Summary
RNA polymerase (RNAP) trapped in intermediate stages of promoter escape, as well as RNAP paused at promoter-proximal σ70-dependent pause sites, gives rise to stable, transcriptionally engaged stalled complexes that can limit promoter function and present potential sites for transcription regulation. To investigate the prevalence of such intermediates, we screened 118 Escherichia coli candidate promoters for RNAP stalling at or near the promoter, using in vivo KMnO4 mapping of RNAP on chromosomal DNA. Of 34 active promoters, the seven preceding lacZ, tnaA, cspA, cspD, rplK, rpsA and rpsU harboured stalled RNAP in vivo; this finding suggests that RNAP stalling after initiation is widespread in E. coli. Consistent with the characteristics of both abortive and promoter-proximal σ70-dependent paused complexes, RNAP trapping at most of the newly identified stall sites was eliminated by the rpoDL402F σ70 mutational alteration and by site mutations, and was enhanced by GreA deficiency. In addition to promoter-proximal RNAP trapping, we observed transcription-dependent DNA modifications spanning the tnaA and cspA leader regions up to 100 bp downstream of the transcription start site.
Introduction
Rate-limiting steps in transcription, and thus potential sites of regulation, can occur after initiation of RNA chain synthesis but close to the promoter. First, the process of promoter escape itself can be limiting, as evidenced by the process of abortive initiation detected in vitro. Abortive initiation is caused by persistent binding of σ70 to core promoter sequences, resulting in failure of an emergent RNA chain to be continued. During this initial RNA synthesis, energy is stored as the initial transcription bubble is enlarged and DNA is ‘scrunched’ out of the active site channel, allowing the active centre to advance along the template while the promoter sigma bonds remain intact (Kapanidis et al., 2006; Revyakin et al., 2006); it is believed that successful (as opposed to abortive) initiation uses this energy to break the σ70-promoter bonds. Importantly, the scrunched intermediate is stable in vitro, with a half-life of about 5 s for one promoter (Margeat et al., 2006; Revyakin et al., 2006). The activity of Gre proteins to inhibit abortive initiation implies that the lifetime may be substantially less in vivo, but there is an example of an artificial promoter for which an apparently stable scrunched state occurs in vivo (Ellinger et al., 1994). This stable state that precedes both elongation and release of an abortive product presents a potential target for regulation.
A second potential rate-limiting transcription stage near the promoter is induced by σ70 binding a nearby reiteration of the −10 promoter sequence after the initial σ70-promoter bonds are broken, thus inducing a transcription pause. Such pauses have been found both in vitro and in vivo in λ family bacteriophage late genes, occurring 16–25 nucleotides after initiation at the phage pR′ promoters; these paused complexes provide a substrate for phage antiterminator protein Q to bind and become a subunit of the elongating complex (Roberts et al., 1998). A similar pause was found in vitro in the lac operon, and a possibly more widespread occurrence has been discussed (Mooney and Landick, 2003; Brodolin et al., 2004; Nickels et al., 2004). These early σ70-dependent paused complexes have structures similar to the open promoter complex, except for the presence of transcript, which destabilizes the binding of σ70 region 4 to the RNA polymerase (RNAP) beta subunit flap structure (Murakami et al., 2002; Nickels et al., 2005; 2006). An important similarity is that both open promoter complexes undergoing abortive initiation and the σ70-dependent paused complexes involve a scrunched state of the transcription bubble (Marr and Roberts, 2000). Whereas relaxation of the scrunched state of abortive complexes results in release of the short transcript and return of the complex to the pre-initiation state, relaxation of the paused complex yields a backtracked complex (Marr and Roberts, 2000), undoubtedly because the longer RNA is stabilized through the interactions that normally stabilize transcription elongation complexes – namely RNA binding to DNA in the templating hybrid and to elements of the RNAP core subunits.
The DNA sequence that elicits the λ pR′ σ70-dependent pause resembles the −10 consensus, with positions 2 and 6 of the TATAAT −10 consensus present in all examples (Fig. 1). Mutation of these sites, +2A or +6T in λ, and analogous positions in others (Fig. 1), strongly reduces RNAP pausing. In addition, the three consecutive G-C base pairs following the ANNNT motif significantly contribute to the pause, largely (like the −10 consensus) through non-template strand interactions in the paused complex (Ring and Roberts, 1994), likely to σ70 region 1.2 (Feklistov et al., 2006; Haugen et al., 2006).
Fig. 1.
Sequence alignment of the extended −10 consensus with σ70-dependent pause-inducing sequences in λ and the lambdoid phages φ80, 21 and 82. Boxes indicate the most highly conserved bases of the −10 consensus, which are absolutely required for signalling the RNAP pause in phage; the transcription start site is in lower case.
Two genetic tools are available to characterize and identify stable promoter-proximal RNAP complexes. First, a mutation in σ70 (L402F) in the RNAP core binding face specifically reduces or eliminates the promoter-proximal σ70-dependent pause, while leaving a sufficient (although somewhat reduced) σ70 activity for promoter function (Ko et al., 1998). In addition to destabilizing this pause, σ70 L402F also reduces abortive initiation at the promoter (Ko et al., 1998). Both of these effects could derive from weakening the σ70 region 2 interaction with the DNA indirectly through weakened σ70-RNAP core interaction, or they could reflect a subtler perturbation of elements that stabilize the scrunched state of the RNAP complex. Second, the lifetime of the σ70-dependent pause associated with the lambda late gene promoters is decreased by Gre activity both in vivo and in vitro (Marr and Roberts, 2000). For the σ70-dependent pause site, the effect of Gre is attributed to rescue of a persistently backtracked (arrested) state of the complex, through the well-established RNAP active centre-mediated cleavage mechanism (Borukhov et al., 1993; Orlova et al., 1995). Gre proteins also inhibit abortive initiation, but the mechanistic basis for this activity is not understood (Feng et al., 1994; Hsu et al., 1995).
