Abstract
Targeting of nanocarriers has long been sought after to improve the therapeutic indices of anticancer drugs. Here we provide the proof of principle for a novel approach of nanocarrier targeting through their fusion with target-specific phage coat proteins. The source of the targeted phage coat proteins are landscape phage libraries—collections of recombinant filamentous phages with foreign random peptides fused to all 4,000 copies of the major coat protein. We exploit in our approach the intrinsic physico-chemical properties of the phage major coat protein as a typical membrane protein. Landscape phage peptides specific for specific tumors can be obtained by affinity selection and purified fusion coat proteins can be assimilated into liposomes to obtain specific drug loaded nanocarriers. As a paradigm for inceptive experiments, a streptavidin-specific phage peptide selected from a landscape phage library was incorporated into ~100 nm liposomes. Targeting of liposomes was proved by their specific binding to streptavidin-coated beads.
Keywords: fusion phage, tumor targeting, streptavidin, liposomes
1. Introduction
The concept of using targeted pharmaceutical nanocarriers to enhance the efficiency of anti-cancer drugs has been proven over the past decade both in pharmaceutical research and clinical settings. Examples of a successful realization of this concept are listed in numerous reviews [1–4]. In particular, it is commonly accepted that selectivity of drug delivery systems can be increased by their coupling with peptide and protein ligands targeted to differentially expressed receptors [5–7]. A new challenge, within the frame of this concept, is to develop highly selective, stable, active and physiologically acceptable ligands that would navigate the encapsulated drugs to the site of disease and control unloading of their toxic cargo inside the cancer cells. The development of tumor-specific ligands that respond to above criteria may turn into a routine procedure due to the progress in combinatorial chemistry and phage display (a long list of different target-specific peptides identified by phage display can be found in recent reviews, [8–11]). Detailed protocols for obtaining phages and peptides internalizing into cancer cells [12–18], and phage homing to tumors of human patients [19] have been developed.
A unique extension of phage display technology was the concept of landscape phages in which the phage constructs display the guest peptide on every one of the 4,000 pVIII subunits [20]. It was shown earlier that the tumor-specific peptides fused to the major coat protein pVIII can be affinity selected from multibillion clone landscape phage libraries by their ability to bind very specifically to cancer cells [17, 21, 45, 46] demonstrating a high potential of landscape phage libraries as a source of tumor specific ligands.
To serve as a targeting interface, selected ligands, once identified have to be harnessed into suitable drug delivery platforms to ensure that their targeting qualities are utilized for a tumor selective chemotherapy. The development of stealth liposomes and further end-functionalized PEG derivatives made liposomal formulations a very attractive platform to which target specific ligands could be coupled to endow the liposomes with specific homing propensities. Several examples of liposomal targeting using phage display-derived peptides exist [22–26]. In conjunction with the successful realization of peptide targeted liposomes, the technology required for conjugating the peptides to liposomes has also evolved. A variety of coupling strategies based on different PEG derivatives have been developed, such as pyridylditiopropionoylamino (PDP)-PEG [27], hydrazide (Hz)-PEG [28], maleimido (Mal)-PEG [29], and p-nitro-phenylcarbonyl (pNP)-PEG-PE [30] with their relative merits and demerits. Also, a ‘post insertion technique’ involving transfer of ligand coupled PEG molecules from micellar prepartions directly into pre-formed liposomes has been described [31, 32], thereby decoupling the conjugation step from the liposome preparation and allowing for a combinatorial synthesis of targeted nanoparticles. In these studies, selected peptides have been coupled to the anchor molecules using various techniques, including formation of a disulfide bond, cross-linking between primary amines, reactions between a carboxylic acid and primary amine, between maleimide and thiol, between hydrazide and aldehyde, or between a primary amine and free aldehyde [33].
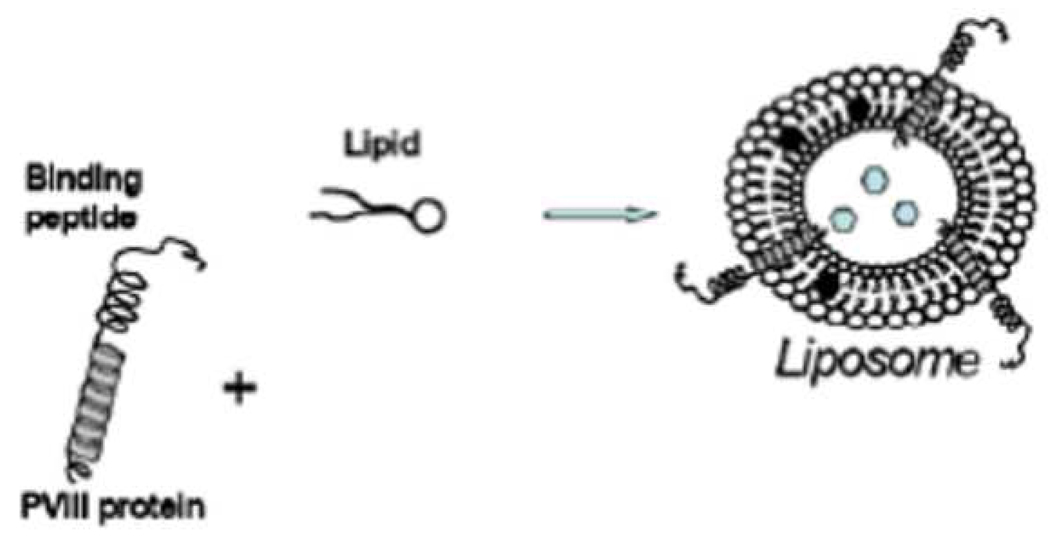
Chemical conjugation procedures have allowed for a successful realization of the concept of targeted vesicles for tumor chemotherapy. However, they would be prohibitive for a volumetric increase in the targeted liposome preparation which is required for clinical studies. These considerations led us to study the phage coat proteins as targeting ligands for drug loaded liposomes. In our approach, disease-specific peptides, genetically fused to the phage major coat protein pVIII, are isolated from a phage specific for a target organ, tissue or cell and are directly inserted into liposomes exploiting their intrinsic amphiphilic properties (Figure 1). Since the targeting peptide is a natural physical extension of the liposomal membrane-spanning anchor protein, the need for any intervening conjugation procedure is obviated. In essence, the amphiphilic property of the coat protein qualifies the phage fusion protein as a made-to-order liposomal ligand.
