNBS primary amine assay are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ringsdorf H. J. Polym. Sci. Symp. 1975;51:135–153. [Google Scholar]
- 2.Hoste K, De Winne K, Schacht E. Int. J. Pharm. 2004;277:119–131. doi: 10.1016/j.ijpharm.2003.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Duncan R, Cable R, Lloyd HC, Rejmanova P, Kopecek J. Makromol. Chem. 1983;184:1997–2008. [Google Scholar]
- 4.Kratz F, Beyer U, Schütte MT. Crit. Rev. Ther. Drug Carrier Syst. 1999;16:245–288. doi: 10.1615/critrevtherdrugcarriersyst.v16.i3.10. [DOI] [PubMed] [Google Scholar]
- 5.Ulbrich K, Subr V. Adv. Drug Delivery Rev. 2004;56:1023–1050. doi: 10.1016/j.addr.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 6.Linsdell P, Hanrahan JW. Am. J. Physiol. Cell Physiol. 1998;275:C323–C326. doi: 10.1152/ajpcell.1998.275.1.C323. [DOI] [PubMed] [Google Scholar]
- 7.Smith CV, Jones DP, Guenther TM, Lash LH, Lauterberg BH. Toxicol. Appl. Pharmacol. 1996;140:1–12. doi: 10.1006/taap.1996.0191. [DOI] [PubMed] [Google Scholar]
- 8.Moad G, Rizzardo E, Thang SH. Aust. J. Chem. 2005;58:379–410. [Google Scholar]
- 9.Moad G, Rizzardo E, Thang SH. Aust. J. Chem. 2006;59:669–692. [Google Scholar]
- 10.Lowe AB, McCormick CL. Prog. Polym. Sci. 2007;32:283–351. [Google Scholar]
- 11.Gauthier MA, Gibson MI, Klok H-A. Angew. Chem. Int. Ed. 2009;48:48–58. doi: 10.1002/anie.200801951. [DOI] [PubMed] [Google Scholar]
- 12.Lutz J-F, Börner HG. Prog. Polym. Sci. 2008;33:1–39. [Google Scholar]
- 13.Nicolas J, Mantovani G, Haddleton DM. Macromol. Rapid Commun. 2007;28:1083–1111. [Google Scholar]
- 14.Kulkarni S, Schilli C, Grin B, Muller AHE, Hoffman AS, Stayton PS. Biomacromolecules. 2006;7:2736–2741. doi: 10.1021/bm060186f. [DOI] [PubMed] [Google Scholar]
- 15.York AW, Scales CW, Huang F, McCormick CL. Biomacromolecules. 2007;8:2337–2341. doi: 10.1021/bm700514q. [DOI] [PubMed] [Google Scholar]
- 16.Yanjarappa MJ, Gujraty KV, Joshi A, Saraph A, Kane RS. Biomacromolecules. 2006;7:1665–1670. doi: 10.1021/bm060098v. [DOI] [PubMed] [Google Scholar]
- 17.De P, Gondi SR, Sumerlin BS. Biomacromolecules. 2008;9:1064–1070. doi: 10.1021/bm701255v. [DOI] [PubMed] [Google Scholar]
- 18.Boyer C, Bulmus V, Liu J, Davis TP, Stenzel MH, Barner-Kowollik C. J. Am. Chem. Soc. 2007;129:7145–7154. doi: 10.1021/ja070956a. [DOI] [PubMed] [Google Scholar]
- 19.Heredia KL, Nguyen TH, Chang C-W, Bulmus V, Davis TP, Maynard HD. Chem. Commun. 2008:3245–3247. doi: 10.1039/b804812f. [DOI] [PubMed] [Google Scholar]
- 20.Boyer C, Bulmus V, Davis TP. Macromol. Rapid Commun. 2009;30:493–497. doi: 10.1002/marc.200800708. [DOI] [PubMed] [Google Scholar]
- 21.York AW, Zhang Y, Holley AC, Guo Y, Huang F, McCormick CL. Biomacromolecules. 2009;10:936–943. doi: 10.1021/bm8014768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia Z, Wong L, Davis TP, Bulmus V. Biomacromolecules. 2008;9:3106–3113. doi: 10.1021/bm800657e. [DOI] [PubMed] [Google Scholar]
- 23.Wong L, Boyer C, Jia Z, Zareie HM, Davis TP, Bulmus V. Biomacromolecules. 2008;9:1934–1944. doi: 10.1021/bm800197v. [DOI] [PubMed] [Google Scholar]
- 24.York AW, Kirkland SE, McCormick CL. Adv. Drug Deliv. Rev. 2008;60:1018–1036. doi: 10.1016/j.addr.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Boyer C, Bulmus V, Davis TP. J. Polym Sci., Part A: Polym. Chem. 2009;47:4302–4313. [Google Scholar]
- 26.Li M, De P, Gondi SR, Sumerlin BS. Macromol. Rapid Commun. 2008;29:1172–1176. [Google Scholar]