Besides the phage σ70-dependent pause, there is little evidence for promoter-proximal RNAP stalling in vivo. In this study, we look for such structures on E. coli chromosomal genes in vivo. By screening of an essentially random set of promoters, we have identified several that harbour transcriptionally engaged RNAP complexes stalled at or near the promoter. We have also identified promoters with transcription-dependent signals – possible distal stalled RNAP complexes – occurring up to 108 bases downstream of the transcription start site. We find that RNAP stalling at or near the promoter is reduced or eliminated by the σ70L402F mutation, as expected for either type of stalled complex. Furthermore, most of the promoter-proximal stalled complexes provide a substrate for GreA in vivo, suggesting similarities to abortive or paused complexes. Finally, we show that σ70-dependent RNAP stalling can present a barrier to transcription elongation, and we discuss its potential biological significance.
Results
KMnO4 footprinting can be used to detect RNAP on chromosomal DNA
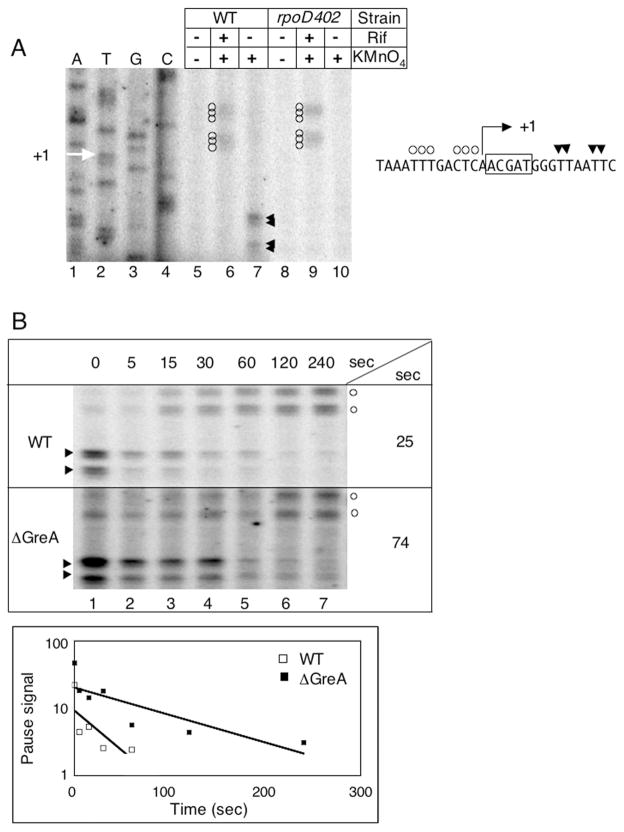
Open promoter complexes and paused elongation complexes have been detected by KMnO4 footprinting of plasmid DNA in vivo (Sasse-Dwight and Gralla, 1989; Kainz and Roberts, 1992). We have adapted the KMnO4 footprinting protocol to use chromosomal DNA as template to facilitate screening, and to eliminate potential artefacts that may accompany the use of multicopy plasmid templates. To demonstrate this procedure, we carried out KMnO4 footprinting on an E. coli strain harbouring a single copy of the λ prophage in its genome (Fig. 2). As expected, the pR′ pause is absent in the rpoD402 strain (Fig. 2A, compare lanes 7 and 10). In the presence of rifampicin, a drug that binds RNAP and prevents initiation of RNA chain synthesis, the appearance of open complex signal (Fig. 2A, lanes 6 and 9) indicates that the pR′ promoter is active in both strains, and therefore that the lack of pause signal in rpoD402 is due specifically to the effect of this mutation on the σ70-dependent pause.
Fig. 2.
Detection of RNAP pausing on chromosomal DNA using KMnO4 footprinting.
A. KMnO4 footprinting of the pR′ promoter region in WT and rpoD402 cells lysogenic for λ phage. Cells were untreated (lanes 5 and 8), treated with rifampicin (rif) for 4 min prior to KMnO4 treatment (lanes 6 and 9), or treated with KMnO4 alone (lanes 7 and 10). A sequencing ladder (lanes 1–4) reflects the sequence of the pR′ template strand, and the transcription start site is marked on the gel image with a white arrow. The sequence of the pR′ promoter non-template strand is indicated to the right of the gel. In this sequence, the transcription start site is underlined, and boxed bases indicate the pause-inducing sequence. On both the gel image and the sequence, KMnO4-modified bases in the open complex are marked with open circles, while KMnO4-modified bases in the paused complex are marked with arrows.
B. KMnO4 footprinting time-courses of the pR′ promoter region in WT and ΔgreA cells lysogenic for λ phage. Cells were treated with KMnO4 alone (lane 1), or rifampicin for 5, 15, 30, 60, 120 or 240 s prior to KMnO4 treatment (lanes 2–7 respectively). KMnO4-modified bases in the open complex are marked with open circles, while KMnO4-modified bases in the paused complex are marked with arrows; one marker may indicate a group of adjacent modified bases. In the plot below, pause signal decay (in arbitrary units) with time is shown for WT (open squares) and ΔgreA (closed squares) cells, and estimated pause half-lives derived from the plots are indicated to the right of each gel.
Pause half-life can be estimated using this technique by treating cells with KMnO4 first in the absence of rifampicin (Fig. 2B, lane 1) to detect complexes during equilibrium growth, and then at intervals following rifampicin addition (Fig. 2B, lanes 2–7); the pause signal decays as RNAP trapped at the pause site is released, while simultaneous appearance of the promoter signal indicates that a second round of transcription is blocked by rifampicin. The pause signal decay rate provides an estimate of pause half-life (Fig. 2B).
The pattern of KMnO4 modification at the λ pR′ pause, as well as the pause half-life measured on chromosomal DNA, are in good agreement with measurements on plasmid DNA (Kainz and Roberts, 1992; 1995; Marr and Roberts, 2000). Specifically, KMnO4-reactive bases at the pause site appear at the leading edge of the transcription bubble at positions +10, +11, +14 and +15, and the estimated pause half-life is 25 s, comparable to the previously reported in vivo half-life of 23 s (Fig. 2). In addition, the pR′ pause duration in vivo is enhanced in a greA null strain by about threefold (Fig. 2B), comparable to the fourfold effect seen previously when a plasmid copy of the pR′ promoter was used (Marr and Roberts, 2000).
