Abstract
Despite the constant exposure to genomic insults that may lead to malignancy, cancer is surprisingly a relatively rare occurrence, and this is largely credited to an elaborate network of endogenous tumor suppression. Many effectors of tumor suppression have been identified, and their functions when activated in damaged cells have in large part been elucidated. What is less clear is whether there are common target gene(s) of tumor suppression, whose expression must be ablated in order to block transformation and preserve cellular homeostasis. Fresh experimental evidence suggests that silencing of the mitotic regulator and cell death inhibitor, survivin, is a universal requirement for successful tumor suppression in humans.
Keywords: survivin, PTEN, tumor suppressor, apoptosis, gene regulation
Survivin is the smallest member of the Inhibitor of Apoptosis (IAP) gene family.1 Originally described as cell survival factors that target caspases, we now know that IAPs have a much broader portfolio of functions, encompassing signaling pathways, cell division, metabolism and adaptation to unfavorable environments.1 Survivin embodies this multifunctional diversity, and compelling data accumulated over a decade have elucidated many of its essential roles as a regulator of mitosis, a broad cytoprotective factor, and an effector of cellular adaptation to stress.2 These disparate functions rely on hosts of regulated interactions that involve survivin and multiple protein partners, including tubulin and various chromosomal passenger proteins in the control of mitosis, other IAP family members to counteract apoptosis, and Heat Shock Proteins in the modulation of the cellular stress response.2
These ‘survivin networks’ are dramatically exploited in cancer, and survivin is unanimously viewed as one of the most prominent cancer genes.2 Overexpressed in virtually every human tumor, but undetectable or present at very low levels in most normal adult tissues, survivin expression has been consistently associated with disease progression, metastatic dissemination, resistance to therapy and death from disease.3 Although not a canonical drug target, efforts to disable the survivin networks for novel cancer therapeutics have now reached the clinic, and some encouraging responses have been noted in early clinical trials.2,3
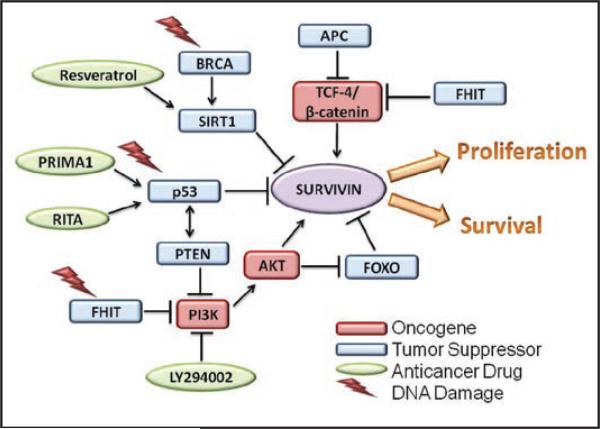
Because of the intrinsic interest in survivin biology, and the potential clinical benefits of survivin-directed therapeutics, extensive efforts have been devoted to understand how survivin becomes so sharply differentially expressed in tumors, compared to most normal tissues. From these studies, a prevailing consensus has emerged that a ‘tumor-specific’ expression of survivin2 is predominantly dictated at the level of transcription, and that survivin gene expression may be globally ‘deregulated’ in tumors, in vivo. Accordingly, survivin promoter activity is basically silent in normal cells, but strongly expressed in tumor cells,4 and this occurs independently of cellular heterogeneity, mitotic status, or genetic makeup. The differential expression of the survivin gene in normal versus tumor cells is so dramatic that therapeutic strategies to drive tumor-specific expression of suicidal genes under the control of the survivin promoter have now advanced to preclinical stages in a number of settings.5,6 Supporting this model, complementary studies identified a number of oncogenic gene expression pathways, for instance initiated by activated STAT3,7 NFκB8 and potentially, Myc,9 that converge on the survivin promoter to stimulate vigorous transcription selectively in tumor cells (Fig. 1). Developmental gene expression pathways exploited in tumorigenesis, for instance Notch10 or Wnt/β catenin11 also earmark the survivin gene for differential expression in transformed cells (Fig. 1). Available evidence suggests that these mechanisms are direct, and discrete binding sites for various oncogenic transcriptional activators, including NFκB, STAT3, Notch and TCF4/β-catenin have been identified in the proximal survivin promoter.