Figure 1.
Drug-loaded liposome targeted by the pVIII protein. The hydrophobic helix of the pVIII spans the lipid layer and binding peptide is displayed on the surface of the carrier particles. The drug molecules are shown as hexagons.
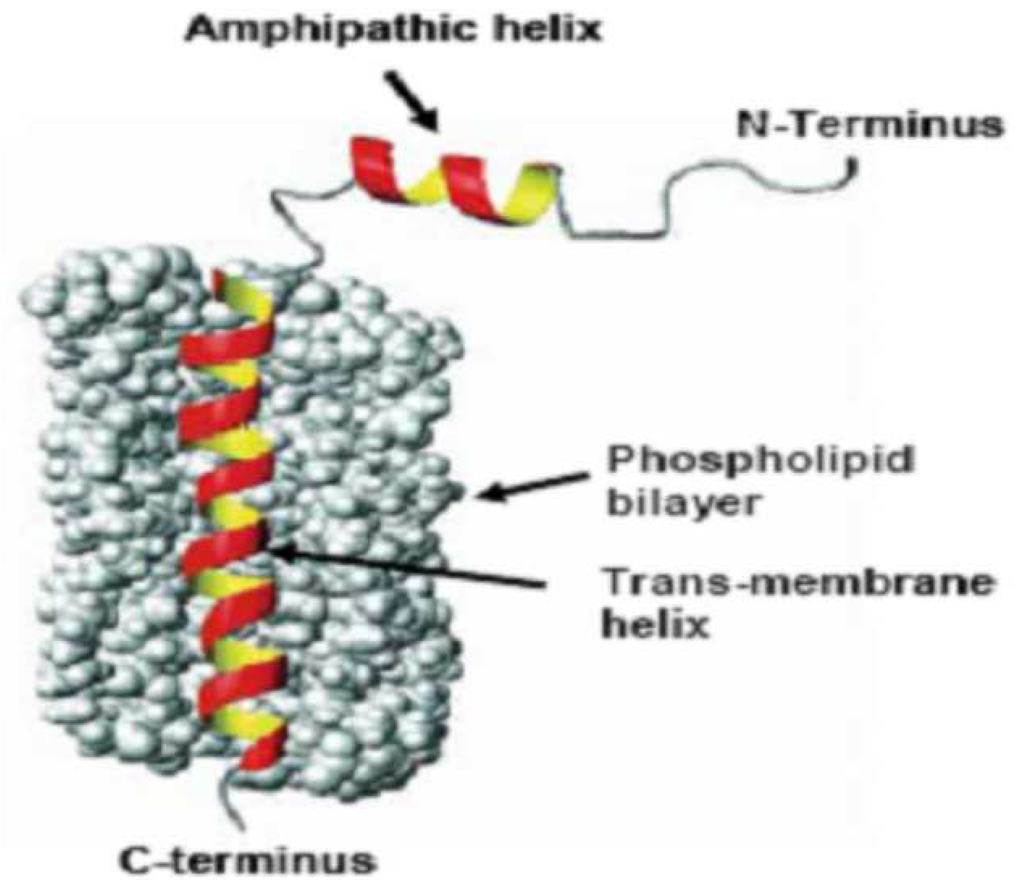
The ability of the major coat protein pVIII to incorporate into micelles and liposomes emerges from its intrinsic function as membrane protein judged by its biological, chemical, and structural properties [34]. The major coat protein pVIII is a 6.5 kDa protein with 55 amino acids in landscape phages used in this study. Numerous studies have investigated the propensity of the “wild type” pVIII to insert into micelles and phospholipid bi-layers as well its behavior in the same [35–38]. Figure 2 depicts the phospholipid bilayer spanning conformation of a phage major coat protein. The 16 Å long amphipathic helix rests on the membrane surface, while the 35 Å long transmembrane helix crosses the membrane at an angle of 26° upto residue Lys40 where the helix tilt changes. The helix tilt accommodates the thickness of the phospholipid bilayer, which is 31 Å for typical lipids used in liposome construction. The usual size of liposomes being 100 nm phage fusion proteins would be suitable targeting ligands for liposomes. To this end, we investigated whether target-specific fusion pVIII units of landscape phage retains their targeting properties as components of PEGylated liposomes. For our model experiments we chose to use commercial DOXIL® as a source of lipids because of the ease of availability, standard composition and the possibility of using constituent doxorubicin as a fluorescent marker. Accordingly, pVIII coat protein units of a streptavidin specific phage VPEGAFSS (here and after the phage is designated by the structure of the fusion peptide) were isolated in cholate solution, purified and allowed to interact with lipid components of DOXIL® under reconstitution conditions. The resulting liposomes were then tested for streptavidin specificity in a functional test using streptavidin-conjugated colloidal gold particles which demonstrated that the modified liposomes acquire streptavidin-targeting properties by virtue of the incorporated fusion proteins.
Figure 2.
