Summary
An AAA+ ATPase, DnaC delivers DnaB helicase at the E. coli chromosomal origin by a poorly understood process. This report shows that mutant proteins bearing alanine substitutions for two conserved arginines in a motif named box VII are defective in DNA replication, but this deficiency does not arise from impaired interactions with ATP, DnaB or single-stranded DNA. Despite their ability to deliver DnaB to the chromosomal origin to form the prepriming complex, this intermediate is inactive. Quantitative analysis of the prepriming complex suggests that the DnaB-DnaC complex contains three DnaC monomers per DnaB hexamer, and that the interaction of primase with DnaB and primer formation triggers the release of DnaC but not the mutants from DnaB. The interaction of primase with DnaB, and the release of DnaC mark discrete events in the transition from initiation to the elongation stage of DNA replication.
Keywords: DnaC, DnaB, primase, initiation
Introduction
In a process that is tightly coupled with cell growth (Nielsen and Lobner-Olesen, 2008), DnaA initiates DNA replication from the E. coli replication origin, oriC, by promoting the assembly of the enzymatic machinery that duplicates the bacterial chromosome. Biochemically, the process that leads to replisome assembly can be separated into discrete steps (Kaguni, 2006). Bound to ATP, DnaA first binds to the DnaA boxes, I- and τ-sites within oriC (Leonard and Grimwade, 2009). Aided by HU or IHF, DnaA then unwinds an AT-rich region carrying three 13-mer motifs (Bramhill and Kornberg, 1988), and loads one DnaB hexamer from the DnaB-DnaC complex onto each of the separated strands within oriC to form the prepriming complex (Carr and Kaguni, 2001; Fang et al., 1999). After DnaC dissociates from DnaB, the helicase translocates on the parental template DNA and interacts via its N-terminal domain with primase, leading to the formation of primers (Tougu and Marians, 1996b; Wu et al., 1992). Several recent studies provide insight into how DnaB and primase coordinate their functions (Bailey et al., 2007a; Corn et al., 2008). A dimeric DNA polymerase III holoenzyme at each replication fork then extends the primers to duplicate the chromosome (McHenry, 2003; Pomerantz and O'Donnell, 2007). Comparing the mechanism of E. coli DNA replication with that of eukaryotes, it is remarkable that organisms in separate kingdoms utilize similar biochemical mechanisms (Nielsen and Lobner-Olesen, 2008; Robinson and Bell, 2005).
DnaB is the replicative helicase for the E. coli chromosome and for many bacterial plasmids. Six identical DnaB protomers assemble into a ring-like structure through which the single-stranded DNA template is thought to pass during DNA unwinding (Lo et al., 2009; Yu et al., 1996). For DnaB to load at oriC, it must be complexed to DnaC, which regulates the helicase function of DnaB (Davey et al., 2002; Wahle et al., 1989b). Whereas DnaB can bind to naked ssDNA (Biswas et al., 2004; Davey et al., 2002), the helicase cannot bind to the unwound region of oriC without DnaC (Sutton et al., 1998). X-ray crystallography of a gram-positive DnaB and cryo-electron microscopy of E. coli DnaB reveal that each DnaB protomer has a larger C-terminal domain containing a RecA-like fold, and a smaller N-terminal domain (Bailey et al., 2007a; San Martin et al., 1998). DnaC bound to the larger domain, possibly as a maximal set of three dimers (Galletto et al., 2003), forms the DnaB-DnaC complex. The N-terminal region of DnaC is necessary for it to interact with DnaB (Ludlam et al., 2001).
DnaC is a member of the AAA+ family of ATPases that contain specific amino acid sequence motifs proposed to coordinate enzyme function with ATP binding and its hydrolysis (Koonin, 1993). For DnaC, ATP mediates several important functions. First, ATP increases the affinity of DnaC for ssDNA, which stimulates DnaC's ATPase activity synergistically with DnaB (Biswas et al., 2004; Davey et al., 2002). Second, ATP or ATPγS, an ATP analogue that is poorly hydrolyzed, stabilizes DnaC complexed to DnaB to arrest DnaB as a DNA helicase (Allen and Kornberg, 1991; Wahle et al., 1989b). In support that ATP binding is essential for DnaC function, mutant DnaCs that are defective in ATP binding due to substitutions in the Walker A box, a nucleotide binding motif, do not inhibit DnaB in vivo, and fail to deliver DnaB to oriC in vitro (Davey et al., 2002; Ludlam et al., 2001). The conundrum is that nucleotide binding apparently is not required for DnaC to interact with DnaB (Biswas and Biswas-Fiss, 2006; Davey et al., 2002; Galletto et al., 2003). Also, the comparable affinity of DnaC for DnaB in the presence of ATP or ADP suggests that ATP hydrolysis by DnaC is not required for its dissociation from DnaB (Davey et al., 2002; Galletto et al., 2003). These contradictory observations, which overlook the possible effect of oriC on ATP metabolism by DnaC, raise several questions. Why does DnaC require ATP to deliver DnaB to oriC, and what induces the dissociation of DnaC from DnaB?
Of the AAA+ motifs of DnaC, the box VII motif contains two conserved arginines. Based on the X-ray crystallographic structure of Aquifex aeolicus DnaC (residues 43-235) bound to ADP, one of the arginines of E. coli DnaC is proposed to interact with the γ phosphate of ATP bound in the pocket formed either between adjacent monomers of a DnaC oligomer (Mott et al., 2008), or between neighboring DnaC protomers in the DnaB-DnaC complex. This residue called the “arginine finger” is thought to promote and to coordinate ATP hydrolysis with a conformational change (Erzberger and Berger, 2006). If ATP hydrolysis by DnaC causes it to dissociate from DnaB at oriC, a prediction is that substitution of this arginine with a nonfunctional residue should lead to DnaC remaining complexed to DnaB, blocking DnaB function to inhibit initiation. Because substitution of the arginine finger residue of HslU/ClpY (Song et al., 2000), or of a conserved arginine in the box VII motif of DnaA obstructs formation of a self-oligomer (Felczak and Kaguni, 2004), the second arginine at residue 216 of DnaC may similarly act in the oligomerization of DnaC monomers in the DnaB-DnaC complex.
We tested these predictions, and showed that substitution of these conserved arginines with alanine substantially impairs DnaC activity in DNA replication in vivo and in vitro. Compared with wild type DnaC, the inactivity of the mutant DnaC proteins in DNA replication is not explained by defects in binding to ATP as measured in a crosslinking assay, or to ssDNA or DnaB as measured respectively by surface plasmon resonance or a protein overlay assay. We show that, like the wild type enzyme, both mutant DnaCs are able to deliver DnaB to oriC and are retained there. We also demonstrate that primase induces the release of DnaC, but not of the mutant DnaCs from DnaB in the prepriming complex whereas DNA polymerase III holoenzyme is only partially effective. Moreover, ATPγS stabilizes the mutant DnaC bearing an alanine substitution for arginine 216 in the prepriming complex whereas ATP does not, suggesting that aberrant ATP hydrolysis by this mutant facilitates its dissociation from DnaB.
