Abstract
p53 is one of the most mutated tumor suppressors in human cancers and as such has been intensively studied for a long time. p53 is a major orchestrator of the cellular response to a broad array of stress types by regulating apoptosis, cell cycle arrest, senescence, DNA repair and genetic stability. For a long time it was thought that these functions of p53 solely rely on its function as a transcription factor, and numerous p53 target genes have been identified [1]. In the last 8 years however, a novel transcription-independent proapoptotic function mediated by the cytoplasmic pool of p53 has been revealed. p53 participates directly in the intrinsic apoptosis pathway by interacting with the multidomain members of the Bcl-2 family to induce mitochondrial outer membrane permeabilization. Our review will discuss these studies, focusing on recent advances in the field.
Keywords: p53, Mitochondria, Apoptosis, Transcription, Bcl-2 family, Pathophysiology, Radiosensitivity, Ischemia
1. Wild-type p53 rapidly translocates to mitochondria in response to apoptosis-inducing stress signals
The first hint of a transcription-independent apoptotic activity of p53 dates back to 1994–1995, when it was shown that p53-dependent apoptosis occurred in the presence of transcriptional or translational inhibitors, and that p53 truncation mutants lacking transcriptional activity can still trigger apoptotic function [2,3]. Later studies suggested that direct p53 signaling is participating in regulating caspase activity [4,5]. However, these studies did not provide any mechanistic explanation and until the beginning of the new century the novel, transcription-independent proapoptotic function of p53 remained enigmatic. Today, the subject is intensely studied. It is clearly established that in response to a stress signal, cytoplasmic p53 rapidly translocates to mitochondria, where it interacts with multi-domain members of the anti- and proapoptotic Bcl-2 family members to either inhibit or activate them. This direct action of p53 results in robust mitochondrial outer membrane permeabilization (MOMP), unleashing the enzymatic apoptotic machinery of caspases and of chromatin degradation.
MOMP is governed by pro- and antiapoptotic Bcl-2 family members. Characteristic of these proteins is the presence of Bcl-2 homology domains (BH). The antiapoptotic members such as Bcl-2, Bcl-xL and Mcl-1 contain BH1, BH2, BH3 and BH4 domains. The proapoptotic members fall into two subclasses: (i) the ultimate MOMP effectors Bax and Bak contain BH1, BH2 and BH3 domains, while the BH3-only class carries a single BH3 domain. Upon activation, Bax and Bak insert into the outer mitochondrial membrane and oligomerize to presumably form dynamic lipid pores that release lethal proteins from the mitochondrial intermembranous space. Functionally, BH3-only proteins (more than one dozen exist) fall into two subgroups — ‘activators’ and ‘derepressors’, according to how they trigger apoptosis. Activators (tBid and Bim) trigger MOMP by direct stimulation of Bax and Bak oligomerization, while derepressors such as Puma, Noxa and Bad inhibit the antiapoptotic Bcl-2 family members to release pro-apoptotic members from their inhibition (e.g. Bax or tBid from Bcl-xL and Bak from Mcl1) (reviewed in Ref. [6]).
The breakthrough study that first showed a direct role of p53 in mitochondrial apoptosis came from Marchenko et al, demonstrating that during p53-dependent apoptosis, a fraction of stress-stabilized wild-type p53 rapidly translocates to the mitochondrial outer membrane. p53 translocation precedes changes of mitochondrial membrane potential, cytochrome c release and caspase activation [7]. As definitive proof, a mitochondrially targeted p53 fusion protein that bypasses the nucleus and has no residual transactivation function was able to induce apoptosis and long-term growth suppression as efficiently as conventional p53 when expressed in several p53-null cancer cell lines [7,8]. Subsequent studies from our lab established that mitochondrial translocation of endogenous wtp53 occurs during the full spectrum of p53-activating cellular stress categories (various types of DNA damage, hypoxia, oncogene deregulation, oxidative damage) in different cell types (human and mouse; primary, immortal and malignant; epithelial, mesenchymal and lymphoid/myeloid) [9–11]. This translocation occurs during p53-dependent apoptosis but not during p53-induced cell cycle arrest or p53-independent apoptosis [7,9–11].
2. Wild-type p53 physically and functionally interacts with multidomain anti- and proapoptotic Bcl-2 members at mitochondria
Mechanistic insights into the mitochondrial function of wtp53 came when it was realized that mitochondrially translocated p53 interacts directly with members of the Bcl-2 family, which are central in governing the induction of mitochondrial outer membrane permeabilization. In response to stress, wtp53 interacts with and neutralizes the anti-apoptotic members Bcl-xL and Bcl-2. This interaction stimulates MOMP and subsequent apoptosis [8] and is associated with disruption of inhibitory complexes between Bcl-xL or Bcl-2 with MOMP inducing members (i.e. BH3-only and Bax/Bak) that pre-exist in unstressed cells [8,12]. Indeed, purified p53 is able to displace tBid and Bax from inhibitory complexes formed with Bcl-xL in vitro [13]. Several structural studies by NMR spectroscopy and other biophysical methods subsequently confirmed the Bcl-xL and Bcl-2 interactions with p53 that had been predicted from protein modeling and mutagenesis structure/function analyses [8,12,14,15]. Only the p53 core domain but not its amino- and carboxyterminal regions interacts [15]. The interaction is electrostatic in nature. It is mediated by the positively charged basic DNA-binding surface of p53 centered around residues 239–248 of loop 3 and extending on both sides, which interacts with the negatively charged acidic ‘underside’ of Bcl-xL/2, which comprises the BH4 domain and the loops between alpha 4/5 and 5/6 of Bcl-xL. (Note: this side is distinct from the hydrophobic BH123 binding pocket located at a different surface [12,14,15]. Significantly, despite high levels of stabilized mutant p53 protein constitutively present at mitochondria, missense mutant p53 proteins are defective in forming complexes with Bcl-xL/2 [8,12].