Escherichia coli genes harbour stalled RNAP complexes proximal and distal to the promoter
Our criterion for an assayable transcription unit was the appearance upon rifampicin treatment of an open complex signal, which is recognizable based on the known extent of the transcription bubble. In addition to weak promoters, those with unstable open complexes are undetectable by this assay; thus, we were unable to detect the very active but unstable rrnP1 promoters (Paul et al., 2004).
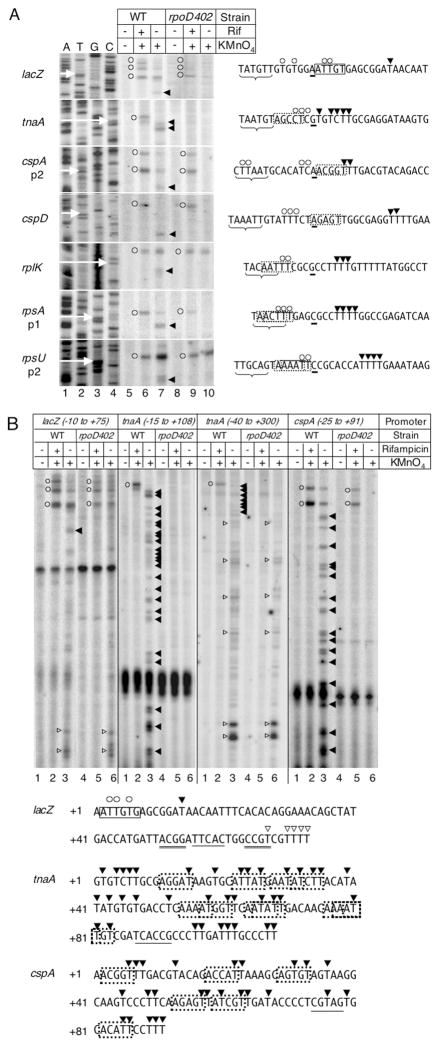
We selected an essentially random set of 118 candidate promoters using three approaches (refer to Experimental procedures), and screened these for evidence of promoter-proximal RNAP arrest using in vivo KMnO4 footprinting. Of the 118 promoters, we found 34 with detectable open complexes. Seven of the 34 harbour RNAP complexes stalled at or near the promoter, as evidenced by promoter-proximal KMnO4 reactivity downstream and distinct from that of the open complex (Fig. 3A). In addition to the promoter-proximal pause at the lacZ promoter described previously in vitro (Brodolin et al., 2004; Nickels et al., 2004), we found that RNAP stalls at or near the tnaA, cspA, cspD, rplK, rpsA and rpsU promoters. Interestingly, in addition to promoter-proximal stalled RNAP complexes, we found transcription-induced KMnO4 reactivity distal to the lacZ, cspA and tnaA promoters up to 108 bases downstream of the transcription start site (Fig. 3B), suggesting the existence of promoter-distal stalled complexes.
Fig. 3.
KMnO4 footprinting of promoter proximal and distal regions with detectable RNAP trapping in WT and rpoD402 cells. A. Cells were untreated (lanes 5 and 8), treated with rifampicin (rif) for 4 min prior to KMnO4 treatment (lanes 6 and 9), or treated with KMnO4 alone (lanes 7 and 10).
A sequencing ladder (lanes 1–4) reflects the template strand sequence for each promoter, and each transcription start site is marked with a white arrow. The non-template strand sequence of each promoter is indicated to the right of each gel image. In each sequence, the underlined base indicates the transcription start site, the brace indicates the promoter −10 sequence, and boxes indicate the known (solid box) or putative (dotted box) pause-inducing sequences. On both the gel image and the promoter sequence, KMnO4-modified bases in the open complex are marked with open circles, while KMnO4-modified bases in the paused complex are marked with arrows; one marker may indicate a group of adjacent modified bases. B. Cells were untreated (lanes 1 and 4), treated with rifampicin (rif) for 4 min prior to KMnO4 treatment (lanes 2 and 5), or treated with KMnO4 alone (lanes 3 and 6). The non-template strand sequence of each leader is indicated beneath the gel image. In each sequence, boxes indicate the known (solid box) or putative (dotted box) pause-inducing sequences, and KMnO4-modified bases in the promoter-distal paused complexes are marked with closed arrows pointing leftward (σ70-dependent) or open arrows pointing rightward (hairpin-dependent). Bases obscured by background bands on the gel (single underline), and bases that may pair to form a putative hairpin (double underline) are indicated.
Most stalled RNAP complexes require wide-type σ70
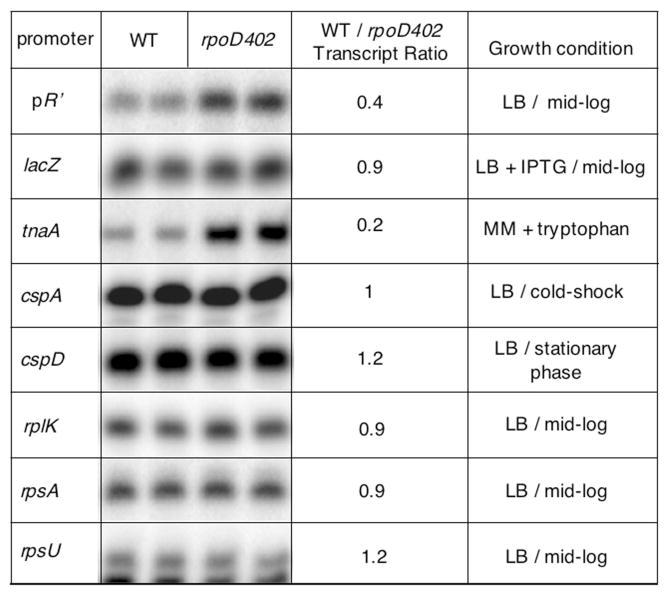
Because the λ pR′ pause provides a well-defined in vivo model of a stalled transcriptionally engaged RNAP complex, we compared its properties with the newly identified in vivo stalled intermediate complexes. We first assessed the role of σ70 by assaying for stalling in rpoD402. Indeed, KMnO4 footprinting revealed that none of the seven promoters have detectable proximal stalled RNAP complexes in rpoD402, indicating a requirement for wild-type (WT) σ70 (Fig. 3A). In addition to the pause signal, the promoter signal for tnaA in rpoD402 is absent following rifampicin treatment; this could mean either that the tnaA promoter is not functional in rpoD402, or that the open complex at the tnaA promoter in rpoD402 is too unstable to detect. Two lines of evidence support the latter possibility: (i) downstream reactivity that aligns plausibly with expected sites of RNAP pausing in the tnaA leader associated with well-characterized Rho-dependent termination sites (Stewart et al., 1986) appears in both WT and rpoD402 strains (Fig. 3B, open arrows) and (ii) tnaA mRNA is detected in both WT and rpoD402 strains using S1 nuclease mapping (Fig. 6).