Figure 1.
Oncogene and tumor suppressor networks targeting the survivin pathway. See text for details.
Although there is little doubt that oncogene signaling activates survivin gene transcription, this is likely not the whole story. Evidence independently contributed by several laboratories seems to point to a reciprocal hypothesis, that it is not only the expression of oncogenes, but also the loss of tumor suppressors programmed to silence the survivin gene that is responsible for the differential expression of survivin in cancer, in vivo (Fig. 1).
In support of this model, pioneering studies of Hoffman and colleagues,12 and Mirza and colleagues13 identified survivin as one of the relatively few known genes that is actively repressed by wild type p53. These studies were universally confirmed by others, discrete p53-resposive elements in the proximal survivin promoter were identified, and monitoring survivin levels is now a routine functional assay to test for wild type p53 activity in cells. In independent experiments, the Altura laboratory found that the retinoblastoma protein (Rb), another pivotal tumor suppressor controlling the G1/S transition of the cell cycle, also acutely silenced survivin gene expression.14 This pathway was also direct in that E2F transcription factors (E2F1, E2F2 and E2F3) intercalated in the Rb pathway bound to discrete sites in the survivin promoter, and repressed its transcription. Other groups independently validated this model, and a role of Rb/E2F in shutting off survivin gene transcription is now viewed as one of the mechanisms that keep survivin levels low in interphase cells. In addition, the Adenomatous Polyposis Coli (APC) protein, a tumor suppressor almost invariably lost in colorectal cancer, and the Fragile Histidine Triad (FHIT) gene, a fragile site with tumor suppressive functions, have also been shown to repress survivin levels, in vivo.15,16 The mechanism(s) of these responses have not been completely elucidated, but it seems likely that both pathways silence the survivin gene directly, potentially via modulation of Akt16 or β-catenin/TCF-4,17 binding to the proximal survivin promoter. And finally, Wang and colleagues linked inhibition of survivin gene expression to one of the main tumor suppression networks in breast and ovarian cancer, i.e., BRCA1.18 This pathway relied on a complex transcriptional network in which BRCA1 bound to the promoter of SIRT1, a NAD-dependent class III histone deacetylase, and stimulated its de novo transcription. In turn, the newly produced SIRT1 bound to the proximal survivin promoter shutting off transcription via epigenetic chromatin modifications involving histone H3.
Altogether these results prompted a broad, but testable experimental question: is survivin a common target of multiple tumor suppression networks in humans? Recent studies from our group tried to address this question by looking at the PTEN pathway, which is arguably one of the most mutated tumor suppressor networks in human cancer.19 The Phosphatase and Tensin homolog deleted from chromosome Ten (PTEN) is a dual specificity phosphatase that controls cell proliferation, cell survival and cell size by removing the D3 phosphate from the lipid second messenger phosphatidylinositol triphosphate. Two of the critical targets of PTEN are the Akt and PDK1 kinases, and shutting off their signaling responses interrupts growth factor-initiated cell proliferation, maintains chromosomal integrity, and further amplifies p53-mediated tumor suppression.20
Against this backdrop, Guha and collaborators found that forced expression of PTEN suppressed survivin levels in tumor cell types, whereas, knocking down PTEN in PTEN wild type tumor cells resulted in increased survivin expression.21 As seen before with other tumor supressors, PTEN expression acutely silenced transcription of the survivin gene, and this response involved a 1,430 nt segment of the proximal survivin promoter.21 But how does PTEN do this? To answer this question, Guha et al. focused on Forkhead transcription factors of the FOXO subfamily, which function as potential effectors of PTEN signaling,22 and were previously implicated in modulating survivin gene expression in endothelial cells.23 In the study by Guha et al. siRNA gene silencing or dominant negative interference of FOXO1 or FOXO3a increased endogenous survivin levels in tumor cells.21 This was a direct transcriptional response as FOXO1 and FOXO3a bound to discrete sites in the proximal survivin promoter, shutting off gene expression in tumor cells. As expected from the role of survivin in tumor maintenance, PTEN-induced downregulation of survivin increased apoptosis in tumor cells, in a reaction reversed by re-expression of recombinant survivin.21 There was also preliminary evidence that this regulatory circuitry was operative in vivo, as expression of survivin and PTEN turned out to be inversely correlated in patient series of glioblastoma and colon cancer, and conditional deletion of PTEN in the mouse prostate resulted in increased survivin expression, before emergence of full blown epithelial dysplasia.21 Almost at the same time, an independent study also reported that FOXO3 signaling silenced survivin gene expression in neuroblastoma models,24 thus similar to the findings of Guha et al.21