The core domain of mitochondrial p53 also directly interacts with pro-apoptotic Bak, which constitutively resides in the outer mitochondrial membrane [8,16]. This interaction liberates Bak from pre-existing inhibitory complexes with the anti-apoptotic Mcl-1 protein [16]. Similarly to Bcl-xL/2 interactions, conserved residues of the p53 core domain participate in Bak interaction [17]. Interestingly, p53 has a 10-fold higher affinity for Bcl-xL/2 than for Bak (with Kd ranging between 160 nM to 16 µM, depending on the methods (solid support or solution) to determine it [12,15,18]. This is likely due to the fact that (Bax and) Bak do not have a very acidic protein surface, nor do they have a BH4 domain, the interacting domain of Bcl-xL/2. This difference in affinity is consistent with sequential binding of p53 first to Bcl-xL and then to Bak [15]. Although the interaction with both proteins is electrostatic in character and involves the p53 core domain, the exact binding surface between p53 and Bak is currently unclear. Fersht and colleagues found no overlap with Bcl-xL binding and suggested a novel Bak-interacting p53 surface consisting of residues located on the surface between p53 monomers that is also formed when two core domains bind DNA [15]. In contrast, Murphy and colleagues find that the conserved H2 helix and the L1 and L3 loops of p53 make contact with Bak [17]. On the other hand, a stable p53–Bax complex does not appear to exist, likely due to structural reasons [15].
In cell free systems with recombinant p53 proteins and isolated healthy unstressed mitochondria, wild-type p53 – but not tumor-derived missense mutants – induces oligomerization of Bak and Bax, the biochemical hallmarks of Bax/Bak lipid pore formation and MOMP activity [8,12,16,19].
In addition (and contributing) to its mitochondrial action, a parallel transcription-independent p53 death pathway was proposed that takes place in the cytosol. Interestingly, it couples nuclear and extranuclear actions of p53. The model holds that in unstressed cells cytosolic p53 is sequestered into an inactive complex by soluble cytosolic Bcl-xL. In response to stress, nuclear p53 first transactivates its target gene Puma. In a second step, induced Puma then liberates p53 from its cytosolic Bcl-xL inhibition by forming a Puma/Bcl-xL complex instead. p53 is then free to activate monomeric Bax in the cytosol [18]. Thus, in this pathway the action of cytosolic p53 requires Puma and Bax. Using isogenic cell systems with defined genetic deficiencies (the HCT116 human colon carcinoma series), we therefore recently characterized to what extent p53's mitochondrial action depends on other key Bcl members for its MOMP release activity. We find that death stimulus-induced translocation of p53 to mitochondria is independent of Puma and Bax. Moreover, mitochondrial p53 is highly efficient and superior to tBid in inducing the release of soluble and insoluble apoptogenic factors by severely disrupting outer and inner membrane integrity. This again does not require Puma or Bax. This MOMP action is associated with wtp53-induced oligomerization not only of Bax and Bak, but also of VDAC, and the formation of a stress-induced endogenous complex between wtp53 and cyclophilin D, normally located at the inner membrane. In contrast, missense mutant p53 proteins have lost the ability to alter the multimeric state of VDAC. Thus, the Puma and Bax independence distinguishes the mitochondrial from the cytosolic p53 death pathway. The data also hints that p53 does not merely act as a BH3-only type protein but induces MOMP differently from tBid [19].
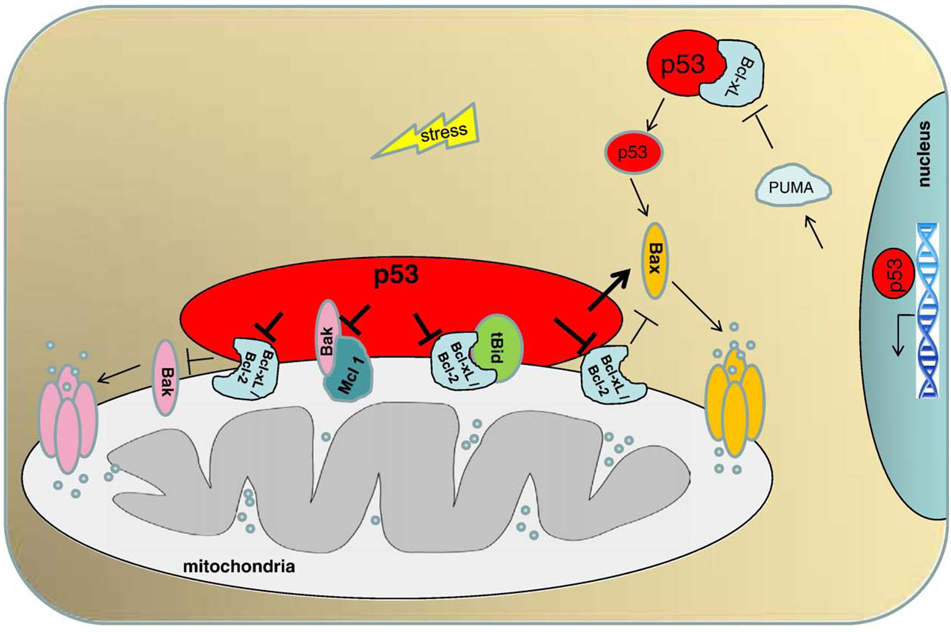
Although a stable p53–Bax interaction was not observed, p53 is able to directly activate Bax in the absence of other proteins [13]. Recombinant p53 and Bax proteins together (but neither of them alone) trigger the permeabilization of model liposomes whose lipid composition mimics that of mitochondrial inner/outer membrane contact sites. This is accompanied by Bax liposome insertion and oligomerization, and it would be due to a “hit and run” mechanism, involving a conformational change of Bax but not a stable interaction between Bax and p53 [13] (Fig. 1).