Fig. 6.
Comparison of mRNA levels generated from the indicated promoters in WT and rpoD402 cells using S1 nuclease mapping. The indicated strains were grown under activating conditions for each promoter prior to the assay. Transcript ratios (WT/rpo402) are listed.
The majority of the putative promoter-distal stalled complexes also require WT σ70, including all of the stalled complexes at cspA and tnaA that appear up to 91 and 108 bp downstream of the transcription start site respectively, as promoter-distal KMnO4 reactivity is absent in rpoD402 (Fig. 3B). Contrarily, the promoter-distal stalled complexes in the lacZ leader and coding region, as well as the known hairpin-dependent pauses that exist beyond position +110 in the tnaA leader (Stewart et al., 1986) persist in rpoD402 (Fig. 3B, open arrows), suggesting that these complexes do not require WT σ70. No hairpin-dependent pauses are known to exist in the lac operon 5′ region, although we have located a putative hairpin that might account for the KMnO4 reactivity at the bottom of the leftmost gel of Fig. 3B.
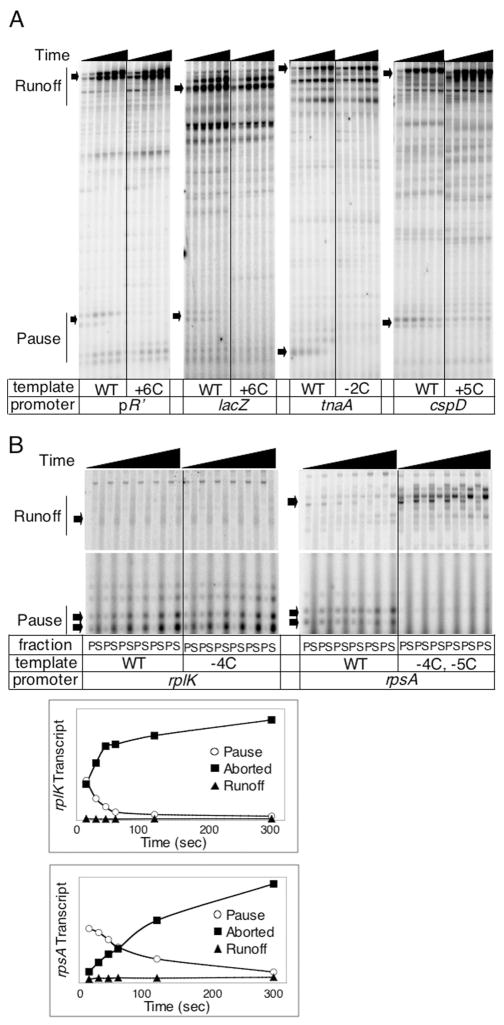
As rpoD402 also reduces abortive initiation, its effect on promoter-proximal reactivity associated with the seven promoters does not distinguish abortive complexes from σ70-dependent downstream paused complexes. The latter should arise, like the λ pR′ pause, from a −10-like sequence in the appropriate position, ~14 bp upstream of the promoter-proximal KMnO4-reactive bases. We did find such sequences near each promoter (Fig. 3A, boxed bases), using the criterion of matches to positions +2 and +6 of the λ pR′ pause-inducing −10 consensus (Fig. 1). To assay for pausing in vitro and to determine its association with these sequences, we cloned each promoter, mutated the putative pause-inducing sequence, and transcribed WT and mutant DNAs in synchronized single-round RNA synthesis. Of the seven promoters, we detected transcript associated with arrested RNAP complexes on five (lacZ, tnaA, cspD, rplK and rpsA).
The transcripts associated with stalled RNAP on lacZ, tnaA and cspD templates chase into the run-off fraction with time, like the transcript associated with paused RNAP at pR′ (Fig. 4A). Furthermore, mutating the −10-like consensus on these templates eliminated the arrested complexes. Thus, these three satisfy the criteria for a σ70-dependent pause in which σ70 binds a −10-like sequence downstream of the promoter −10 sequence.
Fig. 4.
Detection of pause or aborted transcripts using single-round in vitro transcription assays. Single-round in vitro transcription reactions were carried out for 15, 30, 45, 60, 120 or 300 s on WT and indicated mutant templates in solution (A) or immobilized on strepavidin-coated magnetic beads (B). Run-off and pause/aborted transcripts are indicated with arrows. The plots below each gel image in B reflect the changes in pause (open circles), aborted (closed squares) and run-off (closed triangles) transcript with time (P, pellet fraction; S, supernatant fraction).
The transcripts associated with stalled RNAP at rplK and rpsA, for which the putative −10 pause-inducing sequences are located closest to the promoter −10 sequence, do not chase into the run-off, but rather are aborted with time (Fig. 4B). This was determined by in vitro transcription on templates immobilized on magnetic beads, allowing transcripts associated with stalled RNAP in the pellet fraction (P) to be separated from transcripts released in the supernatant (S). As mutation of the −10-like sequence does not eliminate the transcript associated with the rplK promoter, this is probably a long-lived abortive promoter complex (Fig. 4B). The rpsA-related transcript, however, is still a candidate to result from a displaced σ70 recognition sequence like the λ pause, but still aborting because it is too short to be stabilized like the longer RNAs.