Results
The conserved arginines in the box VII motif are necessary for DnaC function in vivo
With derivatives of pACYC184 that encode alanine substitutions for arginine 216 and arginine 220, which reside in the box VII motif of DnaC (Table S1, supplemental data; Figure 1), we used a plasmid exchange assay to show that the dnaC alleles are defective in vivo. This assay relies on a strain that lacks almost the entire dnaC gene, which is complemented by a dnaC-containing plasmid that requires IPTG for its maintenance (Hupert-Kocurek et al., 2007). When this plasmid is lost on media lacking IPTG, we can measure dnaC function encoded by another plasmid. With this method, we observed that a pACYC184 derivative carrying the dnaC+ gene but not the empty vector (pACYC184) maintained the viability of isogenic ΔdnaC strains that encoded either the recA+ or recA::kanR allele (Table S2, supplemental data). Hence, viability requires the dnaC gene, but does not depend on integration of the dnaC plasmid into the bacterial chromosome. In comparison, plasmids encoding the mutant DnaCs (hereafter named R216A and R220A) failed to complement these strains when the resident plasmid was lost. We then performed immunoblot analysis of whole cell lysates prepared from E. coli MC1061 bearing the plasmid-encoded dnaC alleles to confirm that their steady-state levels were sufficient for complementation (data not shown). Whereas the chromosomally-encoded level of wild type DnaC was undetectable in several strains (MC1061, MG1655 and their derivatives; 5 × 108 cells), the abundance of plasmid-encoded R216A and R220A at roughly 3 × 103 molecules per cell was comparable to the level that maintains viability of the ΔdnaC strains by the IPTG-dependent dnaC+ plasmid (pAM34dnaC). As the mutant DnaCs were not proteolytically unstable, these results show that the mutant DnaCs are inactive in this genetic assay. The inactivity of R220A is consistent with a previous study (Mott et al., 2008).
Figure 1. Primary and secondary structure of DnaC, and its amino acid sequence motifs.
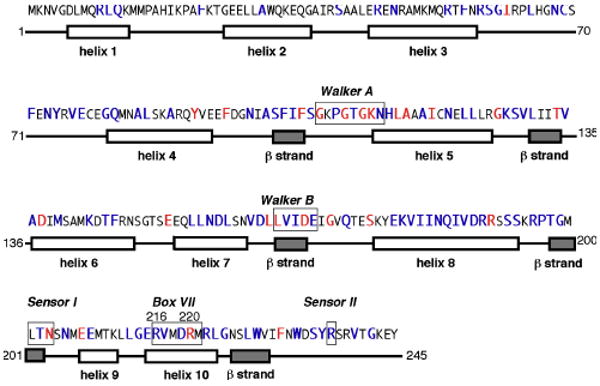
Comparison of the amino acid sequence of E. coli DnaC with its homologs reveals amino acid residues that are conserved (red) and moderately conserved (purple) (Ludlam et al., 2001). Its secondary structure is based on secondary structure prediction (residues 1-70) and a homology model derived from the ATP binding region of A aeolicus DnaC (residues 43-235) (Ludlam et al., 2001; Mott et al., 2008). The numbers at the right correspond to residues of E. coli DnaC. The boxed residues denote Walker A and B, Sensor I and II, and Box VII motifs conserved among AAA+ proteins.
However, the inactivity of R216A encoded by pACYC184 contrasts with the mutant protein's ability to complement the temperature sensitivity of a dnaC2 mutant when this allele was downstream from the araBAD promoter in a separate plasmid (Mott et al., 2008). Hence, we constructed plasmids that bear the box VII mutations under araBAD promoter control (pR216AdnaC and pR220AdnaC), and compared them with the isogenic dnaC+ plasmid (pINCSSD) in complementation of a dnaC2 strain under similar uninduced conditions. Table S3 (supplemental data) shows that the dnaC+ allele but not the allele encoding R220A under control of the araBAD promoter was active in complementation. In contrast, R216A complemented the dnaC2 strain at nonpermissive temperature if the allele was downstream from the araBAD promoter, but not if it was carried in pACYC184. These results, which suggest that R216A is partially active, correlate with biochemical studies described below showing that R216A is modestly active in DNA replication.
By UV crosslinking, R216A and R220A are active in ATP binding
To gain more insight into the specific functions of these arginine residues, we examined the properties of the purified R216A and R220A DnaC mutants (Figure S1, supplemental data) in several biochemical assays. As earlier studies showed that DnaC binds to ATP, albeit weakly (Biswas et al., 2004; Davey et al., 2002; Galletto et al., 2003; Wahle et al., 1989a), we measured the ability of the mutant DnaCs to interact with ATP. The poor affinity of DnaC for ATP obstructed our efforts to measure ATP binding by the nitrocellulose filter retention method. Relying on UV crosslinking, which showed that DnaC is able to bind to ATP (Biswas and Biswas, 1987; Ludlam et al., 2001), we observed similar levels of crosslinked [α-32P] ATP with wild type DnaC and with the mutant proteins but not with bovine serum albumin (BSA) (Figure 2A). As a control, the inclusion of an excess of unlabeled ATP abolished the crosslinking of [α-32P] ATP to wild type DnaC. These observations suggest that R216A and R220A are active in ATP binding. The results with R220A support a recent study, which showed that an alanine substitution of the corresponding amino acid (lysine 210) of Aquifex aeolicus DnaC did not affect ATP binding (Mott et al., 2008).
Figure 2. R216A and R220A are active in ATP binding, which stimulates their ability to interact with DnaB.
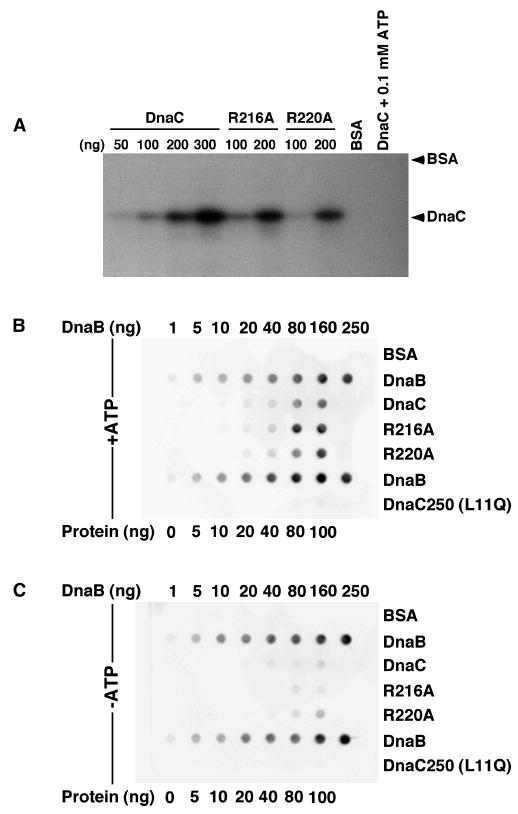
(A) UV crosslinking of [α-32P] ATP (10 μCi) was performed with duplicate samples essentially as described (Ludlam et al., 2001), but only one of each pair is shown because the results are essentially identical. As negative controls, BSA (200 ng) was used in place of wild type DnaC, or wild type DnaC (200 ng) and [α-32P] ATP were incubated with 0.1 mM ATP. The electrophoretic mobilities of BSA and DnaC, which were visualized by staining with Coomassie Brilliant Blue before autoradiography, are indicated at the right. (B, C) Protein overlay analysis was performed to measure the interaction of DnaB with DnaC as described (Ludlam et al., 2001). Values at the top of each panel indicate the amounts of immobilized DnaB in the second and sixth rows that serve as a positive control for the detection method, and a reference to compare the relative amount of binding of DnaB to immobilized DnaC. The bottom of each panel indicates the amounts of BSA, or wild type or mutant DnaC present in the respective column. DnaC250 carries an L11Q substitution, which distrupts the ability of DnaC to interact with DnaB (Ludlam et al., 2001). After protein immobilization using a Bio-Rad dot blot apparatus, the filters (BA-85, Whatman) were incubated with DnaB (1 μg/ml) in buffer (40 mM Hepes-KOH pH 8.0, 20 mM Tris-HCl, pH 7.6, 10 mM magnesium chloride, 4% sucrose, 4 mM DTT, and 0.1 mg/ml BSA) lacking or supplemented with 1 mM ATP. The washed filters were then incubated with affinity-purified antibody that specifically recognizes DnaB. Immune complexes were detected as described in Experimental Procedures.