Fig. 1.
The direct mitochondrial p53 program of apoptosis. Stress-induced mitochondrial translocation of p53 results in interactions with multidomain members (anti- and proapoptotic) of the Bcl-2 family to induce mitochondrial outer membrane permeabilization. p53 interacts with Bcl-xL and Bcl2 and neutralizes their inhibitory effects on proapoptotic Bax and Bak, which are the only members of the Bcl2 family able to oligomerize and form lipid pores on the mitochondrial outer membrane. p53 interaction with Bcl-xL also liberates proapoptotic tBid from its inhibitory complex with Bcl-xL. Moreover, p53 interacts directly with Bak, liberating it from an inhibitory complex with anti-apoptotic Mcl-1. Thus, p53 acts like a ‘super’ BH3-only protein, combining both enabling and activating BH3-only functions. p53 also interacts with Bax in a ‘hit and run’ manner, which stimulates Bax oligomerization and pore formation. In an additional cytosolic p53 pathway, p53 first transactivates Puma, which then liberates p53 from a pre-existing cytosolic Bcl-xL complex to activate monomeric Bax in the cytosol.
In sum, the DNA-binding domain (DBD) of p53 has a dual function: it mediates its nuclear function for driving p53's transcriptional programs as well as its extranuclear functions for interacting with MOMP-inducing/inhibiting proteins. Therefore, mutations in the DBD of p53 in human tumors constitute ‘double hits’ that impair both the mitochondrial and transcriptional apoptotic activities of p53 [8,12].
Recently, the role of p53 oligomerization in the regulation of its pro-apoptotic mitochondrial function was examined, but so far no firm conclusion could be reached. Clearly, the p53 C-terminus containing the oligomerization domain does not contact Bcl-xL or Bak [15]. Also, a C-terminally truncated version of a mitochondrially targeted p53 is as efficient as wtp53 to induce apoptosis in p53 null cancer cells [7]. Consistent with this, in dominant-negative heterozygosity simulations by Roemer et al, tumor-derived missense mutant p53 or ΔEx2/3p73 isoforms interfere with stress-induced expression of wtp53-responsive genes as expected, but leave apoptosis by mitochondrial p53 largely unaffected [20]. Of note, in contrast to tetrameric nuclear p53, mitochondrial p53, be it wild-type or mutant, is mostly monomeric in their cross-linking studies. This might explain its resistance against dominant negative interference by mutant p53. In this respect, the extra-nuclear p53-dependent apoptosis may constitute a fail-safe mechanism against dominant inhibition [20]. On the other hand, Murphy et al find that deletion or mutation of p53's oligomerization domain markedly impairs its ability to oligomerize Bak and their cross-linking studies indicate that the majority of p53 localized to mitochondria is in dimeric or higher-order oligomeric form [17].
3. Monoubiquitination of cytoplasmic p53 promotes mitochondrial translocation — a rapid action binary switch from degradation to activation
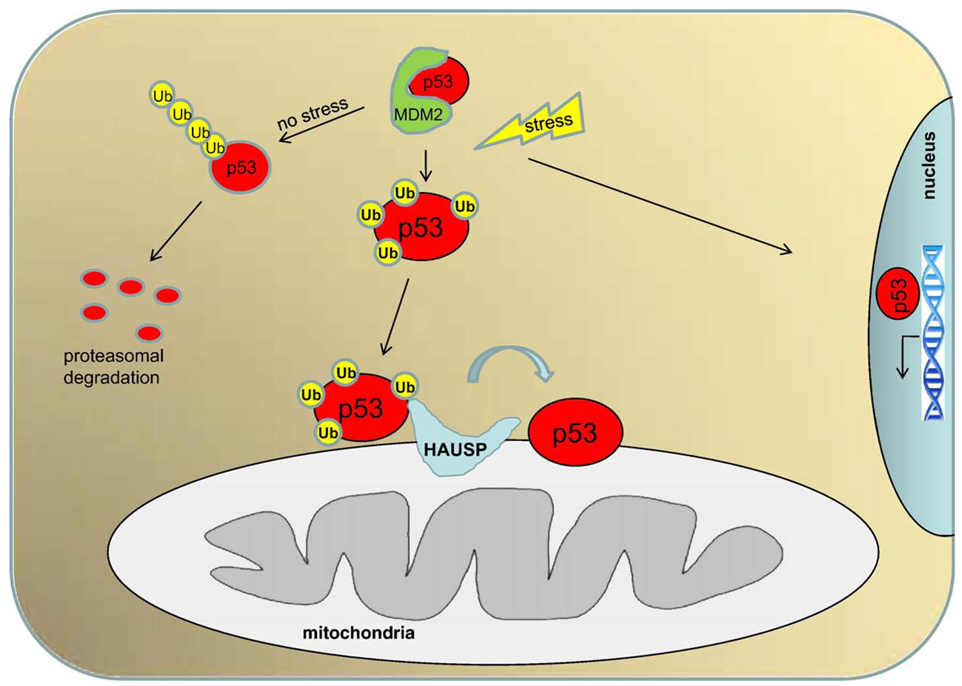
An important question is how translocation of p53 to mitochondria is regulated. In light of two pertinent negatives, i.e. the absence of a mitochondrial translocation motif within the p53 polypeptide sequence and the fact that N- and C-terminal phosphorylation/acetylation modifications play no major role in mitochondrial targeting of p53 [21], our group went on to show that (multi)mono-ubiquitination of p53 promotes its targeting to mitochondria. (Multi)-monoubiquitination has been implicated in processes other than proteosomal destruction of important proteins such as Foxo4 and PTEN, in particular as a signal for intracellular trafficking between compartments [22,23]. (Multi)-monoubiquitinated proteins are stable, since efficient proteasomal degradation minimally requires a multiubiquitin chain that is at least 4 subunits long per individual lysine residue [24]. Importantly, the source of mitochondrially translocated p53 is a separate and distinct stress-stabilized pool in the cytoplasm. Cytoplasmic Mdm2 acts as an E3 ligase but not as a physical shuttler of p53. Upon arrival at mitochondria, p53 undergoes rapid deubiquitination by mitochondrial HAUSP via a stress-induced mitochondrial p53-HAUSP complex, which generates the apoptotically active non-ubiquitinated p53. Taken together, a distinct cytoplasmic pool of monoubiquitinated p53, generated as intermediary in resting cells by basal levels of Mdm2 and similar E3 ligases, is subject to a rapid action binary switch from a fate of inactivation via polyubiquitination and degradation in unstressed cells, to a fate of activation via mitochondrial trafficking ([25] and reviewed in [26] (Fig. 2).