Although there exist −10-like sequences spanning the downstream region of the tnaA and cspA leaders that might account for σ70-dependent promoter-distal stalled complexes (Fig. 3B, boxed sequences), we were unable to detect pauses at the appropriate sites in vitro. If these are in fact σ70-dependent pauses, the failure to find them in vitro might result from short lifetimes, or simply the asynchrony of in vitro transcription that quickly leads to dispersion of the signals when there are multiple pauses. Another possibility is that σ70 dissociates from core more quickly in vitro, so the more distal sites are inactive (Mooney and Landick, 2003). One argument in favour of their σ70 dependence is that promoters lacking such promoter-distal stalled complexes tend to have far fewer ANNNT repeats than the tnaA and cspA leaders (data not shown).
Half-lives of the promoter-proximal stalled complexes are modulated by GreA in most cases
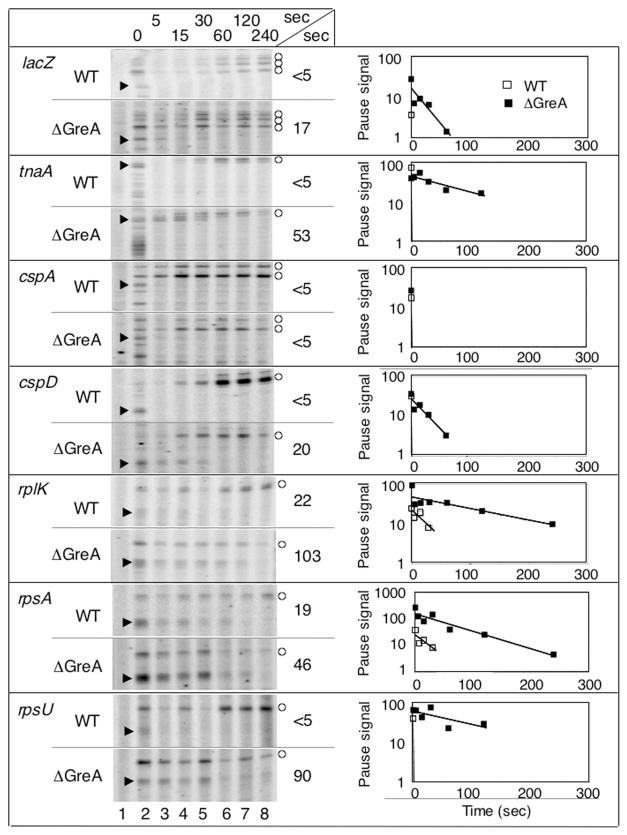
To assess the involvement of Gre proteins in stall duration, we measured the in vivo stall half-life in WT and greA null strains; the greA mutation has stronger effects in vivo than the greB mutation (Marr and Roberts, 2000). Recall that Gre proteins both inhibit abortive initiation and facilitate promoter escape (Borukhov et al., 1993; Feng et al., 1994; Hsu et al., 1995; Orlova et al., 1995), and also reduce the pR′ σ70-dependent pause half-life, presumably by promoting the escape of backtracked RNAP trapped at the pause site (Marr and Roberts, 2000). Indeed, knockout of greA prolongs promoter-proximal stall duration at all sites except cspA, with increases in duration ranging from just over twofold in the case of rpsA, to 19-fold in the case of rpsU (Fig. 5). Interestingly, the greA mutation has no effect on the promoter-distal KMnO4 reactivity in the tnaA and cspA leaders (data not shown).
Fig. 5.
Comparison of RNAP stall duration in WT and ΔgreA cells using KMnO4 footprinting. Cells were untreated (lane 1), treated with KMnO4 alone (lane 2) or treated with rifampicin (rif) for 5, 15, 30, 60, 120 or 240 s prior to KMnO4 treatment (lanes 3–8 respectively). KMnO4-modified bases in the open complex are marked with open circles, while KMnO4-modified bases in the paused complex are marked with arrows; one marker may indicate a group of adjacent modified bases. In the plot to the right of each image, stall signal decay with time is shown for WT (open squares) and ΔgreA (closed squares) cells, and estimated half-lives derived from the plots are indicated.
RNA polymerase stalling at pR′ and tnaA can limit transcription rates in vivo
In addition to a potential role as substrate for regulatory modification analogous to the phage λ σ70-dependent stalled complex, one outcome of promoter-proximal stalling might be to limit promoter function. To determine if RNAP stalling has this effect on the promoters we identified, we used S1 nuclease mapping to measure mRNA levels produced from each promoter in WT and rpoD402 strains grown in conditions that maximally induce each promoter. We find elevated mRNA from the pR′ and tnaA promoters in rpoD402 compared with WT (Fig. 6), suggesting that RNAP stalling limits transcription in vivo from these promoters; for pR′, this confirms previous results with a reporter assay of pR′ activity (Ko et al., 1998). The fact that ratios are near 1 for the other promoters suggests that the rpoD402 mutant does not have a general defect in promoter function.
Discussion
We show here that promoter-proximal RNAP stalling occurs in vivo on E. coli chromosomal genes. In addition to the pause at the lacZ promoter that has been identified by in vitro analysis (Brodolin et al., 2004; Nickels et al., 2004), we have detected RNAP stall sites in vivo associated with the tnaA, cspA, cspD, rplK, rpsA and rpsU promoters. The prevalence of such stalling was assessed by screening an essentially random set of candidate promoters: of the 34 candidate promoters for which we detected an open complex, ~20% harboured proximal stall sites. As some of the remaining 84 promoters of the original set which had no promoter signal and no pause might have had unstable open complexes similar to those of ribosomal RNA promoters, the fraction might be lower; however, it seems reasonable to conclude from this sample that 10–20% of transcription complexes stall at or near the promoter after initiation in E. coli, indicating a widespread phenomenon. Indeed, our results are consistent with the widespread prevalence of RNAP in promoter regions of E. coli detected by formaldhyde DNA–protein cross-linking (Reppas et al., 2006). Furthermore, the existence of ‘poised’ Pol II at eukaryotic promoters bears a striking similarity to the bacterial phenomenon (Saunders et al., 2006; Muse et al., 2007).