Is there a common thread that links these studies? And, if so, what are their broader implications? First, it is now apparent that targeting survivin is a general property of the intrinsic tumor suppression machinery (Fig. 1). Although using different pathways, effectors and DNA recognition sequences, all of the best characterized tumor suppression networks in humans12-16,18,21,24 share the same target gene, survivin, and efficiently silence its transcription (Fig. 1). This leads to a general model that acute lowering of survivin levels may be an indispensable prerequisite to effectively antagonize cellular transformation in humans. In this context, targeting survivin may provide a unique advantage to oppose transformation, by simultaneously disrupting multiple downstream networks of cell proliferation, cytoprotection and adaptive resistance to unfavorable environments.2 By extension, genomic surveillance by multiple tumor suppressor pathways may contribute to maintain low levels of survivin in normal cell types, a straightforward model to explain the differential distribution of survivin in tumor versus normal tissues, in vivo.2 Experimental evidence in transgenic mice seems consistent with this possibility, and has implicated wild type p53 as a pivotal repressor of the survivin gene in most normal adult tissues, in vivo.25
As a corollary argument, these data broaden the traditional model of oncogene-directed survivin gene expression, and suggest that loss of one or more tumor suppressor(s) via inactivating mutations and/or loss of heterozygosity may be just as critical to de-repress transcription of the survivin gene, and contribute to its invariably elevated levels in tumor cells (Fig. 1). Because of the generality of this combined process (Fig. 1), one may tentatively conclude that deregulation of the survivin networks, uncoupled from tumor suppressors, may be required early to initiate the process of malignant transformation, and later to maintain the malignant phenotype of established tumors.2 Consistent with this model, survivin levels are highly elevated in premalignant lesions, for instance of the skin26 or colonic mucosa,27 suggesting that these changes occur early in tumorigenesis, and therapeutic interference with the survivin networks in established tumors cannot be compensated for, and results in apoptosis, proliferative arrest and sensitization to cell death or stress stimuli.2
In summary, recent evidence from various laboratories, including our own, have shown that survivin is a universal target of intrinsic tumor suppression networks. Several questions remain to be answered to position these pathways in their proper pathophysiological context. For instance, is there a preferential aspect of the multiple functions of survivin in cell division, cytoprotection or adaptation that must be absolutely suppressed to antagonize malignancy? Preliminary evidence from transgenic mice suggest that it is the cell cycle-independent, potential anti-apoptotic and adaptive functions of survivin that are predominantly exploited in tumorigenesis,25 but whether the same is true upon loss of tumor suppressor(s) remains to be elucidated. And what is the interplay, if any, between the disparate tumor suppressor networks that converge on the survivin gene? Given the fast pace of survivin research, the answer to some of these questions, and additional ones posed by the new results, will certainly be forthcoming. Regardless, the novel signaling circuitries between survivin gene regulation and tumor suppression further highlights the pivotal and perhaps indispensable role of the survivin networks in cellular transformation and tumorigenicity. This bodes well for the further development of survivin-based therapeutics, which would be expected to mimic the reintroduction of tumor suppressor mechanisms, potentially removing an essential requisite to maintain the malignant phenotype, in vivo.2
Acknowledgements
This work was supported by NIH grants CA78810, CA118005 and CA90917.