- 27.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. J. Control. Release. 2008;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De P, Li M, Gondi SR, Sumerlin BS. J. Am. Chem. Soc. 2008;130:11288–11289. doi: 10.1021/ja804495v. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Lokitz BS, McCormick CL. Angew. Chem. Int. Ed. 2006;45:5792–5795. doi: 10.1002/anie.200602168. [DOI] [PubMed] [Google Scholar]
- 30.He L, Read ES, Armes SP, Adams DJ. Macromolecules. 2007;40:4429–4438. [Google Scholar]
- 31.Zhicheng D, Hassen B, Keshwaree B, Abdelghani H, Nankishoresing C, Ravin N. J. Polym Sci., Part A: Polym. Chem. 2008;46:4984–4996. [Google Scholar]
- 32.Alidedeoglu AH, York AW, McCormick CL, Morgan SE. J. Polym Sci., Part A: Polym. Chem. 2009;47:5405–5415. [Google Scholar]
- 33.Favier A, D'Agosto F, Charreyre M-T, Pichot C. Polymer. 2004;45:7821–7830. [Google Scholar]
- 34.Vosloo JJ, Tonge MP, Fellows CM, D'Agosto F, Sanderson RD, Gilbert RG. Macromolecules. 2004;37:2371–2382. [Google Scholar]
- 35.Eberhardt M, Théato P. Macromol. Rapid Commun. 2005;26:1488–1493. [Google Scholar]
- 36.Li Y, Lokitz BS, McCormick CL. Macromolecules. 2006;39:81–89. [Google Scholar]
- 37.Li Y, Lokitz BS, Armes S, McCormick CL. Macromolecules. 2006;39:2726–2728. [Google Scholar]
- 38.Li RC, Hwang J, Maynard HD. Chem. Commun. 2007:3631–3633. doi: 10.1039/b709304g. [DOI] [PubMed] [Google Scholar]
- 39.Beck JB, Killops KL, Kang T, Sivanandan K, Bayles A, Mackay ME, Wooley KL, Hawker CJ. Macromolecules. 2009;42:5629–5635. doi: 10.1021/ma900899v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang J, Li RC, Maynard HD. J. Control. Release. 2007;122:279–286. doi: 10.1016/j.jconrel.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 41.Scales CW, Convertine AJ, McCormick CL. Biomacromolecules. 2006;7:1389–1392. doi: 10.1021/bm060192b. [DOI] [PubMed] [Google Scholar]
- 42.Jeong JH, Mok H, Oh Y-K, Park TG. Bioconjugate Chem. 2008 doi: 10.1021/bc800278e. ASAP Article. [DOI] [PubMed] [Google Scholar]
- 43.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 44.Dorsett Y, Tuschl T. Nat. Rev. Drug Discovery. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 45.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 46.Scales CW, Huang F, Li N, Vasilieva YA, Ray J, Convertine AJ, McCormick CL. Macromolecules. 2006;39:6871–6881. [Google Scholar]
- 47.Mitsukami Y, Donovan MS, Lowe AB, McCormick CL. Macromolecules. 2001;34:2248–2256. [Google Scholar]
- 48.Perrier S, Takolpuckdee P, Mars CA. Macromolecules. 2005;38:2033–2036. [Google Scholar]
- 49.Perrier S, Takolpuckdee P. J. Polym. Sci., Part A: Polym. Chem. 2005;43:5347–5393. [Google Scholar]
- 50.Chong YK, Moad G, Rizzardo E, Thang SH. Macromolecules. 2007;40:4446–4455. [Google Scholar]
- 51.Carlsson J, Dreven H, Axén R. Biochem. J. 1977;173:723–737. doi: 10.1042/bj1730723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson TJ, Harel S, Arboleda VA, Prunell GF, Shelanski ML, Greene LA, Troy CM. J. Neurosci. 2004;24:10040–10046. doi: 10.1523/JNEUROSCI.3643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. J. Control. Release. 2006;116:123–129. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 54.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Bioconjugate Chem. 2008;19:2156–2162. doi: 10.1021/bc800249n. [DOI] [PubMed] [Google Scholar]
- 55.Matsumoto S, Christie RJ, Nishiyama N, Miyata K, Ishii A, Oba M, Koyama H, Yamasaki Y, Kataoka K. Biomacromolecules. 2008;10:119–127. doi: 10.1021/bm800985e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.