These complexes may exist in distinct states, and the major question is whether they are like the λ pause in deriving from recognition by σ70 of a distinct −10-like sequence downstream from that of the promoter, or whether instead they are complexes stalled in a (potentially) abortive cycle deriving from σ70 still bound to the promoter −10 sequence. All satisfy one criterion for derivation from a downstream −10-like sequence, namely abolition of downstream KMnO4 signal in the rpoD402 mutant in vivo. Five (lacZ, tnaA, cspD, rplK and rpsA) also display a detectable stalled complex in vitro. For three of these, lacZ, cspD and tnaA, mutation of an appropriately spaced downstream sequence does eliminate the paused RNA species detected in vitro, and we suggest that these result from a σ70 recognition event downstream of the promoter. As there can be abortive products as long as the longer paused RNA (18 nucleotides), and as rpoD402 affects abortive initiation, a less likely possibility is that they are abortive products that are reduced coincidentally by the mutation that affects the downstream −10-like consensus.
RNA polymerase stalling on rplK and rpsA is likely to result from stable states of the abortive cycle at the promoter. There is a potential displaced −10-like sequence at rplK, but mutating it had no effect on production of the stalled RNA species in vitro. RpsA might have a downstream sequence displaced by 1 bp from the promoter −10, and mutation of this sequence does eliminate the paused species; however, it seems as likely that the mutation affects the promoter −10 sequence, a change that is expected to affect abortive initiation. Both of these promoters produce stalled complexes that release RNA into a supernatant fraction upon incubation, in contrast to the paused complexes of known authentic σ70-dependent promoter-proximal paused complexes, in which the RNA is bound stably in the complex and is elongated upon incubation. However, this property is not necessarily diagnostic of abortive complexes; presumably, a moderately displaced σ70 binding sequence could produce a paused RNA too short to be effectively stabilized, which then would be released like authentic promoter-dependent abortive RNAs.
We were unable to detect stalled complexes near the rpsU and cspA promoters in vitro, although they have clear signals by KMnO4 footprinting in vivo; these might represent either type of complex. Possibly, the complexes are too short-lived in vitro to detect, or require unknown factors present in the cell. The promoter-distal stalled complexes at tnaA and cspA could be individual σ70-dependent pause sites, particularly for cspA, which retains a promoter signal but lacks all of the pause sites in rpoD402; however, these also could represent some different transcription-dependent structural change in DNA that is sensitive to KMnO4 treatment, for example, extensive unwinding.
Most of the newly discovered arrested complexes provide a substrate for GreA in vivo. The exemplar λ pR′ pause produces two RNA species in vitro: a 17 nt transcript that is sensitive to Gre-mediated cleavage, implying that the complex containing the transcript is in a backtracked state, and a 16 nt transcript that is not sensitive (Marr and Roberts, 2000). As greA knockout greatly enhances pR′ pause half-life in vivo, as determined by a KMnO4 signal that does not distinguish the precise RNA end, we presume that the 17 nt transcript also exists in a backtracked state in vivo and limits promoter escape. Similarly, the promoter-proximal pauses at lacZ, cspD and tnaA are prolonged by the greA mutation, indicating that these also are backtracked. RNAP stall duration at rplK, rpsA and rpsU is also diminished in the presence of GreA, likely as a result of Gre-mediated facilitation of promoter clearance, if these are in fact abortive complexes. This Gre-mediated regulation of ribosomal protein transcription in particular is consistent with a recent global analysis of genes regulated by GreA (Stepanova et al., 2007). That most of the arrested complexes identified in our study provide a substrate for one elongation factor underscores the possibility that other elongation factors may recognize such complexes to regulate transcription.
Promoter-distal KMnO4 modification sites 50–100 bp downstream of the tnaA and cspA transcription start site are absent in rpoD402, and thus are candidates to be σ70-dependent promoter-distal stall sites. The possibility of such σ70-dependent pausing farther downstream than the λ pauses has been discussed, and can be contrived to occur up to 462 bp downstream of the transcription start site in vitro if σ70 is tethered to a core subunit, or if a high concentration of σ70 is provided in vitro (Mooney and Landick, 2003); simply displacing a −10 consensus farther downstream is insufficient (Ring et al., 1996). These results suggest that the absence of downstream σ70-dependent pausing where a consensus exists reflects the absence of σ70 from the elongation complex. It is possible that the sequential ANNNT repeats in the tnaA leader (Fig. 3B) allow σ70-dependent promoter-distal pausing to occur by maintaining σ70 association with the DNA and hence with the elongating RNAP complex over short distances as σ70 hops from one pause-inducing sequence to the next. This model would require that each pause stabilizes the σ70 association in some persistent way, because otherwise stochastic loss of σ70 dependent on time or distance between pauses would not be reduced. It is also possible that the downstream signals from the tnaA and cspA promoters have a transcription-dependent origin but do not reflect transcription complexes at all; for example, they may represent persistent R-loops (Drolet et al., 1995).
The important result is that numerous E. coli transcription units have RNA polymerase paused or stalled in a position near or at the promoter, and sensitive to the transcription modulator GreA; as rate limiting states, they are potential elements of regulation. In one example, osmoregulation in E. coli was reported to function through a stalled RNAP (Lee and Gralla, 2004). Initial attempts to understand their biological significance of our stalled complexes revealed a pause-mediated transcription repression at tnaA that can be observed both in vitro (data not shown) and in vivo (Fig. 6). Such pause-mediated transcription repression might fine-tune gene expression, keeping transcription rates more uniform, and contributing to other types of regulation, such as metabolic control by Crp at the tnaA promoter (Deeley and Yanofsky, 1982) and tryptophan-induced attenuation in the tnaA leader (Konan and Yanofsky, 2000; Gong and Yanofsky, 2001). Potential pause-inducing sequences also span the tnaA leaders of Enterobacter aerogenes SM-18 (Kawasaki, Yokota et al., 1993), Haemophilus influenzae (Martin et al., 1998) and Proteus vulgaris (Kamath and Yanofsky, 1992), suggesting an important conserved function.
Experimental procedures
Strains
MG1655 lysogenic for λ phage was used for all KMnO4 footprinting assays, and single-copy phage integration was confirmed in these strains using a 3-primer polymerase chain reaction (PCR) (Powell et al., 1994). RpoDL402F/Kan was constructed by Marr (2000), and Δ greA::CM was a gift from C. Turnbough (University of Alabama at Birmingham). RpoDL402F/Kan and Δ greA::CM were transduced into MG1655 using P1 phage transduction (Izuhara and Takata, 1990).