The model of pVIII in the lipid environment (adapted from Stopar et al. Protein–lipid interactions of bacteriophage M13 major coat protein, Biochimica et Biophysica Acta (BBA) - Biomembranes, 2003)
2. Methods
Preparation and purification of pVIII coat protein
Streptavidin-targeting phage previously selected from the 8-mer landscape library, f8/8 [20, 39] was solubilized using the procedure outlined by Sprujit et al [38]. Briefly, 350 µl phage in 1× TBS buffer (~1 mg/ml) was mixed with 700 µl 120 mM cholate in 10 mM Tris-HCl, 0.2 mM EDTA and chloroform (2.5% v/v final concentration), pH 8.0. The suspension was incubated at 37°C for 1 h with occasional mixing. The mixture was then applied to a sepharose 6B-CL (Amersham Biosciences, NJ) column (1 cm × 45 cm) and eluted with 10 mM cholate in 10 mM Tris-HCl, 0.2 mM EDTA pH 8.0 to separate major coat protein from viral DNA and traces of bacterial proteins. The chromatographic profile was monitored by Econo UV monitor (Bio-Rad, CA); 5 ml fractions were collected and stored at 4°C. Concentration of protein in samples was determined spectrophotometrically by use of the formula,
and the molar extinction coefficients (8250 ± 412 liters/mole-centimeter) predicted by the program Protean (DNASTAR) [40]. The predicted charge on the protein at pH 8 was +0.5 as determined by the same program.
Analysis of the fusion major coat protein in chromatographic fractions and liposomal preparations by Western Blot
The presence of the fusion major coat protein in relevant fractions/preparations collected was verified by a Western blot using biotinylated rabbit anti-fd IgG. Briefly, samples of fractions were mixed with an equal volume of a sample buffer (8% SDS, 24% glycerol, 0.1 M Tris pH 6.8, 4% 2-Mercaptoethanol, 0.01% Brilliant blue G, 2X) and heated at 95°C for 10 min. 10 µl aliquots of the denatured samples were then loaded onto a 16% Non-Gradient Tris-Tricine gel (Jule Inc. CT). Electrophoresis was carried out for 90 min at 100 v. Proteins in the gel were transferred to an Immobilon-P PVDF membrane (Millipore, MA). The resulting blots were probed with biotinylated anti-fd IgG (0.12 µg/ml, prepared according to [41] followed by incubation with NeutrAvidin™-HRP (PIERCE, IL) and were visualized using a chemiluminescent substrate solution (PIERCE, IL).
Preparation of Targeted Liposomes
As a source of liposomal constituents, we used DOXIL® containing, N-(carbonylmethoxypolyethylene glycol 2000)-1, 2-distearoyl-sn-glycero-3-phosphoethanolamine sodium salt (MPEG-DSPE), 3.19 mg/ml; fully hydrogenated soy phosphatidylcholine (HSPC), 9.58 mg/ml; and cholesterol, 3.19 mg/ml, doxorubicin 2 mg/ml. To incorporate the fusion coat protein into the liposomes, 100 µl of DOXIL® (~1.6 mg total lipids based on the above composition) was first mixed with 100 µl of 120 mM cholate in 10 mM Tris-HCl, 0.2 mM EDTA pH 8.0. An aliquot of the solubilized protein (~24 µg protein in 800 µl of 10 mM cholate, 10 mM Tris-HCl, 0.2 mM EDTA pH 8.0) was added to this mixture and incubated for 2 h at 30°C. The mixture was then dialyzed against a gradually decreasing concentration of cholate as follows (all dialysis steps were carried out in 500 ml of dialysis buffer): 15 mM cholate for 30 min, 10 mM cholate for 30 min, 7.5 mM cholate for 30 min, 5 mM cholate for 30 min, 2.5 mM cholate for 30 min followed finally by an overnight dialysis with buffer containing no cholate. The cholate free liposomes were then purified on a superose 6 (GE Healthcare Bio-Sciences Corp., NJ) column 1 cm × 30 cm using 10 mM Tris-HCl, 0.2 mM EDTA pH 8.0 as eluent. The chromatographic profile was monitored by Econo UV monitor (Bio-Rad, CA); 2.5 ml fractions were collected and stored at 4°C until further analysis.
Functional test of targeted liposomes with streptavidin-conjugated colloidal gold
To determine whether the fusion coat protein integrated into the liposomes retains its binding activity towards its target streptavidin, we adapted a functional test using streptavidin-conjugated colloidal gold nanoparticles. Briefly, Carbon/Formvar-coated electron microscopic grids (Ted Pella Inc., CA) were incubated with drops of relevant fractions containing targeted liposomes or non-targeted DOXIL® liposomes (control) for 20 min. The non-specific binding sites on the grids were then blocked by incubation with separate drops of 2% BSA in 10 mM Tris-HCl, 0.2 mM EDTA pH 8.0 for 20 min. Following the blocking step, the grids were incubated with separate drops of 20 nm streptavidin-conjugated colloidal gold (Ted Pella, Inc., CA) containing 1012 beads/ml in ultra pure water for 60 min. The grids were then washed five times with separate drops of 10 mM Tris-HCl, 0.2 mM EDTA pH 8.0 and dried. The grids were negatively stained with 2% phosphotungstic acid containing 0.1% BSA as wetting agent and dried before being visualized by a Phillips transmission electron microscope (FEI Co., OR).
