Summary
Ischemia and seizure cause excessive neuronal excitation that is associated with brain acidosis and neuronal cell death. However, the molecular mechanism of acidification-triggered neuronal injury is incompletely understood. Here, we show that Asparagine Endopeptidase (AEP) is activated under acidic condition and cuts SET, an inhibitor of Dnase, and triggers DNA damage in brain, which is inhibited by PIKE-L. SET, a substrate of caspases, was cleaved by acidic cytosolic extract independent of caspase activation. Fractionation of the acidic cellular extract yielded AEP that is required for SET cleavage. We found that kainate provoked AEP activation and SET cleavage at N175, triggering DNA nicking in wild-type but not AEP-null mice. PIKE-L strongly bound SET and prevented its degradation by AEP, leading to resistance of neuronal cell death. Moreover, AEP also mediated stroke-provoked SET cleavage and cell death in brain. Thus, AEP might be one of the proteinases activated by acidosis triggering neuronal injury during neuroexcitotoxicity or ischemia.
Introduction
Stroke and seizures are associated with severe cerebral lactic acidosis, which is a key factor leading to permanent brain cell damage. Neuronal death caused by ischemia and seizures, occurs as a result of tremendous increase in the extracellular concentrations of excitatory amino acid (EAA) neurotransmitters, particularly glutamate. The massive release of glutamate activates glutamate receptors resulting in dramatic increases in intracellular Ca2+ (Choi, 1994). The excessive influx of Ca2+ overwhelms Ca2+ homeostasis regulatory mechanisms and leads to cell death. Excitotoxic cell death is often induced experimentally by the administration of kainic acid (KA), a potent agonist of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid/kainate class of glutamate receptors (Schauwecker and Steward, 1997). While the necrotic component of excitotoxicity has been well demonstrated, apoptosis has also been shown to play a role. Kainate injury causes both apoptosis and necrosis, with the injury depending on both the dose of kainate and the age of the culture. The apoptotic component can be selectively reduced by caspase inhibition or cycloheximide (Glassford et al., 2002). Glutamate or KA administration elicits evident pH decrease and intracellular acidification (Deitmer and Schneider, 1997; Wang et al., 1994). However, the molecular mechanism of how acidosis provokes neuronal damage is poorly understood.
PIKE (PI 3-kinase enhancer) was originally identified as a brain specific nuclear GTPase, which binds PI 3-kinase and enhances its lipid kinase activity in a GTP-dependent manner (Ye et al., 2000). To date, three forms of PIKE have been characterized: PIKE-S, PIKE-L and PIKE-A. They are originated from a single gene, CENTG1, by alternative splicing (PIKE-S and PIKE-L) or differential transcription start site (PIKE-A) (Ahn et al., 2004b; Rong et al., 2003). PIKE-S is the initially identified shorter isoform (Ye et al., 2000). PIKE-L, a longer isoform of PIKE gene, differs from PIKE-S by C-terminal extension containing Arf-GAP (ADP ribosylation factor-GTPase Activating Protein) and two ankyrin repeats domains. In contrast to the exclusive nuclear localization of PIKE-S, PIKE-L occurs in both the nucleus and the cytoplasm (Rong et al., 2003). PIKE-A contains the same domains present in PIKE-L but lacks an N-terminal proline-rich domain (PRD), which binds PI 3-kinase and PLC-γ1 (Ahn et al., 2004a; Rong et al., 2003; Ye et al., 2002). We have shown that PIKE-L binds Homer, an adaptor protein for metabotropic glutamate receptor (mGluR). Activation of mGluRIs enhances formation of an mGluRI-Homer-PIKE-L complex, leading to activation of PI 3-kinase activity and prevention of neuronal apoptosis (Rong et al., 2003).
Mammalian asparaginyl endopeptidase (AEP) is a lysosomal cysteine protease that cleaves after asparagine residues. AEP distributes in all mouse tissues, but is particularly abundant in kidney and placenta (Chen et al., 1997; Chen et al., 1998). Like all endocytic proteases, AEP is synthesized as an inactive zymogen, and its activity is regulated by post-translational events. AEP activation is autocatalytic, and requires sequential removal of C- and N-terminal pro-peptides at different pH thresholds. Removal of the N-terminal propeptide requires cleavage after aspartic acid (D) rather than asparagines (N). Cellular processing introduces at least one further cleavage to yield the final mature lysosomal enzyme (Halfon et al., 1998; Li et al., 2003). AEP has been ascribed a role in the initiation of invariant chain processing during MHC class II-mediated antigen presentation (Manoury et al., 1998; Moss et al., 2005). Although the nature of this activity remains controversial, AEP is undoubtedly a key player in lysosomal proteolysis, contributing to the processing of antigenic peptides as well as the processing of the papain family cathepsins. The processing of the lysosomal proteases, cathepsins B, H, and L, from the single-chain forms into the two-chain forms was completely deficient in AEP-null mice (Maehr et al., 2005; Shirahama-Noda et al., 2003).