Candidate promoter selection
We used three main approaches to compile a list of 118 candidate promoters. In the first approach, we used genotype and phenotype arrays to compare gene expression and metabolite utilization (respectively) of WT cells versus rpoD402. Based on these experiments, we chose 50 candidate promoters whose expression or function in metabolic pathways significantly differed between the two strains; lacZ, tnaA and cspAp2 (out of 11 assayable promoters) came from this set. In the second approach, we chose promoters that possess a plausible variant of the λ pause-inducing sequence. Specifically, we examined ~700 known promoters from the promEC index (http://margalit.huji.ac.il/promec/index.html) and BioCyc database (http://www.biocyc.org) and selected 32 that possess a proximal ANNNT consensus followed by three consecutive GC base pairs, with a requirement that the A of the consensus had to fall between +1 and +10 of the transcription start site; no stalled complexes were detected among the seven assayable promoters of this set. As KMnO4 footprinting of these initial candidate promoters revealed that only a small fraction overall (~22%) were assayable (i.e. gave an open complex signal following rifampicin and KMnO4 treatment), we selected an additional 36 candidate promoters that according to spotted array data published in the ASAP database (https://asap.ahabs.wisc.edu/asap/experiment_data.php) produce high levels of gene expression in E. coli MG1655 during mid-log growth in Luria–Bertani (LB); this set yielded an additional three (rpsAp1, rplK, rpsUp2). cspD was picked in order to examine another member of the csp family. A total of 118 candidate promoters were selected, 34 of which (aroK p2, cspA p2, cspD, cspG, cydA, gatY, groE, hns, hupA, icdA, infC p1, lacZ, lpp, manX, mgtA p1, mgtA p2, ompA, ompC, ompF, osmE, pdhR, pnp, ptsI p1, relA, rnc, rplK, rpsA p1, rpsO, rpsU p2, rrnA p2, rrnB p2, rrnC p2, rrnG p2, tnaA) were assayable by in vivo KMnO4 footprinting.
In vivo KMnO4 footprinting
A modified LB medium with 50% bacto-tryptone was used in these experiments to reduce premature quenching of KMnO4. Overnight cultures were diluted 1:50 in modified LB medium, grown to an A600 of 0.4–0.5, and treated with 10 mM KMnO4 for 1 min either alone or following rifampicin treatment (200 μg ml−1) for indicated times. The reaction was quenched by transferring cells to an equal volume of ice-cold 2XSTE buffer [50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 150 mM β-mercaptoethanol]. Genomic DNA was extracted from the cells (Qiagen, as per manufacturer’s instructions), and quantified by measuring A260. Reactive pyrimidines on the non-template strand were mapped by primer extension using 6 μg of genomic DNA and 2.5 pmols of an end-labelled primer (26–30 nucleotides in length) that anneals to the non-template strand 50–150 bases downstream of the promoter of interest. To facilitate identification of modified bases, sequencing reactions were run in parallel using the same end-labelled primer and at least 3 μg of genomic DNA as template. Primer extension products were resolved on a 10% denaturing gel, and reactive pyrimdines in the open and paused complexes were visualized and quantified using a phosphorimager (Typhoon 9400) and ImageQuant software.
Proteins and DNA templates
RNA polymerase holoenzyme was purified as described (Hager et al., 1990). WT transcription templates were PCR-amplified from an MG1655 colony and subjected to agarose gel purification. Mutations were introduced into cloned WT templates using site-directed mutagenesis (Stratagene). To generate biotinylated templates, a forward primer containing a 5′ biotin-TEG moiety was used to PCR-amplify the template from a colony or plasmid.
In vitro transcription
For single-round transcription reactions, transcripts were made by pre-incubating 5 nM DNA template and 50 nM RNAP holoenzyme for 10 min at 37°C in T buffer [20 mM Tris-HCl (pH 8.0), 50 mM potassium glutamate, 0.1 mM EDTA, 0.05 mg ml−1 BSA] and 200 μM each of rATP, rGTP, rCTP, 50 μM rUTP and 0.5 μCi μl−1 Éφ-32P-UTP to form open complex in a final volume of 108 μl. Transcription was initiated by adding 12 μl 10× initiation buffer (40 mM MgCl2 and 100 μg ml−1 rifampicin), and synthesis was stopped at indicated time intervals by placing 19 μl of aliquots of the reaction into 131 μl of ice-cold stop solution [500 mM Tris-HCl (pH 8.0), 20 mM EDTA, 100 μg ml−1 tRNA, and 0.5% SDS]. Transcription reactions were extracted with 150 μl of phenol/chloroform/isoamyl alcohol (25:24:1) and precipitated with 2.5 vols of 100% EtOH. Samples were resuspended in 5 μl of loading buffer (90% formamide, 1xTBE, 0.05% bro-mophenol blue and 0.05% xylene cyanol), and transcripts were resolved on a 12% denaturing polyacrylamide gel. For transcription on beads, a 5′ biotinylated template was pre-incubated with 50 μl of streptavidin-coated magnetic beads (Promega) in SA buffer [10 mM Tris-HCL (pH 8.0), 150 mM NaCl and 0.1 mg ml−1 BSA] for 30 min at room temperature, and the template associated with beads was washed three times in transcription buffer using a magnetic stand prior to the transcription assay. Single-round transcription was carried out as described above, except that reactions were stopped by placing 19 μl of aliquots into empty tubes on ice, and pellet and supernatant fractions were separated prior to the addition of stop solution to a final volume of 150 μl.