3. Results
Isolation and purification of the fusion phage protein
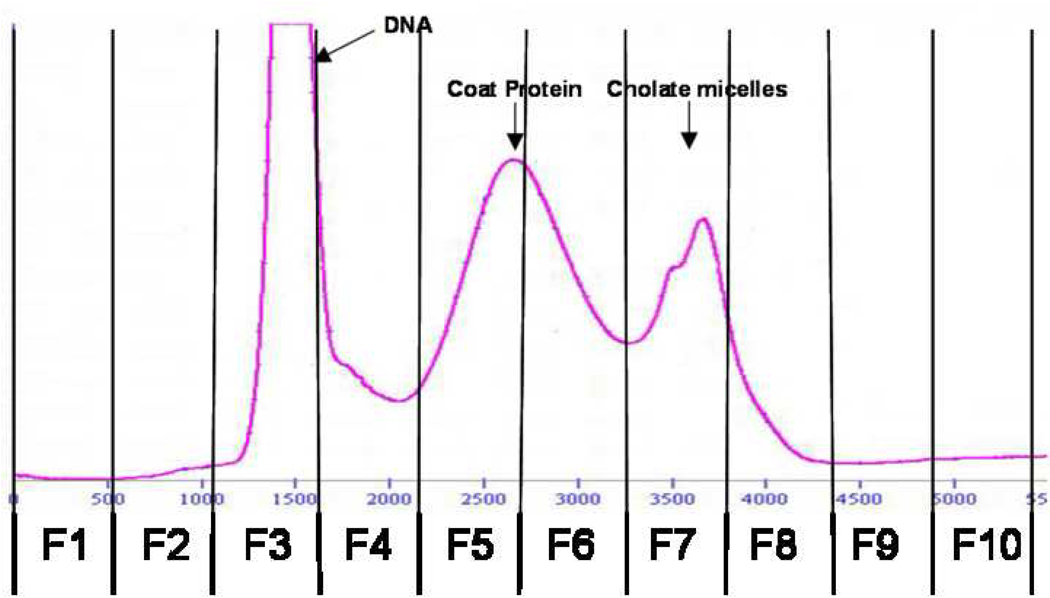
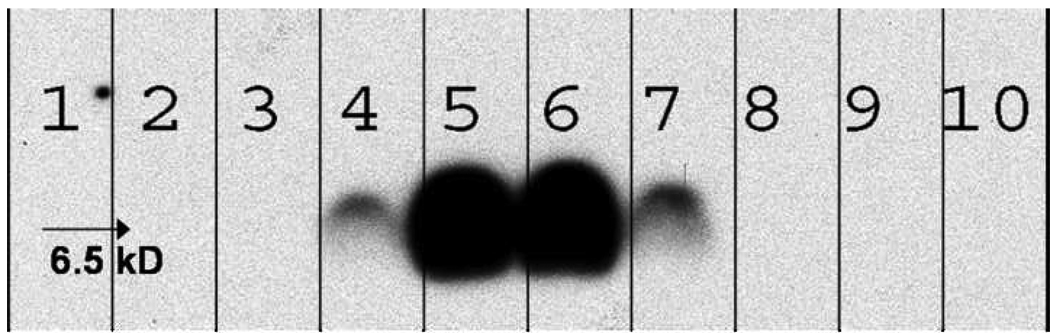
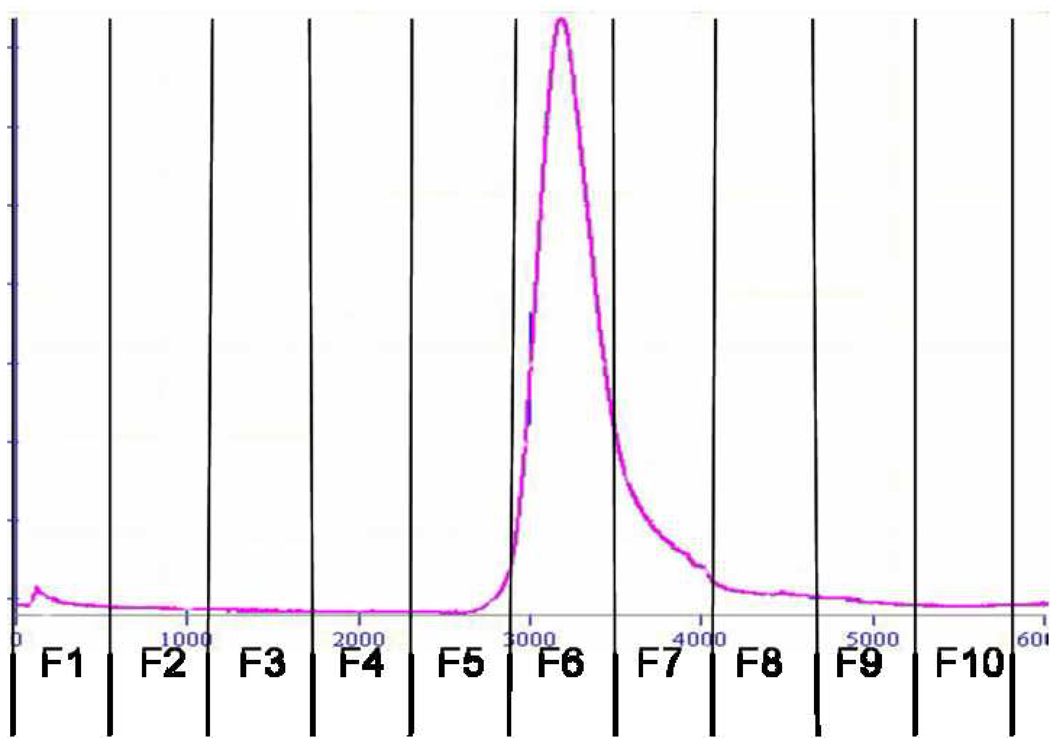
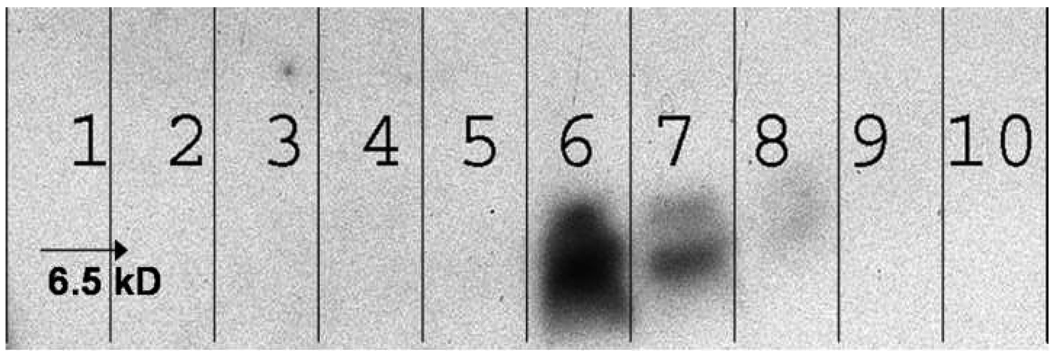
It has been previously demonstrated that deoxycholate-solubilized wild-type bacteriophage f1 can be readily fractionated by gel chromatography into DNA and coat proteins [42]. Furthermore, it was shown that the phage major coat protein pVIII isolated using sodium cholate displays lower aggregation numbers at least in the fresh state [38]. These observations led us to adapt the cholate isolation method for obtaining fusion phage coat protein from landscape phage. Accordingly, phage particles were disrupted using sodium cholate solution containing chloroform and fractionated as outlined in the experimental section. Figure 3 shows the elution profile of disrupted phage particles. As expected, DNA eluted first followed by the phage coat proteins with a clear distinction between the two peaks. The last peak probably corresponds to cholate micelles as assumed previously [38]. Analysis of the different chromatographic fractions by Western blot (Figure 4) revealed the presence of phage coat protein in the fractions 5 and 6 corresponding to the protein peak.
Figure 3.
Gel Exclusion Chromatogram of Cholate Solubilized Streptavidin specific phage.
Streptavidin specific phage (VPEGAFSS) was solubilized in Cholate solution as outlined in the experimental section.
The mixture was then eluted on a Sepharose 6B-CL column. DNA along with contaminating bacterial components eluted initially followed by phage major coat protein with a final peak of cholate micelles. Lines are drawn to specify the schema of fraction collection.
Figure 4.
Western Blot Analysis of different fractions collected during gel chromatography of cholate solubilized phage major coat protein.
Aliquots of successive fractions collected during the sepharose 6B-CL chromatography were mixed with an equal volume of Tricine sample buffer, heated to 950C for 5 min before being loaded onto a 16% Non-gradient Tricine gel. Proteins in the gel were transferred to a PVDF membrane and probed with biotinylated Anti-fd IgG. Visualization was achieved by treatment with Neutravidin-HRP followed by a chemiluminescent substrate. Numbers on the blot indicate the respective fractions to correlate with the fractions shown on Figure 3.