Fig. 2.
Regulation of mitochondrial p53 translocation. The source of mitochondrially translocated p53 is a stress-stabilized pool in the cytoplasm. A two-step regulation of p53 creates a rapid-action binary switch. The product of the first step – monoubiquitinated p53, generated by basal levels of Mdm2-type E3 ligases – can undergo two opposite fates: degradation and inactivation by subsequent polyubiquitination via Mdm2 or E4-type ligases under normal conditions. Alternatively, if stress occurs, this monoubiquitinated p53 intermediate is rapidly stabilized by stress-induced disruption of the p53–Mdm2 complex and diverted to mitochondria. Upon arrival, p53 undergoes rapid deubiquitination by mitochondrial HAUSP via a stress-induced p53–HAUSP complex, which generates the apoptotically active non-ubiquitinated p53. Another major fraction of p53 translocates to the nucleus or is stabilized within to initiate its transcriptional program of target gene activation and repression.
4. The mitochondrial p53 pathway in pathophysiology
The relevance of the direct mitochondrial p53 program to the overall p53-mediated stress response in vivo is underlined by some recent studies in animals and supports a role in the pathophysiologic response to genotoxic, hypoxic and specific toxic insults.
4.1. Acute radiation response
In normal mice exposed to whole body γ-irradiation (5 or 10 Gy) or a single injection of a clinical dose of etoposide, we found that mitochondrial p53 accumulation occurs in radiosensitive organs like thymus, spleen and testis, but not in radioresistant organs like liver and kidney. In radiosensitive organs mitochondrial p53 translocation is rapid (detectable at 30 min in thymus and spleen) and triggers a p53-dependent first wave of caspase 3 activation within the first hour, followed by an early wave of apoptosis (detectable at 2 h in thymus). Importantly, in this system the rapid mitochondrial p53 action precedes p53 target gene activation. The earliest apoptotic p53 target gene product in irradiated thymus is Puma, whose mRNA starts to be minimally induced after 2 h but only peaks at 3 h. (Contributions by other pro-apoptotic p53 targets such as Noxa, Bax and DR5 appear minor or have late kinetics, while Bid, Killer/DR5 and p53DinP1 remain uninduced). Puma induction then coincides with a second increase in Casp 3 activation. Similar biphasic kinetics was seen in wtp53-harboring human cancer cells. Conversely, a lack of mitochondrial p53 accumulation after γIR in vivo correlates with cell cycle arrest and a complete absence of apoptosis in radioresistant kidney and liver. Thus, these results suggest that in radiosensitive organs mitochondrial p53 accumulation in vivo triggers a rapid first wave of apoptosis that is transcription-independent, followed by a slower wave that is transcription-dependent and largely mediated by its target gene Puma [27].
The in vivo relevance of mitochondrial p53 is also highlighted by the recent identification of the small molecule inhibitor Pifithrin-μ (PFTμ) in the Gudkov lab [28]. PFTμ selectively inhibits p53 mitochondrial translocation and reduces its affinity to Bcl-xL and Bcl-2, without interfering with p53's transcription function. Remarkably, PFTμ rescues lethally irradiated mice from bone marrow failure after 8–9 Gy of γIR and strongly reduces γIR-induced thymocyte death [28].
Thus, based on our in vivo radiosensitivity data and the PFTμ protection data from the Gudkov lab, the mitochondrial p53 program might be responsible in great measures for the collateral damage of chemo- and radiotherapy in sensitive normal tissues during cancer treatment and in acute radiation accidents. Thus, it appears promising that inhibition of this program might dampen this damage.