R216A and R220A can interact with DnaB
Previously, we developed a genetic assay to identify mutant DnaC proteins that are defective in interacting with DnaB (Ludlam et al., 2001). We also showed biochemically that specific amino acid substitutions, including an L11Q substitution near the N-terminus of DnaC, abrogate this interaction, thus identifying an N-terminal domain that is necessary to interact with DnaB. However, other amino acid substitutions mapped to the Walker A box of DnaC, suggesting that ATP stabilizes the DnaB-DnaC complex, which is consistent with the conclusion of an earlier study (Wahle et al., 1989a). Hence, we examined the R216A and R220A for their ability to interact with DnaB because of the proposed role of residues in the box VII motif of DnaC in ATP binding and ATP hydrolysis. Using a protein overlay method, we showed that wild type DnaC, R216A and R220A were similar in their ability to bind to DnaB, and that ATP stabilized this interaction (Figure 2B, C). As controls, we failed to detect an interaction between the L11Q-substituted protein or BSA and DnaB. These results indicate that the alanine substitutions do not impair the interaction of DnaC with DnaB. In support, biosensor results confirmed that DnaC+, R216A and R220A interacted with DnaB, and suggest that the affinity of DnaC+ compared with the mutants for DnaB is about two-fold stronger (data not shown). Although the effect of ATP on the mutant DnaCs was not tested, ATP (150 μM) increased the affinity of wild type DnaC for DnaB by about two-fold. In summary, both methods show that the inactivity of the mutant proteins in vivo (Table S2, supplemental data) does not stem from a defect in interacting with DnaB.
R216A and R220A can interact with ssDNA
The interaction of DnaC with single-stranded DNA (ssDNA) has been demonstrated by UV crosslinking, fluorescence polarization, and surface plasmon resonance (Biswas et al., 2004; Davey et al., 2002; Learn et al., 1997; Mott et al., 2008). ATP enhances this interaction (Biswas et al., 2004; Davey et al., 2002), but the mechanism by which DnaC binds ssDNA and how ATP stimulates this process are uncertain. Mott et al, suggested that the assembly of DnaC monomers into a helical structure may play a role in ssDNA binding (Mott et al., 2008). If so, mutation of residues involved in stabilizing the interaction between the monomers, such as arginine 216 or perhaps arginine 220, may disrupt ssDNA binding. To test this possibility, we compared the binding of the mutant proteins with wild type DnaC to an immobilized oligonucleotide by surface plasmon resonance in buffer lacking or supplemented with ATP. We calculated the dissociation constants based on the rates of association and dissociation derived from the binding isotherms. As shown in Figure 3, ATP stimulated ssDNA binding by wild type DnaC by about 2.4-fold, which is comparable to results of an independent study (Biswas et al., 2004). ATP also enhanced ssDNA binding by R220A, but the magnitude of this effect (1.7-fold) was not as great. In contrast, ATP only marginally stimulated ssDNA binding by R216A. Despite these differences, the ability of the mutant DnaCs to bind to ssDNA does not explain their inactivity in vivo.
Figure 3. ATP stimulates the binding of wild type DnaC and R220A but not R216A to ssDNA.
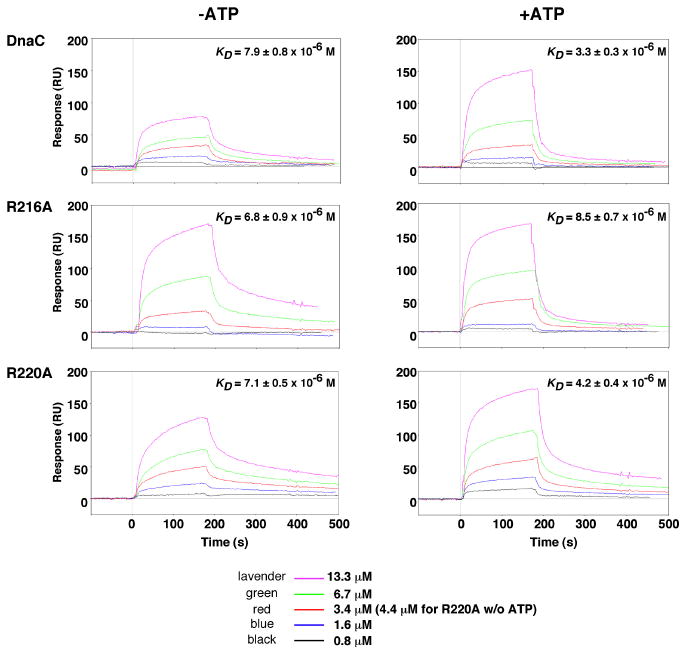
The interaction of wild type or mutant DnaC protein at the concentrations indicated with a 5′-biotinylated oligonucleotide (CATCCAATAAATCATACACAAGGC; 200 resonance units immobilized onto a streptavidin-coated sensor chip) was measured by surface plasmon resonance (Biacore 2000 biosensor, GE Healthcare). Binding was measured in duplicate at 22 °C at a flow rate of 5 μl min-1 in buffer containing 40 mM Hepes-KOH pH 8.0, 40 mM potassium glutamate, 4% sucrose, 0.5 mM magnesium acetate, and 4 mM DTT, and supplemented with 150 μM ATP where indicated. Because both sensorgrams at each concentration were so similar, only one binding isotherm is shown. After each measurement, the residual protein was removed from the chip with 2 M NaCl for 2 min. The background signal from a reference flow cell corresponding to a streptavidin-coated surface was subtracted from each data set. As the data did not conform with a 1:1 Langmuir interaction model, the sensorgrams were fit globally to estimate the on-rate (ka), the off-rate (kd) and the dissociation constants (KD = ka/kd) using BIAevaluation 3.1 software.
The capacity of R220A to bind to ssDNA contrasts with defective ssDNA binding by a mutant DnaC of A. aeolicus (Mott et al., 2008). Measured by fluorescence polarization, substitution of the corresponding “arginine” finger residue (lysine 210) with alanine disrupted binding to poly-dT25. As this mutant DnaC also lacked the first 42 amino acids of the native protein, the defect in ssDNA binding suggests the requirement for the missing N-terminal region.
R216A and R220A are defective in DNA replication in vitro
Although R216A and R220A can interact with ATP, DnaB and ssDNA, we expected these mutant proteins to be defective in DNA replication in vitro because of their apparent inactivity in vivo. In a reconstituted system dependent on a supercoiled oriC-containing plasmid, one DnaB hexamer is loaded onto each of the separated strands within oriC (Carr and Kaguni, 2001; Fang et al., 1999). This loading process may be concerted because fork movement is bidirectional even when DnaB is at a subsaturating level (Baker et al., 1987). By comparison, DNA replication of a ssDNA that carries a DnaA box in a hairpin structure involves the binding of a single DnaB hexamer next to the DnaA box (Carr and Kaguni, 2002). Because it is conceivable that one or both substitutions specifically impair the concerted loading of DnaB at oriC, we examined the mutant proteins in DNA replication of both the oriC plasmid and the ssDNA. With the oriC-containing plasmid, R220A was essentially inert whereas R216A reproducibly supported a marginal level of DNA synthesis (Figure 4A). With the ssDNA, R216A supported a limited level of DNA synthesis that was slightly greater than the level observed with R220A (Figure 4B). These results indicate that the alanine substitutions for these arginine residues substantially reduce the activity of DnaC in both replication systems.
Figure 4. R216A and R220A are defective in DNA replication in vitro.