Preparation of Targeted Liposomes
Targeted liposomes were prepared by reconstitution of DOXIL® liposomes in the presence of phage coat proteins as outlined in the experimental section. Liposomes were initially disrupted in cholate solution, followed by incubation with phage coat protein solution. Controlled uniform integration of phage coat protein into liposomes was achieved by gradient dialysis in decreasing concentration of cholate, conditions that can prevent spontaneous aggregation of coat proteins. The reconstituted liposomes were purified by gel fractionation (Figure 5). The presence of phage coat protein in liposomal fractions, 5 and 6 was demonstrated by Western blot (Figure 6). Since no free protein was observed during chromatography, the protein content of the liposomes (1.5%) was calculated based on input (24 µg phage coat protein/ 1.6 mg lipids) and confirmed by Western blot. Doxorubicin content of the preparations at different stages was monitored by absorbance at 491 nm. Not surprisingly, we lost ~ 95% of the drug during liposome reconstruction that proved the reconstruction mechanism of protein insertion into liposomes.
Figure 5.
Gel Exclusion Chromatogram of Targeted Liposomes.
Liposome carriers in DOXIL® were disrupted using 120 mM cholate, mixed with an aliquot of cholate solubilized purified phage major coat protein and incubated. The mixture was then dialyzed against a decreasing concentration of cholate followed by purification on superose 6 prep grade column. The targeted liposomes eluted as a single peak with a broad tail. Lines are drawn to specify the schema of fraction collection.
Figure 6.
Western Blot Analysis of different fractions collected during gel chromatography of targeted liposomes.
Aliquots of successive fractions collected during the superose 6 prep grade chromatography were mixed with an equal volume of Tricine sample buffer, heated to 95°C for 5 min before being loaded onto a 16% Non-gradient Tricine gel. Proteins in the gel were transferred to a PVDF membrane and probed with biotinylated Anti-fd IgG. Visualization was achieved by treatment with Neutravidin-HRP followed by a chemiluminescent substrate. Numbers on the blot indicate the respective fractions to correlate with the fractions shown on Figure 5.