S1 nuclease mapping
Overnight cultures grown in the indicated medium were diluted 1:100 and grown to an A600 of 0.4–0.5. Cells were boiled in an equal volume of pre-warmed lysis buffer [2% SDS, 4 mM EDTA (pH 8.0)] for 10 min, and RNA was extracted twice in an equal volume of buffered phenol at 67°C, and once in an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) at room temperature. RNA was precipitated in 2.5 vols of EtOH, pelleted, and subjected to DNase digestion to remove contaminating DNA. RNA quantity and purity were assessed by measuring A260 and A280 Equal amounts of total RNA (10 μg–50 μg) were mixed with 2.5 pmols of end-labelled primer in hybridization solution [1 M NaCl, 0.17 M HEPES (pH 7.5) and 0.3 mM EDTA (pH 8.0)] to a final volume of 30 μl. The primers used were 30–40mers that anneal to the non-template strand 50–150 bases downstream of the indicated promoter. The mixtures were overlayed with one drop of mineral oil, and incubated first at 75°C for 10 min to denature RNA, and then at 50°C overnight for hybridization. The hybridization mixtures were then diluted 10-fold in S1 nuclease buffer [40 mM sodium-acetate (pH 4.5), 300 mM NaCl and 2 mM ZnSO4] and 20 μg ml−1 single-stranded calf-thymus DNA and digested with 100 U S1 nuclease at 37°C for 1 h. The nuclease reactions were stopped by adding 5 mM EDTA (pH 8.0) and 10 μg of tRNA, and samples were precipitated in 2.5 vols of 100% EtOH. Samples were resuspended in 10 μl of loading buffer (90% formamide, 1xTBE, 0.05% bromophenol blue and 0.05% xylene cyanol), and transcripts were resolved on a 12% denaturing polyacrylamide gel.
Acknowledgments
We thank members of the laboratory for reading the manuscript and the NIH (Grant GM21941) for support.
References
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Brodolin K, Zenkin N, Mustaev A, Mamaeva D, Heumann H. The sigma 70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nat Struct Mol Biol. 2004;11:551–557. doi: 10.1038/nsmb768. [DOI] [PubMed] [Google Scholar]
- Deeley MC, Yanofsky C. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J Bacteriology. 1982;151:942–951. doi: 10.1128/jb.151.2.942-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger T, Behnke D, Bujard H, Gralla JD. Stalling of Escherichia coli RNA polymerase in the [plus]6 to [plus]12 region in vivo is associated with tight binding to consensus promoter elements. J Mol Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, et al. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Feng GH, Lee DN, Wang D, Chan CL, Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- Gong F, Yanofsky C. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J Biol Chem. 2001;276:1974–1983. doi: 10.1074/jbc.M008892200. [DOI] [PubMed] [Google Scholar]
- Hager DA, Jin DJ, Burgess RR. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by non-optimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Hsu LM, Vo NV, Chamberlin MJ. Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuhara M, Takata R. Site-directed insertion mutagenesis with cloned fragments in Escherichia coli by P1 phage transduction. Mol Gen Genet. 1990;220:339–340. doi: 10.1007/BF00260506. [DOI] [PubMed] [Google Scholar]
- Kainz M, Roberts J. Structure of transcription elongation complexes in vivo. Science. 1992;255:838–841. doi: 10.1126/science.1536008. [DOI] [PubMed] [Google Scholar]
- Kainz M, Roberts JW. Kinetics of RNA polymerase initiation and pausing at the lambda late gene promoter in vivo. J Mol Biol. 1995;254:808–814. doi: 10.1006/jmbi.1995.0657. [DOI] [PubMed] [Google Scholar]
- Kamath AV, Yanofsky C. Characterization of the tryptophanase operon of Proteus vulgaris. Cloning, nucleotide sequence, amino acid homology, and in vitro synthesis of the leader peptide and regulatory analysis. J Biol Chem. 1992;267:19978–19985. [PubMed] [Google Scholar]
- Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Yokota A, Oita S, Kobayashi C, Yoshikawa S, Kawamoto S, Takao S, Tomita F. Cloning and characterization of a tryptophanase gene from Enterobacter Aerogenes SM-18. J Gen Microbiol. 1993;139:3275–3281. doi: 10.1099/00221287-139-12-3275. [DOI] [PubMed] [Google Scholar]
- Ko DC, Marr MT, Guo J, Roberts JW. A surface of Escherichia coli sigma 70 required for promoter function and antitermination by phage lambda Q protein. Genes Dev. 1998;12:3276–3285. doi: 10.1101/gad.12.20.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konan KV, Yanofsky C. Rho-dependent transcription termination in the tna operon of Escherichia coli: roles of the boxA sequence and the rut site. J Bacteriol. 2000;182:3981–3988. doi: 10.1128/jb.182.14.3981-3988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Gralla JD. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol Cell. 2004;14:153–162. doi: 10.1016/s1097-2765(04)00202-3. [DOI] [PubMed] [Google Scholar]
- Margeat E, Kapanidis AN, Tinnefeld P, Wang Y, Mukhopadhyay J, Ebright RH, Weiss S. Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys J. 2006;90:1419–1431. doi: 10.1529/biophysj.105.069252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr M. PhD thesis. Cornell University; 2000. Protein nucleic acid interactions in the promoter and early transcribed region of lambdoid late genes. [Google Scholar]
- Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- Martin K, Morlin G, Smith A, Nordyke A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Landick R. Tethering sigma70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma70-dependent pausing at promoter-distal locations. Genes Dev. 2003;17:2839–2851. doi: 10.1101/gad.1142203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A. The sigma 70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat Struct Mol Biol. 2004;11:544–550. doi: 10.1038/nsmb757. [DOI] [PubMed] [Google Scholar]
- Nickels BE, Garrity SJ, Mekler V, Minakhin L, Severinov K, Ebright RH, Hochschild A. The interaction between sigma 70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci USA. 2005;102:4488–4493. doi: 10.1073/pnas.0409850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Roberts CW, Roberts JW, Hochschild A. RNA-mediated destabilization of the sigma(70) region 4/beta flap interaction facilitates engagement of RNA polymerase by the Q antiterminator. Mol Cell. 2006;24:457–468. doi: 10.1016/j.molcel.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Powell BSMP, Rivas DL, Court Y, Nakamura, Turnbough Jr. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring BZ, Roberts JW. Function of a nontranscribed DNA strand site in transcription elongation. Cell. 1994;78:317–324. doi: 10.1016/0092-8674(94)90300-x. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Yarnell WS, Roberts JW. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, et al. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S, Gralla JD. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biolog Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Stepanova E, Lee J, Ozerova M, Semenova E, Datsenko K, Wanner BL, et al. Analysis of promoter targets for E. coli transcription elongation factor GreA in vivo and in vitro. J Bacteriol. 2007;189:8769–8771. doi: 10.1128/JB.00911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166:217–223. doi: 10.1128/jb.166.1.217-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]