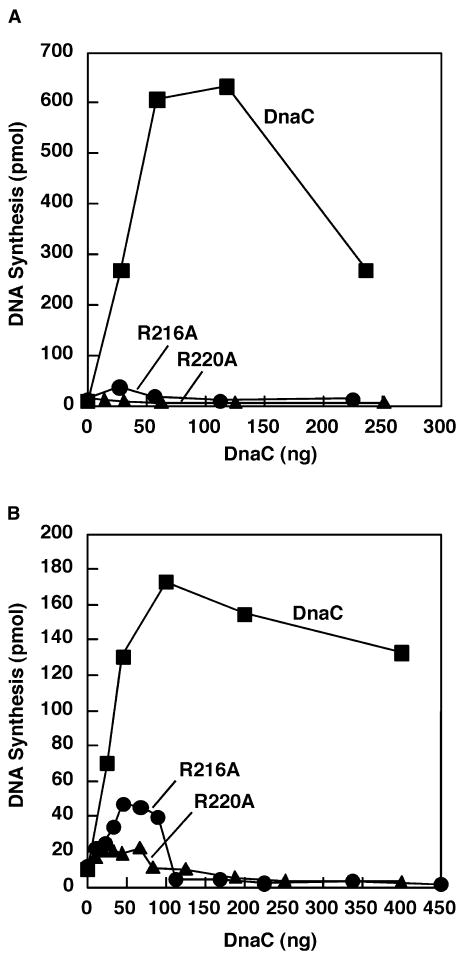
DNA replication with the indicated amounts of DnaC, R216A, or R220A was measured in a reconstituted system of purified proteins with either M13oriC2LB5 supercoiled DNA (A) or M13 A-site ssDNA (B) as described in Experimental Procedures.
R216A and R220A are able to deliver DnaB to oriC
The O'Donnell laboratory reported that a mutant DnaC bearing an arginine substitution for the conserved lysine in the Walker A box was inert in ATP binding (Davey et al., 2002). Despite its ability to form a complex with DnaB, the mutant DnaC's failure to guide the helicase to oriC suggests the requirement for ATP binding. Because of the proposed role of arginine 220 and perhaps arginine 216 in ATP binding and its hydrolysis, we considered the possibility that our mutant DnaCs may fail to deliver DnaB to oriC. As an assay, we measured the assembly of the prepriming complex, which forms by the binding of DnaA to oriC in a supercoiled plasmid followed by the HU-stimulated unwinding of a region within oriC (Chodavarapu et al., 2008). At the step of unwinding, DnaA complexed to ATP or ATPγS but not ADP is active (Sekimizu et al., 1987). DnaA then mediates the binding of DnaB from the DnaB-DnaC complex to the resultant unwound region to form the prepriming complex (Marszalek and Kaguni, 1994).
We assembled the prepriming complex with wild type DnaC or the mutants and other necessary components, and then separated the unbound proteins from the prepriming complex by gel filtration chromatography (see Experimental Procedures). In several essentially identical experiments, such as that of Figure 5A, we showed that the isolated complex assembled with wild type DnaC was active upon addition of primase, DNA polymerase III holoenzyme, DNA gyrase, and other reagents that are required during the subsequent stage of DNA synthesis, recovering 70-90% of the activity in the sample before isolation. Hence, this complex is a stable intermediate of the initiation process. In contrast, the isolated complex assembled with R220A was essentially inactive whereas that formed with R216A had only about 15% of the activity of the isolated complex assembled with wild type DnaC.
Figure 5. R216A and R220A are able to deliver DnaB to oriC, but the prepriming complex is inactive.
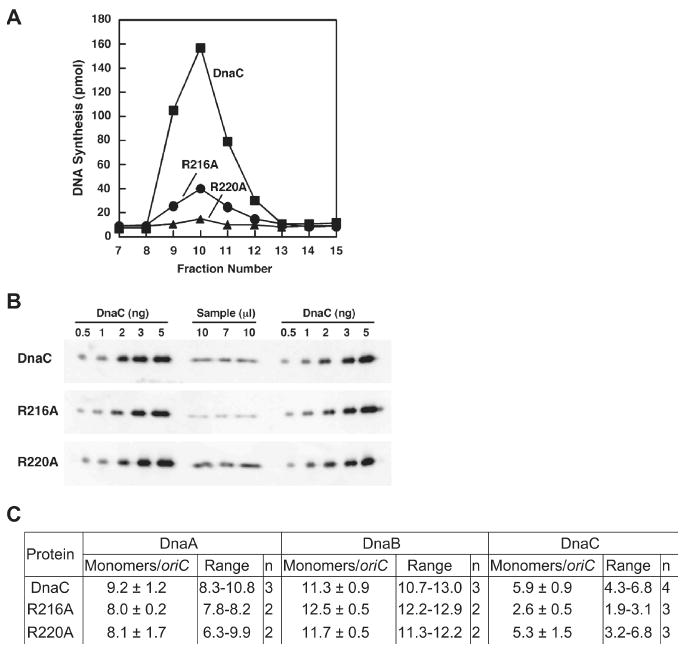
The prepriming complex was assembled with DnaC, R216A or R220A and isolated as described in Experimental Procedures. (A) Portions (20 μl) of the eluted fractions were assayed for DNA replication activity after incubation at 30°C for 30 min with CTP, GTP, UTP, dNTPs, SSB, primase, DNA polymerase III holoenzyme and DNA gyrase at levels used in standard replication reactions. (B) In parallel, the amount of DnaC+, R216A, or R220A in the respective isolated prepriming complex at the volumes indicated was measured by quantitative immunoblotting relative to a standard curve obtained with known amounts of DnaC, which were analyzed in duplicate. (C) The amounts of DnaA, DnaB, and DnaC (wild type or mutants) were quantified and are reported relative to the amount of the oriC-containing plasmid (see Experimental Procedures). The column labeled “n” indicates the number of experiments.
We then quantified the amount of the oriC plasmid in the isolated complex by agarose gel electrophoresis (data not shown). In parallel, we determined the amounts of DnaA, DnaB and DnaC by quantitative immunoblot analysis, and calculated the stoichiometry of these proteins relative to the oriC plasmid (Figure 5B, C). In the prepriming complex assembled with wild type DnaC, we observed nine DnaA monomers and eleven DnaB monomers per oriC plasmid, which substantiate previous results of ten DnaA monomers (Carr and Kaguni, 2001; Chodavarapu et al., 2008; Felczak and Kaguni, 2004; Felczak et al., 2005), and two DnaB hexamers bound to oriC (Carr and Kaguni, 2001; Fang et al., 1999). Previous work showed that the loading of DnaB requires DnaC (Funnell et al., 1987; Davey et al., 2002). In agreement, we determined that the omission of DnaC under conditions that otherwise assemble the prepriming complex (as in Figure 5) led to the absence of DnaB on the oriC-containing plasmid (data not shown). We also showed that DnaB was not retained on the DNA when we replaced the oriC-containing plasmid (M13oriC2LB5) with the plasmid vector (M13ΔE101). Hence, DnaB requires DnaC and the oriC sequence for the helicase to load at oriC in a supercoiled plasmid.
For the prepriming complex assembled with R216A or R220A, the stoichiometry of DnaA and DnaB corresponds within experimental error with the results obtained with wild type DnaC (Figure 5). Thus, despite the marginal activity of R216A and inactivity of R220A in oriC plasmid replication, both mutant DnaCs are able to deliver two DnaB hexamers to oriC. We conclude that their biochemical defect is at a step after the delivery of DnaB.
Interestingly, we calculated about six DnaC monomers in the prepriming complex assembled with wild type DnaC and 5 mM ATP (Figure 5B, C; see below). The inclusion of all four ribonucleotides or subsets at 0.5 mM each in combination with 5 mM ATP did not lead to the dissociation of DnaC (data not shown). When we replaced the oriC plasmid with the empty vector (M13ΔE101) as a control, the ratio of 0.7 DnaC monomers per DNA indicates that nonspecific binding of DnaC to DNA is negligible. The simplest explanation for these results is that each DnaB hexamer at oriC is bound by three DnaC monomers, but other ratios cannot be excluded. Relative to the plasmid, we also calculated a ratio of 5.3 R220A monomers, which is similar to that for DnaC, or 2.6 R216A monomers. That R216A is able to interact with DnaB (Figure 2), and assembles two DnaB hexamers at oriC suggests that some other defect leads to its lower ratio per oriC plasmid. We excluded the possibility that impaired ATP binding explains its lower stoichiometry because ATP binding by DnaC is not required for its interaction with DnaB (Biswas and Biswas-Fiss, 2006; Davey et al., 2002; Galletto et al., 2003). Moreover, R216A (and R220A) are active in ATP binding (Figure 2). Because DnaC is reported to hydrolyze ATP weakly in the presence of ssDNA (Davey et al., 2002), we considered the possibility that R216A aberrantly hydrolyzes ATP, which causes it to partially dissociate from DnaB at oriC. However, we were unable to measure ATP hydrolysis by wild type DnaC under the conditions described. Our observations are consistent with other studies that failed to detect an ATPase activity for DnaC (Biswas and Biswas, 1987), but these experimental conditions may not contain the proper effector necessary to elicit ATPase activity.