Functional activity of targeted liposomes
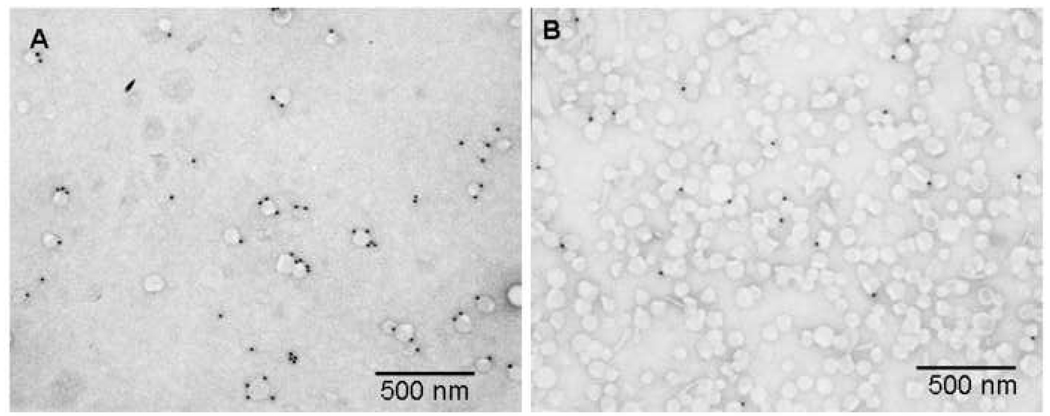
To validate our postulate that decoration of liposomes with target specific phage proteins enables target recognition by the liposomes, we exposed liposomes targeted with streptavidin-specific fusion proteins to streptavidin-conjugated colloidal gold nanoparticles. Non-targeted liposomes were used as a control. According to figure 7, targeted liposomes bind streptavidin-coated beads at a much higher level than non-targeted liposomes, as was measured by calculating the ratio of particles to liposomes which was 1.8 for targeted liposomes and 0.06 for non-targeted liposomes. The functionality of the streptavidin-specific fusion phage protein in the liposomes as effective targeting ligands has been demonstrated previously by flow cytometry and microarray tests [43].
Figure 7.
Functional test for targeted liposomes with streptavidin conjugated colloidal gold nano-particles.
Targeted liposomes prepared by co-incubation of purified major coat protein and stealth liposomal carriers of DOXIL® were tested using streptavidin conjugated colloidal gold particles for the specificity and functionality of the incorporated phage coat protein. Targeted liposomes were loaded onto formvar/carbon coated E/M grids, incubated with the colloidal gold particles followed by visualization with a Phillips transmission electron microscope (FEI Co., OR). Panel A shows targeted liposomes studded with streptavidin conjugated colloidal gold particles with a particle to liposome ratio of 1.8. Panel B shows non-targeted DOXIL® liposomes subjected to the same assay with a particle to liposome ratio of 0.06.
4. Discussion
Phage display libraries have been shown to be the reservoirs of specific peptide binders to tumor vasculature, cancer cells or their isolated receptors [44]. We have in our laboratory isolated phage peptides specific for glioma, breast and prostate cancer cell lines [17, 45,46]. Several peptides selected from phage-displayed peptide libraries have been converted into therapeutics, which validates peptide-targeting potential [5]. In these and other studies, selected peptides have been coupled to the surface of the nanocarriers using various chemical techniques mentioned in brief in the introduction. Although these approaches in anchoring peptide ligands on the liposome surface may provide target selectivity in vitro and in vivo, the cost and reproducibility of these derivatives in quality and quantity sufficient for pharmaceutical applications are challenging problems.
Herein we provide the proof of principle for a novel approach for targeting of liposomes through their fusion with purified coat proteins of a target-specific landscape phage, capitalizing on the structural and biochemical properties of phage coat protein. The “membranophilic” nature of phage coat proteins drives them to integrate into liposomes and micelles. Thus, coat protein derived from a tumor-specific landscape phage can be used to create tumor-specific liposomes by co-incubation of the relevant constituents. The drug of choice can then be loaded into the liposomes using any one of the available techniques [47]. This modular approach to the production of targeted therapeutic liposomes provides for a combinatorial setup in which different components can be added in alternate combinations to obtain different drugs. As a continuum of the current study, phage fusion peptides specific for MCF-7 breast cancer cell line were used to prepare targeted liposomes containing Rhodamine-T and such liposomes were demonstrated to selectively bind cognate target cells (Torchilin et al, unpublished). Current studies involve also loading anti-cancer drug doxorubicin into targeted liposomes and monitoring the stability of the formulations. Therapeutic liposomes targeted to the different cancer cell lines by cognate phage fusion peptides will be prepared and their targeting as well as cytotoxic potential will be evaluated. The simplicity of the approach as demonstrated by our results leads us to believe that this approach has definite potential to contribute to not only a tumor-specific but a patient-specific chemotherapeutic profile.