References
- 1.Srinivasula SM, Ashwell JD. IAPs: what's in a name? Mol Cell. 2008;30:123–35. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 3.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 4.Li F, Altieri DC. The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression. Cancer Res. 1999;59:3143–51. [PubMed] [Google Scholar]
- 5.Ulasov IV, Zhu ZB, Tyler MA, Han Y, Rivera AA, Khramtsov A, et al. Survivin-driven and fiber-modified oncolytic adenovirus exhibits potent antitumor activity in established intracranial glioma. Hum Gene Ther. 2007;18:589–602. doi: 10.1089/hum.2007.002. [DOI] [PubMed] [Google Scholar]
- 6.Chen JS, Liu JC, Shen L, Rau KM, Kuo HP, Li YM, et al. Cancer-specific activation of the survivin promoter and its potential use in gene therapy. Cancer Gene Ther. 2004;11:740–7. doi: 10.1038/sj.cgt.7700752. [DOI] [PubMed] [Google Scholar]
- 7.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007;104:7391–6. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawakami H, Tomita M, Matsuda T, Ohta T, Tanaka Y, Fujii M, et al. Transcriptional activation of survivin through the NFkappaB pathway by human T-cell leukemia virus type I tax. Int J Cancer. 2005 doi: 10.1002/ijc.20954. [DOI] [PubMed] [Google Scholar]
- 9.Cosgrave N, Hill AD, Young LS. Growth factor-dependent regulation of survivin by c-myc in human breast cancer. J Mol Endocrinol. 2006;37:377–90. doi: 10.1677/jme.1.02118. [DOI] [PubMed] [Google Scholar]
- 10.Lee CW, Raskett CM, Prudovsky I, Altieri DC. Molecular dependence of estrogen receptor-negative breast cancer on a notch-survivin signaling axis. Cancer Res. 2008;68:5273–81. doi: 10.1158/0008-5472.CAN-07-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim PJ, Plescia J, Clevers H, Fearon ER, Altieri DC. Survivin and molecular pathogenesis of colorectal cancer. Lancet. 2003;362:205–9. doi: 10.1016/S0140-6736(03)13910-4. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–57. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 13.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–22. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Y, Saavedra HI, Holloway MP, Leone G, Altura RA. Aberrant regulation of survivin by the RB/E2F family of proteins. J Biol Chem. 2004;279:40511–20. doi: 10.1074/jbc.M404496200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T, Otevrel T, Gao Z, Ehrlich SM, Fields JZ, Boman BM. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001;61:8664–7. [PubMed] [Google Scholar]
- 16.Semba S, Trapasso F, Fabbri M, McCorkell KA, Volinia S, Druck T, et al. Fhit modulation of the Akt-survivin pathway in lung cancer cells: Fhit-tyrosine 114 (Y114) is essential. Oncogene. 2006;25:2860–72. doi: 10.1038/sj.onc.1209323. [DOI] [PubMed] [Google Scholar]
- 17.Weiske J, Albring KF, Huber O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc Natl Acad Sci USA. 2007;104:20344–9. doi: 10.1073/pnas.0703664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1 and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–85. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 20.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guha M, Plescia J, Leav I, Li J, Languino LR, Altieri DC. Endogenous tumor suppression mediated by PTEN involves survivin gene silencing. Cancer Res. 2009;69:4954–8. doi: 10.1158/0008-5472.CAN-09-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–6. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 23.Daly C, Wong V, Burova E, Wei Y, Zabski S, Griffiths J, et al. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1). Genes Dev. 2004;18:1060–71. doi: 10.1101/gad.1189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obexer P, Hagenbuchner J, Unterkircher T, Sachsenmaier N, Seifarth C, Bock G, et al. Repression of BIRC5/survivin by FOXO3/FKHRL1 sensitizes human neuroblastoma cells to DNA damage-induced apoptosis. Mol Biol Cell. 2009;20:2041–8. doi: 10.1091/mbc.E08-07-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia F, Altieri DC. Mitosis-independent survivin gene expression in vivo and regulation by p53. Cancer Res. 2006;66:3392–5. doi: 10.1158/0008-5472.CAN-05-4537. [DOI] [PubMed] [Google Scholar]
- 26.Grossman D, McNiff JM, Li F, Altieri DC. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol. 1999;113:1076–81. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 27.Boman BM, Walters R, Fields JZ, Kovatich AJ, Zhang T, Isenberg GA, et al. Colonic crypt changes during adenoma development in familial adenomatous polyposis: immunohistochemical evidence for expansion of the crypt base cell population. Am J Pathol. 2004;165:1489–98. doi: 10.1016/S0002-9440(10)63407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]