SET (also known as PHAPII, TAF-Iβ, I2 PP2A) was first identified as a translocated gene that is fused to the CAN gene in a patient with acute undifferentiated leukemia (von Lindern et al., 1992). SET is a 39-kDa phosphoprotein widely expressed in various tissues and localizes predominantly in the nucleus (Adachi et al., 1994a; Adachi et al., 1994b; Fan et al., 2002). SET, identical to the template-activating factor I, stimulates DNA replication of the adenovirus genome. SET has been also identified as a potent inhibitor of the protein phosphatase 2A (Li et al., 1996; Neviani et al., 2005), for which the N-terminus is necessary. Moreover, SET binds to nucleosomal histones and protects histones from acetylation by histone acetyl transferases (Seo et al., 2001), regulating chromatin condensation and transcription (Brown et al., 2000; Kuo and Allis, 1998). Recently, SET has also been found as a component of a 270–420 kDa complex residing in the ER and mediates granzyme A (GzmA)-triggered caspase-independent nuclease activity (Fan et al., 2003). So far, at least 6 proteins have been identified in this SET complex including SET, HMG-2, NM23-H1, Ape1/ref-1, pp32 and TREX1. Gzm A induces a caspase-independent cell death pathway characterized by single-stranded DNA nicking and other features of apoptosis. NM23-H1 binds to SET and is released from inhibition by GzmA cleavage of SET. After GzmA loading or CTL attack, SET and NM23-H1 translocate to the nucleus and SET is degraded, allowing NM23-H1 to nick chromosomal DNA (Fan et al., 2003). The exonuclease TREX1 is the most recently identified SET complex member, and it acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death (Chowdhury et al., 2006). In addition to blocking the DNAse activity of NM23-H1, SET is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain (Madeira et al., 2005). Moreover, SET protein has been shown to be a substrate of caspases in vitro and in vivo (Morita et al., 2000).
In this report, we demonstrate a crucial role for AEP in mediating kainic acid (KA) or stroke-elicited neurotoxicity. We showed that PIKE-L specifically associated with SET in the nucleus and protected SET from proteolytic cleavage by AEP in vitro and in vivo. KA is neural excitotoxic and elicits pH decreases in brain. We found that KA activated AEP in mouse brain and triggered SET degradation, DNA nicking and neuronal cell death, which were diminished by PIKE-L overexpression. By contrast, in AEP deficient mice, KA or stroke failed to provoke DNA nicking or neuronal cell death.