Acknowledgments
This work was supported by NIH grant # NIH-1 R21 AI055645 (to VAP) and Animal Health And Disease Research grant 2006-9 (College of Veterinary Medicine Auburn University) to VAP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Everts M. Targeted therapies directed to tumor-associated antigens. Drugs of the future. 2005;30:1067–1076. [Google Scholar]
- 2.Noble CO, Kirpotin DB, Hayes ME, Mamot C, Hong K, Park JW, et al. Development of ligand-targeted liposomes for cancer therapy. Expert Opinion on Therapeutic Targets. 2004;8:335–353. doi: 10.1517/14728222.8.4.335. [DOI] [PubMed] [Google Scholar]
- 3.Torchilin VP. Drug targeting. European Journal of Pharmaceutical Sciences. 2000;11:S81–S91. doi: 10.1016/s0928-0987(00)00166-4. [DOI] [PubMed] [Google Scholar]
- 4.Vasir JK, Labhasetwar V. Targeted drug delivery in cancer therapy. Technol Cancer Res Treat. 2005;4:363–374. doi: 10.1177/153303460500400405. [DOI] [PubMed] [Google Scholar]
- 5.Krumpe L, Mori T. The Use of Phage-Displayed Peptide Libraries to Develop Tumor-Targeting Drugs. International Journal of Peptide Research and Therapeutics. 2006;12:79–91. doi: 10.1007/s10989-005-9002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori T. Cancer-Specific Ligands Identified from Screening of Peptide-Display Libraries. Current Pharmaceutical Design. 2004;10:2335–2343. doi: 10.2174/1381612043383944. [DOI] [PubMed] [Google Scholar]
- 7.Shadidi M, Sioud M. Selective targeting of cancer cells using synthetic peptides. Drug Resistance Updates. 2003;6:363–371. doi: 10.1016/j.drup.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Aina OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66:184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson F, Tarli L, Viti F, Neri D. The use of phage display for the development of tumour targeting agents. Advanced Drug Delivery Reviews. 2000;43:165–196. doi: 10.1016/s0169-409x(00)00068-5. [DOI] [PubMed] [Google Scholar]
- 10.Romanov VI. Phage display selection and evaluation of cancer drug targets. Curr Cancer Drug Targets. 2003;3:119–129. doi: 10.2174/1568009033482010. [DOI] [PubMed] [Google Scholar]
- 11.Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 12.Brigati JR, Samoylova TI, Jayanna PK, Petrenko VA. Phage Display for Generating Peptide Reagents. In: Coligan JE, Dunn BM, Speicher DW, Wingfield PT, editors. Current Protocols in Protein Science. vol. 18.9. New Jersey: John Wiley & Sons, Inc.; 2008. pp. 1–27. [DOI] [PubMed] [Google Scholar]
- 13.Hong FD, Clayman GL. Isolation of a Peptide for Targeted Drug Delivery into Human Head and Neck Solid Tumors. Cancer Res. 2000;60:6551–6556. [PubMed] [Google Scholar]
- 14.Ivanenkov VV, Felici F, Menon AG. Uptake and intracellular fate of phage display vectors in mammalian cells. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1999;1448:450–462. doi: 10.1016/s0167-4889(98)00162-1. [DOI] [PubMed] [Google Scholar]
- 15.Ivanenkov VV, Felici F, Menon AG. Targeted delivery of multivalent phage display vectors into mammalian cells. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1999;1448:463–472. doi: 10.1016/s0167-4889(98)00163-3. [DOI] [PubMed] [Google Scholar]
- 16.Ivanenkov VV, Menon AG. Peptide-Mediated Transcytosis of Phage Display Vectors in MDCK Cells. Biochemical and Biophysical Research Communications. 2000;276:251–257. doi: 10.1006/bbrc.2000.3358. [DOI] [PubMed] [Google Scholar]
- 17.Samoylova TI, Petrenko VA, Morrison NE, Globa LP, Baker HJ, Cox NR. Phage probes for malignant glial cells. Mol Cancer Ther. 2003;2:1129–1137. [PubMed] [Google Scholar]
- 18.Zhang J, Spring H, Schwab M. Neuroblastoma tumor cell-binding peptides identified through random peptide phage display. Cancer letters. 2001;171:153–164. doi: 10.1016/s0304-3835(01)00575-4. [DOI] [PubMed] [Google Scholar]
- 19.Krag DN, Shukla GS, Shen G-P, Pero S, Ashikaga T, Fuller S, et al. Selection of Tumor-binding Ligands in Cancer Patients with Phage Display Libraries. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 20.Petrenko VA, Smith GP, Gong X, Quinn T. A library of organic landscapes on filamentous phage. Protein Eng. 1996;9:797–801. doi: 10.1093/protein/9.9.797. [DOI] [PubMed] [Google Scholar]
- 21.Romanov VI, Durand DB, Petrenko VA. Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. Prostate. 2001;47:239–251. doi: 10.1002/pros.1068. [DOI] [PubMed] [Google Scholar]
- 22.Koivunen E, Arap W, Valtanen H, Rainisalo A, Medina OP, Heikkila P, et al. Tumor targeting with a selective gelatinase inhibitor. Nat Biotech. 1999;17:768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- 23.Lee T-Y, Wu H-C, Tseng Y-L, Lin C-T. A Novel Peptide Specifically Binding to Nasopharyngeal Carcinoma For Targeted Drug Delivery. Cancer Res. 2004;64:8002–8008. doi: 10.1158/0008-5472.CAN-04-1948. [DOI] [PubMed] [Google Scholar]
- 24.Medina OP, Soderlund T, Laakkonen LJ, Tuominen EKJ, Koivunen E, Kinnunen PKJ. Binding of Novel Peptide Inhibitors of Type IV Collagenases to Phospholipid Membranes and Use in Liposome Targeting to Tumor Cells in Vitro. Cancer Res. 2001;61:3978–3985. [PubMed] [Google Scholar]
- 25.Pastorino F, Brignole C, Di Paolo D, Nico B, Pezzolo A, Marimpietri D, et al. Targeting Liposomal Chemotherapy via Both Tumor Cell-Specific and Tumor Vasculature-Specific Ligands Potentiates Therapeutic Efficacy. Cancer Res. 2006;66:10073–10082. doi: 10.1158/0008-5472.CAN-06-2117. [DOI] [PubMed] [Google Scholar]