Inasmuch as DnaC can interact with DnaB in the absence of ATP, ATP binding by DnaC is required to deliver the helicase to oriC (Davey et al., 2002). Also, ATP hydrolysis by DnaC is speculated to promote its release (Biswas et al., 2004; Wahle et al., 1989a). Because ATPγS stabilizes DnaC complexed to DnaB (Wahle et al., 1989b), we reasoned that this poorly hydrolysable nucleotide analog should block the dissociation of DnaC from DnaB at oriC. Alternatively if a factor is necessary to facilitate the release of DnaC and is absent in the reaction to assemble the prepriming complex, the ratio of DnaB to DnaC will be similar to that obtained with ATP. Figure 6A shows that the ratios of wild type DnaC, R216A, and R220A in the prepriming complex assembled and isolated with ATPγS are comparable; DnaC was retained at about 6 monomers per oriC plasmid. DnaB at about 9-11 monomers is consistent with two DnaB hexamers bound to oriC, validating the results of Figure 5 and previous results (Carr and Kaguni, 2001; Fang et al., 1999). Compared with Figure 5C, we measured slightly lower ratios for DnaA, which presumably reflect experimental variation. Nevertheless, these results strongly suggest that each DnaB hexamer is complexed with three DnaC monomers, and that aberrant ATP hydrolysis by R216A leads to its lower ratio in the prepriming complex assembled with ATP. However, the mutant DnaC is still able to deliver DnaB at oriC.
Figure 6. Primase induces the dissociation of DnaC but not R216A or R220A from DnaB.
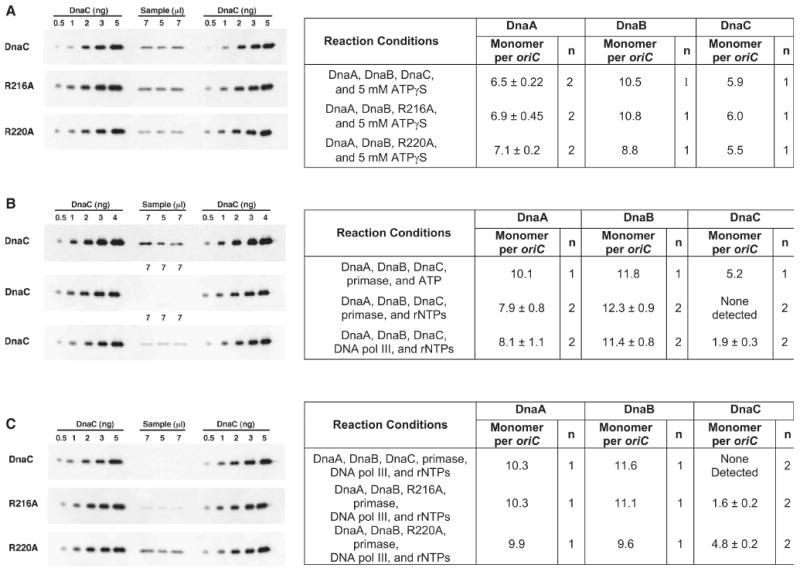
In each panel at the left, the indicated volumes of the isolated prepriming complex were analyzed concurrently with known amounts of DnaC that were used for a standard curve. (A) The prepriming complex was assembled with DnaC, R216A or R220A as described in Experimental Procedures, but in buffer containing 5 mM ATPγS instead of ATP. After isolation in this buffer, the stoichiometry of DnaA, DnaB, and either DnaC, R216A or R220A per oriC plasmid was calculated. (B) Reactions to assemble the prepriming complex also contained ATP (5 mM) or all four ribonucleotides (0.5 mM each except ATP at 5 mM), and primase (1.1 μg) or DNA polymerase III holoenzyme (DNA pol III) reconstituted with the β subunit (0.5 μg) and DNA polymerase III* (3 μg) as indicated at ten-fold greater levels than those used in standard replication reactions. The immunoblots at the left show the amount of DnaC in the isolated complex assembled with primase (top and middle) or DNA polymerase III holoenzyme (bottom). (C) Reactions for prepriming complex formation contained DnaC, R216A or R220A, all four ribonucleotides, and both primase and DNA polymerase III holoenzyme at the amounts described above. The immunoblots at the left show DnaC, R216A, and R220A in the isolated complex.
Primer formation by primase induces the dissociation of DnaC from DnaB in the prepriming complex
DnaC attenuates the ATPase and helicase activity of DnaB (Allen and Kornberg, 1991; Wahle et al., 1989b). That DnaC persists in the prepriming complex suggests a mechanism that induces its dissociation so that DnaB can unwind the parental DNA to advance the replication fork. If so, the mutant DnaCs may remain complexed to DnaB because they fail to respond to the dissociating signal.
After the delivery of DnaB at oriC, primase interacts with DnaB as the helicase translocates on the parental DNA, and forms primers for leading and lagging strand DNA synthesis (Tougu and Marians, 1996a; Wu et al., 1992). The primers are only 10-12 nucleotides long (Kitani et al., 1985; Yoda and Okazaki, 1991), and the template sequence used to initiate and direct primer synthesis is not very specific in vitro (Bhattacharyya and Griep, 2000). These observations suggest that the binding of primase to DnaB in the prepriming complex may induce DnaC to dissociate. To test this possibility, we assembled the prepriming complex but also included primase, and found that DnaC (5.2 DnaC monomers per oriC plasmid) was still retained (Figure 6B). As the reaction contained ATP, the capacity of DnaB or DnaC to hydrolyze ATP is insufficient to liberate DnaC. Interestingly, we failed to detect DnaC in the isolated complex assembled as above but supplemented with both primase and either all four ribonucleotides (Figure 6B) or only ATP, CTP and UTP that should support the synthesis of primers of shorter average length (data not shown). As described above, the inclusion of all four ribonucleotides but without primase during assembly did not lead to the dissociation of DnaC. Because DNA polymerase III holoenzyme via its τ subunit interacts with DnaB (Kim et al., 1996), this DNA polymerase may also induce the release of DnaC from the prepriming complex. We found that this enzyme was only partially effective based on the ratio of 1.9 DnaC molecules per oriC plasmid (Figure 6B). These results suggest that the interaction of primase with the N-terminal region of DnaB and primer synthesis induce a conformational change that promotes the release of DnaC from the larger C-terminal region of DnaB where it was once bound.
Primer formation by primase fails to liberate R216A and R220A from the prepriming complex
We then examined the mutant DnaCs in reactions supplemented with primase, ribonucleotides to support primer formation, and the DNA polymerase. We reasoned that the retention of a mutant DnaC at the same level as when primase and the DNA polymerase were absent would indicate that the mutant resists both proteins. We found that wild type DnaC was not retained (Figure 6C), but R216A and R220A remained at about the same stoichiometry as when these components were omitted (Figure 5C). These results with wild type DnaC agree with those of Figure 6B and an earlier study in which less than one DnaC molecule was retained in prepriming complex assembled under similar conditions (Fang et al., 1999). The failure of primase to induce the release of R220A or R216A indicates that their complete dissociation from DnaB is necessary in the transition from the initiation to the elongation stage of DNA replication, and provides an explanation for their inactivity in DNA replication.