4.2. Ischemic cerebral injury
Evidence for an important role of the mitochondrial p53 death pathway specifically in ischemic cerebral injury is also starting to accumulate: Mitochondrial – rather than nuclear – accumulation of p53 in hypoxia-sensitive hippocampal CA1 neurons strongly correlates with CA1 cytochrome c release and neuronal death within the first 24 h in a transient global cerebral ischemia model in rat. Occasional nuclear p53 staining was seen only later, at 72 h. Conversely, pan-p53 small molecule inhibitor PFTα given i.v. suppressed p53 mitochondrial translocation and blocked apoptosis by 70% in vivo. On the other hand, mitochondrial p53 accumulation was not seen in hypoxia-resistant CA3 neurons [29,30]. Moreover, in C6 glioma cells and primary rat astrocyte cultures exposed to oxidative H2O2 stress for 1 h followed by 6–24 h recovery, p53 levels continue to rise. Of note, its localization becomes overwhelmingly mitochondrial, coinciding with caspase 3 activation and apoptosis [31]. Conversely, a neuron-protective effect from ischemic pre-conditioning correlates with suppressed mitochondrial p53 translocation during subsequent ischemia-reperfusion challenge [32].
4.3. Ischemia-reperfusion injury of the kidney
Another pathophysiologic setting where the mitochondrial p53 program likely contributes to apoptosis is ischemia-reperfusion injury of the kidney. This condition ends in ‘acute tubular necrosis’ (ATN) manifested as acute renal failure, a very common and serious clinical problem in shock syndromes, severe hypotension and thromboembolic events [33]. This injury is modeled by transient bilateral renal artery occlusion. It is characterized by death of tubular epithelial cells mainly in the cortex and outer medulla that undergo p53-dependent apoptosis of the proximal convoluted tubules and the thick ascending tubules (as well as p53-independent necrosis of the proximal straight tubules. Both components contribute to renal failure). Depletion of intracellular ATP/GTP pools is an important and specific inducer of tubular apoptosis (but not of necrosis), and p53 is its critical downstream effector [33,34]. In human kidney samples, p53 stabilization in tubular epithelial cells is a reliable marker for ischemic and nephrotoxic ATN [35].
Current evidence from in vivo experiments in rats is based on p53 immunolocalization and protection by the pan-p53 inhibitor PFTα, which blocks p53 transcription and mitochondrial translocation [28]. In ischemia-reperfusion injured tubules of the outer medulla, p53 stabilization is to a large extent cytoplasmic rather than nuclear, strongly co-localizes with mitochondria and correlates with apoptosis of tubular cells [33]. Conversely, intraperitoneal PFTα prevents tubular apoptosis in the outer medulla and protects renal function in mice. Of note, this PFTα effect was found to be largely due to its ability to prevent mitochondrial p53 translocation. Similarly, mitochondrial p53 translocation and apoptosis occurred in cultured primary proximal tubular epithelial cells from rat subjected to chemical anoxia with antimycin A, again blocked by PFTα [33].
4.4. Liver injury
As discussed above, upon stress-induced p53 translocation, mitochondrial p53 directly interacts with and activates pro-apoptotic Bak in many cell types [16]. p53 competes with Mcl-1 for Bak interaction and this disrupts the preexisting anti-apoptotic Mcl-1/Bak complex, resulting in Bak oligomerization and MOMP. Conversely, the inverse scenario is the molecular basis for the profound refractoriness of the liver towards p53-induced apoptosis after genotoxic stress (e.g. doxorubicin or cisplatin). In the liver, activated p53 transcriptionally induces the IGFBP1 prosurvival factor, which in part translocates to mitochondria and binds to Bak. This impairs the mitochondrial p53 activity by preventing its binding to Bak. IGFBP1−/− liver shows spontaneous apoptosis associated with mitochondrial p53–Bak complexes. Conversely, overexpression of IGFBP1 attenuates the apoptotic response to cisplatin and doxorubicin even in non-hepatic cells. On the other hand, treatment with PFTμ (which selectively inhibits p53 mitochondrial function without interfering with p53's transcription function) significantly reduces cisplatin-induced liver cell apoptosis [36].
In sharp contrast, the fulminant and often fatal liver ‘necrosis’ that occurs after α-amanitin poisoning by the deathcap mushroom Amanitis phalloides and claims several hundred victims world-wide every year is overwhelmingly due to the mitochondrial p53 program. α-amanitin is a potent transcriptional inhibitor by blocking RNA polymerase II (shutting down virtually all gene transcription including that of p53), but kills cells by inducing mitochondrial p53 translocation and p53-dependent MOMP in liver and other cells [9,36]. Accordingly, p53−/− or Bak−/− mice are largely protected from α-amanitin-induced liver damage, while wild-type mice undergo organ destruction [36].
5. Implications for cancer therapy — exploiting the shortest circuit of p53 death signaling
Strategies to treat cancer by restoring p53 function are a highly promising field. In tumors with loss of p53 function, which is an extremely common clinical situation, sustained inactivation of the p53 tumor suppressor pathway is required for tumor maintenance. This concept was recently proven by several conditional mouse models, where restoration of p53 function leads to tumor regression in vivo, either via induction of apoptosis or senescence programs, depending on the tumor type [37–39]. Thus, this paradigm represents a mirror image of the concept that tumors are addicted to dominant oncogenes and regress after their shut down.