- 26.Slimani H, Guenin E, Briane D, Coudert R, Charnaux N, Starzec A, et al. Lipopeptide-based liposomes for DNA delivery into cells expressing neuropilin-1. Journal of Drug Targeting. 2006;14:694–706. doi: 10.1080/10611860600947607. [DOI] [PubMed] [Google Scholar]
- 27.Allen TM, Brandeis E, Hansen CB, Kao GY, Zalipsky S. A new strategy for attachment of antibodies to sterically stabilized liposomes resulting in efficient targeting to cancer cells. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1995;1237:99–108. doi: 10.1016/0005-2736(95)00085-h. [DOI] [PubMed] [Google Scholar]
- 28.Zalipsky S. Synthesis of an end-group functionalized polyethylene glycol-lipid conjugate for preparation of polymer-grafted liposomes. Bioconjugate Chem. 1993;4:296–299. doi: 10.1021/bc00022a008. [DOI] [PubMed] [Google Scholar]
- 29.Kirpotin D, Park JW, Hong K, Zalipsky S, Li W-L, Carter P, et al. Sterically Stabilized Anti-HER2 Immunoliposomes: Design and Targeting to Human Breast Cancer Cells in Vitro. Biochemistry. 1997;36:66–75. doi: 10.1021/bi962148u. [DOI] [PubMed] [Google Scholar]
- 30.Torchilin VP, Levchenko TS, Lukyanov AN, Khaw BA, Klibanov AL, Rammohan R, et al. p-Nitrophenylcarbonyl-PEG-PE-liposomes: fast and simple attachment of specific ligands, including monoclonal antibodies, to distal ends of PEG chains via p-nitrophenylcarbonyl groups. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2001;1511:397–411. doi: 10.1016/s0005-2728(01)00165-7. [DOI] [PubMed] [Google Scholar]
- 31.Holig P, Bach M, Volkel T, Nahde T, Hoffmann S, Muller R, et al. Novel RGD lipopeptides for the targeting of liposomes to integrin-expressing endothelial and melanoma cells. Protein Engineering, Design and Selection. 2004;17:433–441. doi: 10.1093/protein/gzh055. [DOI] [PubMed] [Google Scholar]
- 32.Ishida T, Iden DL, Allen TM. A combinatorial approach to producing sterically stabilized (Stealth) immunoliposomal drugs. FEBS Letters. 1999;460:129–133. doi: 10.1016/s0014-5793(99)01320-4. [DOI] [PubMed] [Google Scholar]
- 33.Nobs L, Buchegger F, Gurny R, Allemann E. Current methods for attaching targeting ligands to liposomes and nanoparticles. J Pharm Sci. 2004;93:1980–1992. doi: 10.1002/jps.20098. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Mesleh MF, Opella SJ. Structure and dynamics of a membrane protein in micelles from three solution NMR experiments. J Biomol NMR. 2003;26:327–334. doi: 10.1023/a:1024047805043. [DOI] [PubMed] [Google Scholar]
- 35.Chamberlain BK, Nozaki Y, Tanford C, Webster RE. Association of the major coat protein of fd bacteriophage with phospholipid vesicles. Biochim Biophys Acta. 1978;510:18–37. doi: 10.1016/0005-2736(78)90127-x. [DOI] [PubMed] [Google Scholar]
- 36.Haigh NG, Webster RE. The major coat protein of filamentous bacteriophage f1 specifically pairs in the bacterial cytoplasmic membrane. Journal of Molecular Biology. 1998;279:19–29. doi: 10.1006/jmbi.1998.1778. [DOI] [PubMed] [Google Scholar]
- 37.Ohkawa I, Webster R. The orientation of the major coat protein of bacteriophage f1 in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 1981;256:9951–9958. [PubMed] [Google Scholar]
- 38.Sprujit RB, Wolfs CJAM, Hemminga M. Aggregation-Related Conformational Change of the Membrane-Associated Coat Protein of Bacteriophage M13. Biochemistry. 1989;28:9158–9165. doi: 10.1021/bi00449a030. [DOI] [PubMed] [Google Scholar]
- 39.Petrenko VA, Smith GP. Phages from landscape libraries as substitute antibodies. Protein Eng. 2000;13:589–592. doi: 10.1093/protein/13.8.589. [DOI] [PubMed] [Google Scholar]
- 40.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochemistry. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 41.Barbas CF, Barton RB, Scott JK, Silverman GJ. Phage Display: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 42.Makino S, Woolford JL, Jr, Tanford C, Webster RE. Interaction of deoxycholate and of detergents with the coat protein of bacteriophage f1. J. Biol. Chem. 1975;250:4327–4332. [PubMed] [Google Scholar]
- 43.Petrenko VA, Ustinov SN, Chen I. Fusion Phage as a Bioselective Nanomaterial: Evolution of the concept. Technical Proceedings of the European Nano Systems 2006; December 14–15; Paris. 2006. pp. 89–94. [Google Scholar]
- 44.Craig R, Li S. Function and Molecular Mechanism of Tumor-Targeted Peptides for Delivering Therapeutic Genes and Chemical Drugs. Mini Reviews in Medicinal Chemistry. 2006;6:757–764. doi: 10.2174/138955706777698615. [DOI] [PubMed] [Google Scholar]
- 45.Fagbohun OA, Bedi D, Jayanna PK, Deinnocentes P, Bird RC, Petrenko VA. Landscape phage probes for Breast cancer cells. Technical Proceedings of the 2008 Nanotechnology Conference and Trade Show, NSTI Nanotech; June 1–5; Boston. 2008. pp. 461–464. [Google Scholar]
- 46.Jayanna PK, Deinnocentes P, Bird RC, Petrenko VA. Landscape phage probes for PC3 prostate carcinoma cells. Technical Proceedings of the 2008 Nanotechnology Conference and Trade Show, NSTI Nanotech; June 1–5; Boston. 2008. pp. 457–460. [Google Scholar]
- 47.Fenske DB, Maurer N, Cullis PR. Encapsulation of weakly basic drugs, antisense oligonucleotides, and plasmid DNA within large unilamellar vescicles for drug delivery applications. In: Torchilin VP, Weissing V, editors. Liposomes: A Practical Approach. 2nd edn ed. New York: Oxford University Press; 2003. pp. 173–180. [Google Scholar]