Discussion
Arginine 216 of DnaC stabilizes the interaction of DnaC with DnaB
DnaC has two conserved arginines in the box VII motif (Figure 1). Based on studies of mutant proteins of HslU/ClpY and DnaA (Felczak and Kaguni, 2004; Song et al., 2000), we speculated that the R216A substitution may disrupt the interaction between DnaC protomers to impede the assembly of the DnaB-DnaC complex. Alternatively, this substitution may weaken the association between mutant DnaC molecules in the DnaB-DnaC complex after helicase loading at oriC. In the first case, the mutant DnaC should fail to deliver DnaB into the prepriming complex. In the second, R216A in the prepriming complex may be unstable. Our results show that R216A delivers DnaB to oriC to exclude the first possibility. Supporting the second possibility, the partial dissociation of R216A and not wild type DnaC from the prepriming complex assembled with ATP suggests that arginine 216 stabilizes DnaC molecules that are complexed with DnaB. Interestingly, R216A was retained at a ratio comparable with wild type DnaC in the presence of ATPγS but at a lower ratio with ATP, suggesting that this mutant improperly hydrolyzes ATP that leads to its partial dissociation. As the isolated complex assembled with DnaC and not with R216A is fully active in DNA replication, these results also suggest that after R216A delivers DnaB to oriC, the few molecules of R216A that remain bound to DnaB inhibits the helicase.
In response to primase, arginine 220 of DnaC coordinates the release of DnaC from DnaB
Based on the crystal structure of the ATP binding region of Aquifex aeolicus DnaC complexed to ADP with BeF3, a widely used substitute for the γ phosphate of ATP (Petsko, 2000), homology modeling predicts that arginine 220 of E. coli DnaC is the arginine finger residue. In AAA+ proteins, the arginine finger is proposed to promote ATP hydrolysis and to coordinate a conformational change during this process. Regardless of whether the prepriming complex was assembled with ATP, or ATPγS (Figure 5, 6), R220A was retained at a level comparable with DnaC+. With all four ribonucleotides, or with ATP, CTP and UTP to support primer formation, primase induced the release of DnaC+ but not R220A. In contrast, ATP as the sole ribonucleotide was insufficient. These observations suggest that arginine 220 acts to transduce the signal generated by the interaction of primase with DnaB and primer formation, which leads to the release of DnaC from DnaB, and implicates ATP hydrolysis by DnaC in the dissociation process. That DNA polymerase III holoenzyme, which also interacts with DnaB via its τ subunit (Kim et al., 1996), caused the partial release of DnaC suggests a separate pathway to favor that DnaB remains uninhibited by DnaC at the replication fork.
DnaC at oriC
Previous studies have attempted to detect DnaC in the prepriming complex with varying results. The Kornberg laboratory visualized proteins in the oriC prepriming complex by electron microscopy by antibody labeling and Protein A conjugated to colloidal gold, and failed to detect DnaC (Funnell et al., 1987). However, other proteins or DNA may have occluded DnaC from the antibody. Our finding corroborates previous studies, which found that DnaC is retained in the prepriming complex (Marszalek and Kaguni, 1994; Wold et al., 1998).
Different forms of the DnaB-DnaC complex
Our observations of about six DnaC monomers and two DnaB hexamers bound to oriC suggest that each DnaB hexamer is complexed to three DnaC monomers. This conclusion appears at odds with several studies that document the formation of the DnaB6-DnaC6 complex (Galletto et al., 2003; Kobori and Kornberg, 1982b). Despite the maximum capacity of six DnaC molecules in the DnaB-DnaC complex, biophysical studies show fewer DnaC molecules in complexes that are predicted to exist in vivo (Galletto et al., 2003). Our results suggest that the DnaB6-DnaC3 complex is the active form at oriC.
Interestingly, the N-terminal domain of DnaC, which interacts with DnaB (Ludlam et al., 2001), is similar in amino acid sequence with the corresponding region of P protein encoded by bacteriophage λ (Nakayama et al., 1987). This region of λ P protein presumably interacts with DnaB in forming the DnaB-λ P protein complex in a process whereby bacteriophage λ appropriates DnaB helicase to the λ replication origin. Thus, λ P protein is the viral analogue to DnaC, but it outcompetes with DnaC in binding to DnaB and also displaces DnaC from the DnaB-DnaC complex (Mallory et al., 1990). Like the DnaB6-DnaC3 complex at oriC, three λ P monomers interact with DnaB to form the DnaB6-[λ P protein]3 complex. These observations, which add support to the validity of the DnaB6-DnaC3 complex, suggest a common mechanism at the step of helicase loading at the respective replication origins.
A model of the switch from the stage of initiation to elongation of DNA replication
In the hexamer of E. coli DnaB, each protomer (470 residues) has two domains joined by a linker that may function as a flexible hinge (Miles et al., 1997; Nakayama et al., 1984). The N-terminal domain (residues 15 to 126) is required for helicase activity, and the structure of this domain has been solved by NMR and X-ray crystallography (Fass et al., 1999; Weigelt et al., 1999). The X-ray crystallographic structure of hexameric Bacillus stearothermophilus DnaB, or monomeric Thermus aquaticus DnaB has been solved also (Bailey et al., 2007a; Bailey et al., 2007b). During fork movement, the N-terminal domain faces away from the replication fork (Jezewska et al., 1998a; Jezewska et al., 1998b; Jezewska et al., 1998c) and transiently interacts with primase during primer formation (Bird et al., 2000; Chang and Marians, 2000; Oakley et al., 2005; Tougu and Marians, 1996a; Tougu and Marians, 1996b). The larger C-terminal domain (residues 172 to 470) is a hexamer containing a RecA-like fold, and bears the ATPase and DNA-binding sites (Bird et al., 2000; Nakayama et al., 1984). DnaC appears to bind to the larger domain (Barcena et al., 2001; Galletto et al., 2003). In the dynamic interactions that establish the replication fork machinery at oriC, the results of our study support a model that the interaction of primase with the N-terminal domain of DnaB induces the dissociation of DnaC from the C-terminal domain of DnaB.
The results of our study support the following model (Figure 7). First, DnaA bound to DnaA boxes, I- and τ-sites within oriC unwinds a region near the left border, and then directs the loading of a DnaB6-DnaC3 complex on each of the separated strands of DNA. In the DnaB-DnaC complex, DnaB helicase is inactive and awaits the release of DnaC. Next, primase interacts with the C-terminal region of DnaB to induce the dissociation DnaC. We suggest that this step is coupled with the translocation of DnaB on the parental DNA strand and primer synthesis. Primase then synthesizes the first primer (in red; arrows denote the 3′-end) that is used for leading strand synthesis by DNA polymerase III holoenzyme whereas subsequent primers (as represented by the primer in blue) are used for Okazaki fragment synthesis. Thus, the transition from the stage of initiation to the elongation phase of chromosomal DNA replication is marked by a discrete event: the interaction of primase with DnaB.
Figure 7. The interaction of primase with DnaB defines the switch from initiation to the elongation phase of chromosomal DNA replication.
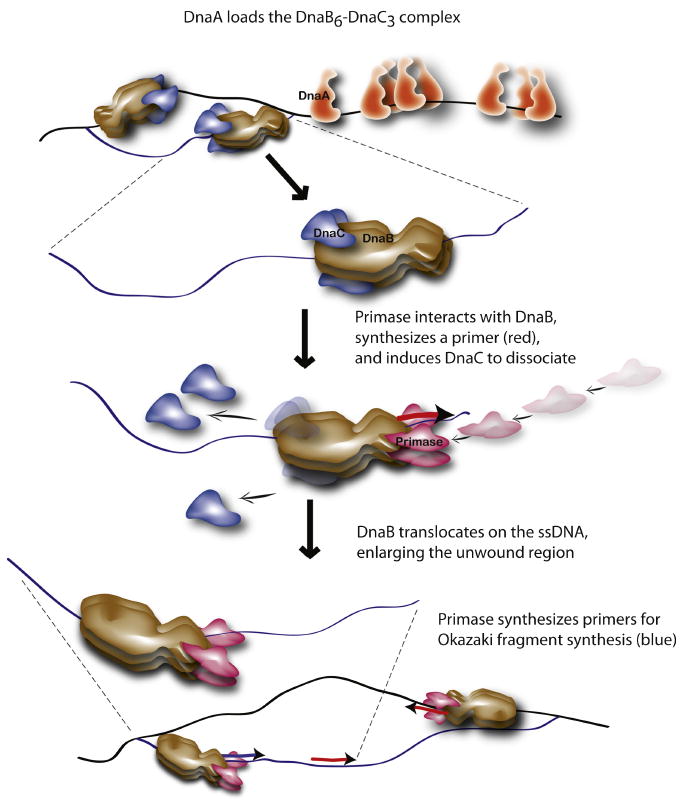
Please see the Discussion for details.