In p53-based therapeutics research, up to now efforts focused on restoring transcription-mediated p53 apoptosis. One venue tries to identify small molecules such as CP-31398 (Pfizer) and PRIMA that structurally rescue tumor-derived p53 mutant proteins. Alternatively, in tumors retaining wild-type p53, compounds such as Nutlin (Roche) or RITA try to activate wtp53 by liberating it from inhibitory Hdm2 interaction. Another important venue focuses on supplying ‘conventional’ wtp53 via adenoviral type 5 delivered gene therapy [40–44]. Currently this holds the biggest promise and in late 2003 was approved by the Chinese Food and Drug Administration for treatment of head and neck squamous cell carcinoma in combination with radiotherapy [41]. Ad5–wtp53 is currently also in clinical trials in China and the West for other malignancies. Both the wtp53 and small molecule approach have in common that they focus on p53's transcriptional action and thus rely on a putatively preserved ability of tumor cells to respond with transcriptional activation of p53 target genes. However, many human cancers have lost this prerequisite due to global epigenetic deregulation of their genome, leading to broadly aberrant gene silencing patterns [45,46]. Thus, in such cancers wtp53-based therapies might inadvertently dilute p53's tumor killing powers by diverting the majority of its action to a non-productive route. Indeed, wtp53 gene therapy did fail in a large multinational phase IV clinical trial of ovarian cancer [47]. Another disadvantage is that wtp53-based therapy can lead to cytostatic arrest rather than the desired apoptosis as outcome, in particular if tumor cells express p21Waf1, which is typically the case, which raises the threshold for an apoptotic outcome. This desensitizes cells against conventional chemotherapy drugs that preferentially kill proliferating cells.
To exploit the shortest circuit of p53 death signaling, we therefore introduced the concept of delivering p53 into cancer cells that is specifically targeted to mitochondria. We generated surface-restricted and non-restricted (imported) versions of mitochondrially targeted human wild-type p53 fusion proteins in different viral vectors: i) surface-restricted p53CTB and p53CTM, by fusing the transmembrane domains of Bcl-xL (CTB) and Bcl-2 (CTM) to the C-terminus of wtp53 [8,48,49] and ii) non-restricted Lp53wt, by fusing the import leader sequence of ornithine transcarbamylase to the N-terminus of wtp53. Lp53wt strongly interacts with surface Bcl-xL [8]. In addition, it is imported into the mitochondrial matrix (presumably via the TOM/TIM complex), where the leader is cleaved off by endopeptidases prior to p53 redistribution within the submitochondrial compartments [7]. All p53 fusions target exclusively to mitochondria and are completely devoid of transcriptional activity. Importantly, they are sufficient to induce apoptosis and long-term colony suppression in a transcription-independent fashion in all tested p53-null human cancer cell lines [7,8].
We then did a series of pre-clinical proof of principle experiments with promising outcomes. We first used retroviral gene transfer to compare in vivo efficacy of mitochondrially targeted wild-type p53 with ‘conventional’ wtp53 or empty vector in the cMyc transgenic Burkitt lymphoma model. Using intravenous tumor cell transfer into syngeneic recipients and an in vivo competition protocol against admixed parental tumor cells, we found that mitochondrially-targeted p53 effectively kills p53-null and ARF-null primary B-lymphomas in vivo. Moreover, this proof-of-principle experiment showed that exclusive reliance on the direct mitochondrial program exerts a significant tumor suppressor activity in vivo that is equipotent to ‘conventional’ wild-type p53 [49]. Since human tumors typically express highly elevated levels of missense mutant p53 protein, we next could show that retroviral targeting of p53 to mitochondria does indeed confer a significant growth disadvantage even in B-lymphomas that express vastly stabilized mutant p53 proteins. Interestingly, killing efficacy of Lp53wt proofed superior to that of p53CTB and p53CTM [48]. Finally, in solid tumor xenografts, we used intratumoral adenoviral delivery of p53. In classic nude mouse assays with the human colon carcinoma line HCT116 p53−/−, we generated replication-deficient Ad5–GFP constructs to compare tumor cell killing by mitochondrially targeted wtp53 with that of wtp53. Reproducibly, mitochondrial Ad5–Lp53wt was equipotent to conventional Ad5–wtp53 which is currently clinically used in vivo and in vitro [50].
Thus, the advantage of mitochondrially targeted p53-based gene therapy appears three-fold: it bypasses the need for transcriptional reactivation, is focused to directly activate the mitochondrial death program and eliminates cell cycle arrest/senescence outcomes in the tumor cell targets. Together, these data provide encouragement to further explore the potential of mitochondrial p53-based cancer therapeutics and exploit the shortest circuit of p53 death signaling.
6. Additional homeostatic functions of endogenous p53 in mitochondria?
Evidence for additional functions of endogenous p53 in mitochondria is also emerging, although their significance remains to be fully characterized. In addition to localizing to the outer membrane (described above), a subfraction of p53 localizes inside mitochondria and complexes with the major mitochondrial import proteins mthsp70 and mthsp60, present in the matrix [7,51].
This p53 might have a role in normal mitochondrial homeostasis and/or non-apoptotic outcomes after stress. Intramitochondrial p53 was found to interact with mitochondria-specific DNA polymerase gamma (Pol γ) and enhance its processivity in vitro. Since the genetic stability of mtDNA depends on the accuracy of Pol γ, this suggests a role for p53 in maintaining the stability of the mitochondrial genome. This notion is supported by the finding that loss of p53 results in significant sensitivity of mtDNA to genotoxic stress [52,53]. Moreover, a complex between mitochondrial transcription factor A (mtTFA) and stress-induced mitochondrial p53 was demonstrated, suggesting that p53 influences the repair capacity of mtTFA upon mtDNA damage and supporting a role for p53 in maintaining the stability of the mitochondrial genome. [54].
An open question is whether p53 serves as a transcription factor for the mitochondrial genome. Major mitochondrial transcription factors are mtTFA and TFBM1 and 2, which regulate the HSP and LSP promoters [55]. In addition, transcription factors from the steroid receptor family have been suggested to regulate transcription of mitochondrial DNA [56,57]. p53-deficient mouse embryos show a reduction in mitochondrial 16S rRNA transcript levels during embryonal development [58]. Moreover, the use of a mitochondria-targeted dominant-negative p53 miniprotein correlated with inhibition of 16S rRNA expression in NIH3T3 cells [59]. Andrews et al. identified 7 sequences within mitochondrial DNA as putative p53-recognition sequence candidates. Several of them are located within the 12S and 16S rDNA region and the NADH dehydrogenase genes [60]. A subsequent study found one of these sequences, located within the 16S rDNA, to be p53-responsive in human cancer cells. However, although Southwestern blotting suggested a direct interaction of p53 with this sequence, ChIP assays failed to confirm this. Thus, more studies are necessary to determine whether p53 directly associates with mitochondrial DNA or whether the effect of p53 on mitochondrial transcription is indirect [61].
References
- 1.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, manypossibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 2.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 3.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 4.Ding HF, McGill G, Rowan S, Schmaltz C, Shimamura A, Fisher DE. Oncogene-dependent regulation of caspase activation by p53 protein in a cell-free system. J. Biol. Chem. 1998;273:28378–28383. doi: 10.1074/jbc.273.43.28378. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb E, Oren M. p53 facilitates pRb cleavage in IL-3-deprived cells: novel pro-apoptotic activity of p53. EMBO J. 1998;17:3587–3596. doi: 10.1093/emboj/17.13.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 7.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 8.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 9.Arima Y, Nitta M, Kuninaka S, Zhang D, Fujiwara T, Taya Y, Nakao M, Saya H. Transcriptional blockade induces p53-dependent apoptosis associated with translocation of p53 to mitochondria. J. Biol. Chem. 2005;280:19166–19176. doi: 10.1074/jbc.M410691200. [DOI] [PubMed] [Google Scholar]
- 10.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3 — transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25:4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 11.Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488:110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- 12.Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, Pan H, Kessler H, Pancoska P, Moll UM. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J. Biol. Chem. 2006;281:8600–8606. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- 13.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 14.Petros AM, Gunasekera A, Xu N, Olejniczak ET, Fesik SW. Defining the p53 DNA-binding domain/Bcl-x(L)-binding interface using NMR. FEBS Lett. 2004;559:171–174. doi: 10.1016/S0014-5793(04)00059-6. [DOI] [PubMed] [Google Scholar]
- 15.Sot B, Freund SM, Fersht AR. Comparative biophysical characterization of p53 with the pro-apoptotic Bak and the anti-apoptotic BCL-XL. J. Biol. Chem. 2007 Oct 5;282(40):29193–29200. doi: 10.1074/jbc.M705544200. [DOI] [PubMed] [Google Scholar]
- 16.Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat. Cell Biol. 2004;6:443–450. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- 17.Pietsch EC, Perchiniak E, Canutescu AA, Wang G, Dunbrack RL, Murphy ME. Oligomerization of BAK by p53 utilizes conserved residues of the p53 DNA binding domain. J. Biol. Chem. 2008;283:21294–21304. doi: 10.1074/jbc.M710539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 19.Wolff S, Erster S, Palacios G, Moll UM. p53's mitochondrial translocation and MOMP action is independent of Puma and Bax and severely disrupts mitochondrial membrane integrity. Cell Research. 2008 Jul;18(7):733–744. doi: 10.1038/cr.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyne K, Schmitt K, Mueller D, Armbruester V, Mestres P, Roemer K. Resistance of mitochondrial p53 to dominant inhibition. Mol. Cancer. 2008;7:54. doi: 10.1186/1476-4598-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemajerova A, Erster S, Moll UM. The post-translational phosphorylation and acetylation modification profile is not the determining factor in targeting endogenous stress-induced p53 to mitochondria. Cell Death Differ. 2005;12:197–200. doi: 10.1038/sj.cdd.4401526. [DOI] [PubMed] [Google Scholar]
- 22.Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr. Top. Microbiol. Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- 24.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6:1718–1723. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- 27.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol. Cell. Biol. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat. Chem. Biol. 2006;2:474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 29.Endo H, Kamada H, Nito C, Nishi T, Chan PH. Mitochondrial translocation of p53 mediates release of cytochrome c and hippocampal CA1 neuronal death after transient global cerebral ischemia in rats. J. Neurosci. 2006;26:7974–7983. doi: 10.1523/JNEUROSCI.0897-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endo H, Saito A, Chan PH. Mitochondrial translocation of p53 underlies the selective death of hippocampal CA1 neurons after global cerebral ischaemia. Biochem. Soc. Trans. 2006;34:1283–1286. doi: 10.1042/BST0341283. [DOI] [PubMed] [Google Scholar]