The molecular events in the initiation of DNA replication in E. coli are strikingly similar to those that occur at a eukaryotic replication origin. Indeed, DnaA and DnaC are comparable in structure to Cdc6 and Orc1, a component of the origin recognition complex (Erzberger et al., 2002). Like DnaC, Cdc6 in a process dependent on its ability to hydrolyze ATP with Ctd1 loads the eukaryotic Mcm2-7 complex, which likely functions as the replicative DNA helicase, at chromosomal origins (Randell et al., 2006). Like DnaB, the Mcm2-7 complex is recruited in an inactive form and must be activated after loading (Borlado and Mendez, 2008; Francis et al., 2009; Labib and Gambus, 2007; Sheu and Stillman, 2006). Thus, the work described herein has broader implications, and may provide insight into the process of initiation in eukaryotic cells.
Experimental Procedures
DNAs, strains, proteins, and reagents
Plasmids and E. coli K12 strains are described in Table S1 (supplemental data). The 5′-biotinylated oligonucleotide for biosensor experiments was synthesized by an in-house facility. Replication proteins including a mutant DnaC carrying an L11Q substitution have been described (Ludlam et al., 2001; Marszalek and Kaguni, 1994). Mutant DnaC proteins were purified as described in Figure S1 (supplemental data). Commercial reagents were BSA, horseradish peroxidase conjugated to goat anti-rabbit IgG (Jackson Laboratories), ribo- and deoxyribonucleotides (Pharmacia P-L Biochemicals), [α-32P] ATP (Amersham Biosciences), and [methyl-3H] dTTP (MP Biochemicals). Affinity-purified rabbit polyclonal antibodies for DnaB and DnaC, and rabbit antiserum that specifically recognizes the C-terminal region of DnaA (amino acids 370-467) were prepared in our laboratory.
DNA replication assays
Reactions (25 μl) with M13 A-site ssDNA or with M13oriC2LB5 supercoiled DNA were incubated at 30 °C for 15 or 30 min, respectively, to measure DNA synthesis, and were assembled with DnaA (55 ng) and the indicated amounts of wild type DnaC, R216A, or R220A as described (Chodavarapu et al., 2008; Marszalek and Kaguni, 1994). DNA synthesis was measured by liquid scintillation spectrometry of acid-insoluble radioactivity.
Isolation of the prepriming complex
Except where noted, reactions (85 μl) contained 10-fold greater amounts of proteins and DNA in a standard reaction for oriC plasmid replication, and included M13oriC2LB5 or M13ΔE101 (2 μg), SSB (200 ng), HU (α dimer; 50 ng), DnaA (920 ng), DnaB (1 μg), and wild type or mutant DnaC (500 ng) as indicated in buffer containing 40 mM Hepes-KOH pH 8.0, 40 mM potassium glutamate, 4% sucrose, 0.5 mM magnesium acetate, 5 mM ATP or ATPγS as indicated, 4 mM DTT, and 1 mg/ml BSA. The components for priming and DNA replication (primase, β clamp and DNA polymerase III* to reconstitute DNA polymerase III holoenzyme, and CTP, GTP, and UTP each at 0.5 mM) were omitted unless indicated. After incubation for 10 min at 37°C, the prepriming complex was isolated at 22°C by gel filtration chromatography (Sepharose 4B, 0.7 × 8.6 cm, Amersham Biosciences) in the buffer described above. Aliquots (20 μl) of the void volume fractions (120 μl) were placed on ice, magnesium acetate was adjusted to 10 mM, and components for priming and DNA replication were added at the standard amounts for oriC plasmid replication to a final volume of 25 μl. After incubation at 30°C for 30 min. DNA synthesis was measured as described above.
Quantitative analysis of the prepriming complex
The amount of M13oriC2LB5 DNA in the isolated prepriming complex was quantified by agarose gel electrophoresis relative to known amounts of this DNA as described (Carr and Kaguni, 2001). Concurrently, the amounts of DnaA, DnaB, and wild type or mutant DnaC protein were quantified by immunoblotting relative to known amounts of the respective proteins, which were analyzed in parallel to prepare standard curves. The filters (BA-85, Whatman) were probed with either antiserum that specifically recognizes DnaA protein (Chodavarapu, HU 2008), or with affinity-purified polyclonal antibody to detect DnaB or DnaC. The chemiluminescent signal (SuperSignal, Pierce) of immune complexes detected with horseradish peroxidase conjugated to the secondary antibody was quantified with a Kodak 4000R Image Station.
Supplementary Material
Acknowledgments
This research was supported by grant GM33992 from the National Institutes of Health and by the Michigan Agricultural Experiment Station. We thank Dr. Magdalena M. Felczak for her expert assistance in this work. The authors do not have a financial conflict of interest that might be construed to influence the results presented in the manuscript, or the interpretation of these results.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen GJ, Kornberg A. Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J Biol Chem. 1991;266:22096–22101. [PubMed] [Google Scholar]
- Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007a;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- Bailey S, Eliason WK, Steitz TA. The crystal structure of the Thermus aquaticus DnaB helicase monomer. Nucleic Acids Res. 2007b;35:4728–4736. doi: 10.1093/nar/gkm507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Funnell BE, Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987;262:6877–6885. [PubMed] [Google Scholar]
- Barcena M, Ruiz T, Donate LE, Brown SE, Dixon NE, Radermacher M, Carazo JM. The DnaB.DnaC complex: a structure based on dimers assembled around an occluded channel. EMBO J. 2001;20:1462–1468. doi: 10.1093/emboj/20.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Griep MA. DnaB helicase affects the initiation specificity of Escherichia coli primase on single-stranded DNA templates. Biochemistry. 2000;39:745–752. doi: 10.1021/bi991555d. [DOI] [PubMed] [Google Scholar]
- Bird LE, Pan H, Soultanas P, Wigley DB. Mapping protein-protein interactions within a stable complex of DNA primase and DnaB helicase from Bacillus stearothermophilus. Biochemistry. 2000;39:171–182. doi: 10.1021/bi9918801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SB, Biswas EE. Regulation of dnaB function in DNA replication in Escherichia coli by dnaC and lambda P gene products. J Biol Chem. 1987;262:7831–7838. [PubMed] [Google Scholar]
- Biswas SB, Biswas-Fiss EE. Quantitative analysis of binding of single-stranded DNA by Escherichia coli DnaB helicase and the DnaB × DnaC complex. Biochemistry. 2006;45:11505–11513. doi: 10.1021/bi060118d. [DOI] [PubMed] [Google Scholar]
- Biswas SB, Flowers S, Biswas-Fiss EE. Quantitative analysis of nucleotide modulation of DNA binding by the DnaC protein of Escherichia coli. Biochem J. 2004;379:553–562. doi: 10.1042/BJ20031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlado LR, Mendez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis. 2008;29:237–243. doi: 10.1093/carcin/bgm268. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Carl PL. Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol Gen Genet. 1970;109:107–122. doi: 10.1007/BF00269647. [DOI] [PubMed] [Google Scholar]
- Carr KM, Kaguni JM. Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J Biol Chem. 2001;276:44919–44925. doi: 10.1074/jbc.M107463200. [DOI] [PubMed] [Google Scholar]
- Carr KM, Kaguni JM. Escherichia coli DnaA protein loads a single DnaB helicase at a DnaA box hairpin. J Biol Chem. 2002;277:39815–39822. doi: 10.1074/jbc.M205031200. [DOI] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Marians KJ. Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork. J Biol Chem. 2000;275:26187–26195. doi: 10.1074/jbc.M001800200. [DOI] [PubMed] [Google Scholar]