- 31.Bonini P, Cicconi S, Cardinale A, Vitale C, Serafino AL, Ciotti MT, Marlier LN. Oxidative stress induces p53-mediated apoptosis in glia: p53 transcription-independent way to die. J. Neurosci. Res. 2004;75:83–95. doi: 10.1002/jnr.10822. [DOI] [PubMed] [Google Scholar]
- 32.Racay P, Tatarkova Z, Drgova A, Kaplan P, Dobrota D. Effect of ischemic preconditioning on mitochondrial dysfunction and mitochondrial p53 translocation after transient global cerebral ischemia in rats. Neurochem.Res. 2007;32:1823–1832. doi: 10.1007/s11064-007-9437-3. [DOI] [PubMed] [Google Scholar]
- 33.Kelly KJ, Plotkin Z, Vulgamott SL, Dagher PC. P53 mediates the apoptotic response to GTP depletion after renal ischemia-reperfusion: protective role of a p53 inhibitor. J. Am. Soc. Nephrol. 2003;14:128–138. doi: 10.1097/01.asn.0000040596.23073.01. [DOI] [PubMed] [Google Scholar]
- 34.Dagher PC. Apoptosis in ischemic renal injury: roles of GTP depletion and p53. Kidney Int. 2004;66:506–509. doi: 10.1111/j.1523-1755.2004.761_7.x. [DOI] [PubMed] [Google Scholar]
- 35.McLaren BK, Zhang PL, Herrera GA. P53 protein is a reliable marker in identification of renal tubular injury. Appl. Immunohistochem. Mol. Morphol. 2004;12:225–229. doi: 10.1097/00129039-200409000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Leu JI, George DL. Hepatic IGFBP1 is a prosurvival factor that binds to BAK, protects the liver from apoptosis, and antagonizes the proapoptotic actions of p53 at mitochondria. Genes Dev. 2007;21:3095–3109. doi: 10.1101/gad.1567107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 39.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim DS, Bae SM, Kwak SY, Park EK, Kim JK, Han SJ, Oh CH, Lee CH, Lee WY, Ahn WS. Adenovirus-mediated p53 treatment enhances photodynamic anti-tumor response. Hum. Gene Ther. 2006 Mar;17(3):347–352. doi: 10.1089/hum.2006.17.347. [DOI] [PubMed] [Google Scholar]
- 41.Peng Z. Current status of gendicine in China: recombinant human Ad-p53 agent for treatment of cancers. Hum. Gene Ther. 2005;16:1016–1027. doi: 10.1089/hum.2005.16.1016. [DOI] [PubMed] [Google Scholar]
- 42.Roth JA. Adenovirus p53 gene therapy. Expert Opin. Biol. Ther. 2006;6:55–61. doi: 10.1517/14712598.6.1.55. [DOI] [PubMed] [Google Scholar]
- 43.Sauthoff H, Pipiya T, Chen S, Heitner S, Cheng J, Huang YQ, Rom WN, Hay JG. Modification of the p53 transgene of a replication-competent adenovirus prevents mdm2-and E1b-55kD-mediated degradation of p53. Cancer Gene Ther. 2006 Jul;13(7):686–695. doi: 10.1038/sj.cgt.7700936. [DOI] [PubMed] [Google Scholar]
- 44.Yin S, Goodrich DW. Combination gene therapy with p53 and Thoc1/p84 is more effective than either single agent in an animal model of human pancreatic adenocarcinoma. Int. J. Oncol. 2006;28:781–785. [PubMed] [Google Scholar]
- 45.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer — a mechanism for early oncogenic pathway addiction? Nat. Rev., Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 46.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 47.Zeimet AG, Marth C. Why did p53 gene therapy fail in ovarian cancer? Lancet Oncol. 2003;4:415–422. doi: 10.1016/s1470-2045(03)01139-2. [DOI] [PubMed] [Google Scholar]
- 48.Palacios G, Moll UM. Mitochondrially targeted wild-type p53 suppresses growth of mutant p53 lymphomas in vivo. Oncogene. 2006;25:6133–6139. doi: 10.1038/sj.onc.1209641. [DOI] [PubMed] [Google Scholar]
- 49.Talos F, Petrenko O, Mena P, Moll UM. Mitochondrially targeted p53 has tumor suppressor activities in vivo. Cancer Res. 2005;65:9971–9981. doi: 10.1158/0008-5472.CAN-05-1084. [DOI] [PubMed] [Google Scholar]
- 50.Palacios G, Crawford HC, Vaseva A, Moll UM. Mitochondrially targeted wild-type p53 induces apoptosis in a solid human tumor xenograft model. Cell Cycle 7. 2008 Aug 15;7(16):2584–2590. doi: 10.4161/cc.7.16.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dumont P, Leu JI, Della Pietra AC, III, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 52.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Souza-Pinto NC, Harris CC, Bohr VA. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene. 2004;23:6559–6568. doi: 10.1038/sj.onc.1207874. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. p53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 55.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 56.Enriquez JA, Fernandez-Silva P, Garrido-Perez N, Lopez-Perez MJ, Perez-Martos A, Montoya J. Direct regulation of mitochondrial RNA synthesis by thyroid hormone. Mol. Cell. Biol. 1999;19:657–670. doi: 10.1128/mcb.19.1.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casas F, Daury L, Grandemange S, Busson M, Seyer P, Hatier R, Carazo A, Cabello G, Wrutniak-Cabello C. Endocrine regulation of mitochondrial activity: involvement of truncated RXRalpha and c-Erb Aalpha1 proteins. FASEB J. 2003;17:426–436. doi: 10.1096/fj.02-0732com. [DOI] [PubMed] [Google Scholar]
- 58.Ibrahim MM, Razmara M, Nguyen D, Donahue RJ, Wubah JA, Knudsen TB. Altered expression of mitochondrial 16S ribosomal RNA in p53-deficient mouse embryos revealed by differential display. Biochim. Biophys. Acta. 1998;1403:254–264. doi: 10.1016/s0167-4889(98)00066-4. [DOI] [PubMed] [Google Scholar]
- 59.Donahue RJ, Razmara M, Hoek JB, Knudsen TB. Direct influence of the p53 tumor suppressor on mitochondrial biogenesis and function. FASEB J. 2001;15:635–644. doi: 10.1096/fj.00-0262com. [DOI] [PubMed] [Google Scholar]
- 60.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 61.Heyne K, Mannebach S, Wuertz E, Knaup KX, Mahyar-Roemer M, Roemer K. Identification of a putative p53 binding sequence within the human mitochondrial genome. FEBS Lett. 2004;578:198–202. doi: 10.1016/j.febslet.2004.10.099. [DOI] [PubMed] [Google Scholar]