- Chodavarapu S, Felczak MM, Yaniv JR, Kaguni JM. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol Microbiol. 2008;67:781–792. doi: 10.1111/j.1365-2958.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- Corn JE, Pelton JG, Berger JM. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat Struct Mol Biol. 2008;15:163–169. doi: 10.1038/nsmb.1373. [DOI] [PubMed] [Google Scholar]
- Davey MJ, Fang L, McInerney P, Georgescu RE, O'Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21:4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Davey MJ, O'Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- Fass D, Bogden CE, Berger JM. Crystal structure of the N-terminal domain of the DnaB hexameric helicase. Structure. 1999;7:691–698. doi: 10.1016/s0969-2126(99)80090-2. [DOI] [PubMed] [Google Scholar]
- Felczak MM, Kaguni JM. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J Biol Chem. 2004;279:51156–51162. doi: 10.1074/jbc.M409695200. [DOI] [PubMed] [Google Scholar]
- Felczak MM, Simmons LA, Kaguni JM. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J Biol Chem. 2005;280:24627–24633. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- Galletto R, Jezewska MJ, Bujalowski W. Interactions of the Escherichia coli DnaB Helicase Hexamer with the Replication Factor the DnaC Protein. Effect of Nucleotide Cofactors and the ssDNA on Protein-Protein Interactions and the Topology of the Complex. J Mol Biol. 2003;329:441–465. doi: 10.1016/s0022-2836(03)00435-2. [DOI] [PubMed] [Google Scholar]
- Hupert-Kocurek K, Sage JM, Makowska-Grzyska M, Kaguni JM. Genetic method to analyze essential genes of Escherichia coli. Appl Environ Microbiol. 2007;73:7075–7082. doi: 10.1128/AEM.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DS, Kaguni JM. Purification and characterization of the dnaA46 gene product. J Biol Chem. 1988;263:10625–10632. [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB Helicase? Fluorescence energy transfer studies. J Biol Chem. 1998a;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowski W. Complex of Escherichia coli primary replicative helicase DnaB protein with a replication fork: recognition and structure. Biochemistry. 1998b;37:3116–3136. doi: 10.1021/bi972564u. [DOI] [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowski W. Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1998c;273:9058–9069. doi: 10.1074/jbc.273.15.9058. [DOI] [PubMed] [Google Scholar]
- Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- Kim MH, Hines JC, Ray DS. Viable deletions of the M13 complementary strand origin. Proc Natl Acad Sci U S A. 1981;78:6784–6788. doi: 10.1073/pnas.78.11.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- Kitani T, Yoda K, Ogawa T, Okazaki T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J Mol Biol. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. II. Purification, physical properties, and role in replication. J Biol Chem. 1982a;257:13763–13769. [PubMed] [Google Scholar]
- Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the dnaB- dnaC protein complex. J Biol Chem. 1982b;257:13770–13775. [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Learn BA, Um SJ, Huang L, McMacken R. Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc Natl Acad Sci U S A. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Initiating chromosome replication in E. coli: it makes sense to recycle. Genes Dev. 2009;23:1145–1150. doi: 10.1101/gad.1809909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YH, Tsai KL, Sun YJ, Chen WT, Huang CY, Hsiao CD. The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res. 2009;37:804–814. doi: 10.1093/nar/gkn999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlam AV, McNatt MW, Carr KM, Kaguni JM. Essential amino acids of Escherichia coli DnaC protein in an N-terminal domain interact with DnaB helicase. J Biol Chem. 2001;276:27345–27353. doi: 10.1074/jbc.M101940200. [DOI] [PubMed] [Google Scholar]
- Mallory JB, Alfano C, McMacken R. Host virus interactions in the initiation of bacteriophage lambda DNA replication. Recruitment of Escherichia coli DnaB helicase by lambda P replication protein. J Biol Chem. 1990;265:13297–13307. [PubMed] [Google Scholar]
- Marszalek J, Kaguni JM. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- McHenry CS. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- Miles CS, Weigelt J, Stamford NP, Dammerova N, Otting G, Dixon NE. Precise limits of the N-terminal domain of DnaB helicase determined by NMR spectroscopy. Biochem Biophys Res Commun. 1997;231:126–130. doi: 10.1006/bbrc.1997.6059. [DOI] [PubMed] [Google Scholar]
- Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Arai N, Kaziro Y, Arai K. Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J Biol Chem. 1984;259:88–96. [PubMed] [Google Scholar]
- Nakayama N, Bond MW, Miyajima A, Kobori J, Arai K. Structure of Escherichia coli dnaC. Identification of a cysteine residue possibly involved in association with dnaB protein. J Biol Chem. 1987;262:10475–10480. [PubMed] [Google Scholar]
- Nielsen O, Lobner-Olesen A. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep. 2008;9:151–156. doi: 10.1038/sj.embor.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley AJ, Loscha KV, Schaeffer PM, Liepinsh E, Pintacuda G, Wilce MC, Otting G, Dixon NE. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J Biol Chem. 2005;280:11495–11504. doi: 10.1074/jbc.M412645200. [DOI] [PubMed] [Google Scholar]
- Petsko GA. Chemistry and biology. Proc Natl Acad Sci U S A. 2000;97:538–540. doi: 10.1073/pnas.97.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Robinson NP, Bell SD. Origins of DNA replication in the three domains of life. FEBS J. 2005;272:3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- San Martin C, Radermacher M, Wolpensinger B, Engel A, Miles CS, Dixon NE, Carazo JM. Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure. 1998;6:501–509. doi: 10.1016/s0969-2126(98)00051-3. [DOI] [PubMed] [Google Scholar]
- Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HK, Hartmann C, Ramachandran R, Bochtler M, Behrendt R, Moroder L, Huber R. Mutational studies on HslU and its docking mode with HslV. Proc Natl Acad Sci U S A. 2000;97:14103–14108. doi: 10.1073/pnas.250491797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MD, Carr KM, Vicente M, Kaguni JM. E. coli DnaA protein: the N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J Biol Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- Tougu K, Marians KJ. The extreme C terminus of primase is required for interaction with DnaB at the replication fork. J Biol Chem. 1996a;271:21391–21397. doi: 10.1074/jbc.271.35.21391. [DOI] [PubMed] [Google Scholar]
- Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. J Biol Chem. 1996b;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. I. Formation and properties. J Biol Chem. 1989a;264:2463–2468. [PubMed] [Google Scholar]
- Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J Biol Chem. 1989b;264:2469–2475. [PubMed] [Google Scholar]
- Weigelt J, Brown SE, Miles CS, Dixon NE, Otting G. NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer. Structure. 1999;7:681–690. doi: 10.1016/s0969-2126(99)80089-6. [DOI] [PubMed] [Google Scholar]
- Wold S, Boye E, Slater S, Kleckner N, Skarstad K. Effects of purified SeqA protein on oriC-dependent DNA replication in vitro. EMBO J. 1998;17:4158–4165. doi: 10.1093/emboj/17.14.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Zechner EL, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem. 1992;267:4030–4044. [PubMed] [Google Scholar]
- Yoda K, Okazaki T. Specificity of recognition sequence for Escherichia coli primase. Mol Gen Genet. 1991;227:1–8. doi: 10.1007/BF00260698. [DOI] [PubMed] [Google Scholar]
- Yu X, Jezewska MJ, Bujalowski W, Egelman EH. The hexameric E. coli DnaB helicase can exist in different quaternary states. J Mol Biol. 1996;259:7–14. doi: 10.1006/jmbi.1996.0297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