Results
PIKE interacts with SET
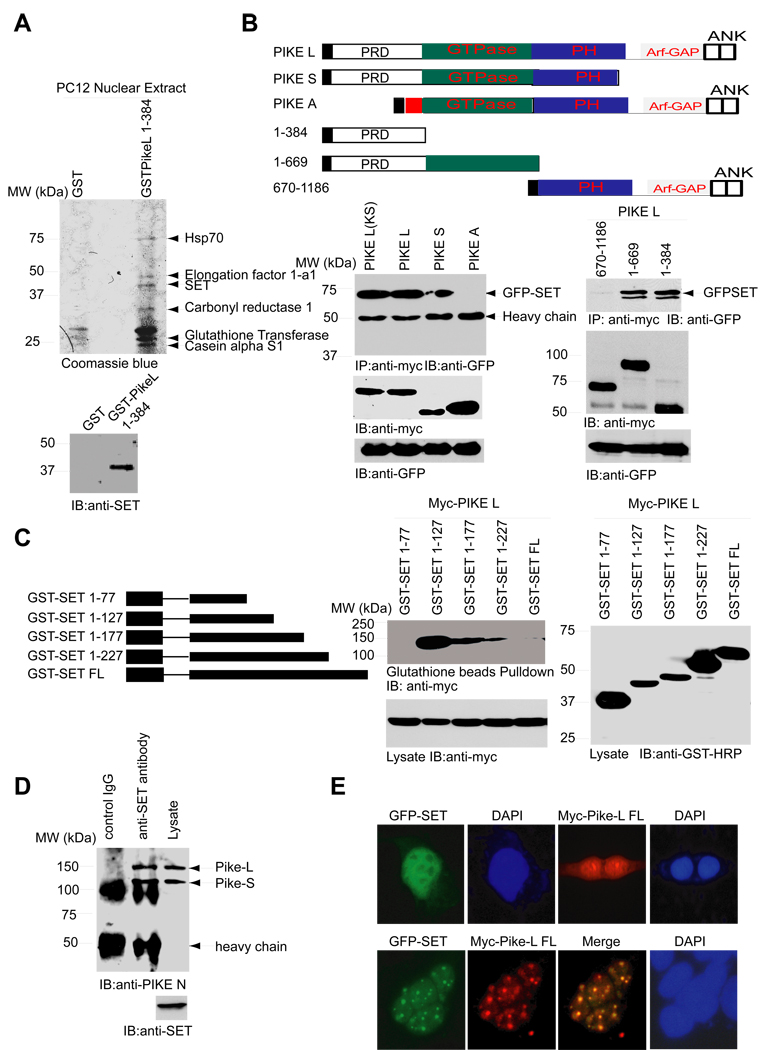
The N-terminus of PIKE-L and –S isoforms (1–384 residues), solely distributing in the nucleus, is required for PI 3-kinase enhancing activity (Ye et al., 2000). To search for its nuclear binding partners, we conducted an in vitro binding assay with nuclear extract of PC12 cells. Compared with GST control, GST-PIKE-1–384 fragment selectively associated with a few targeting proteins. The proteins selectively interacted with the N-terminus of PIKE were characterized by proteomic analysis (Figure 1A). Coimmunoprecipitation demonstrated that both PIKE-S and -L but not PIKE-A interacted with SET. A GTPase deficient form of PIKE-L, PIKE-L-KS, also bound SET, indicating that the association is GTPase independent. Truncation assay revealed that the N-terminal 1–384 region was sufficient and necessary for the binding (Figure 1B). Systematic truncation of the C-terminus of SET elicited progressive association with PIKE-L. The strongest binding occurred to the fragment of 1–127. However, the N-terminal 1–77 region failed to bind PIKE-L (Figure 1C), suggesting that the C-terminus of SET somehow inhibits the complex formation and 77–127 region is required for the interaction. In cortical neurons, SET antibody but not control IgG selectively pulled down both PIKE-L and –S (Figure 1D). Transfected GFP-SET or PIKE-L alone predominantly distributed in the nucleus and diffused throughout the whole nucleoplasm without obvious concentration within a discrete subnuclear organelle. Strikingly, cotransfected SET and PIKE-L precisely colocalized and revealed spotty subnuclear structures in the nucleus (Figure 1E). Immunofluorescent staining with NPM/B23 and SC35, the specific marker for the nucleolus and the nuclear speckle, respectively, revealed that GFP-SET did not reside in either of the subnuclear organelles (data not shown). Thus, these results support that PIKE-L binds SET in vitro and in vivo.
Figure 1. SET binds PIKE L in vitro and in vivo.
(A). Identification of SET. PC12 nuclear extract was incubated with GST-PIKE (1–384 a.a.) and GST control, respectively. The elutes of 1 M NaCl from both columns were analyzed by Coomassie blue staining and proteomics analysis. The proteins binding to GST-PIKE but not control GST were labeled. The 39 kDa band selectively binding to PIKE N-terminus is SET β. (B). PIKE-L and -S but not PIKE A bind to SET. HEK293 cells were cotransfected with GFP-SET and N-terminal tagged myc-PIKE isoforms or PIKE-L fragments. The transfected proteins were immunoprecipitated and analyzed by immunoblotting. PIKE-L and –S but not PIKE-A bound to SET, and the N-terminus of PIKE-L was essential for binding the phosphorylated SET doublet (lower panels). The diagram of different PIKE isoforms (upper panel). Red square stands for PIKE-A specific 1–72 residues. N-terminal blank rectangles stand for PIKE-L and –S Proline-rich domains (PRD). Black rectangles represent Myc tag. (C). The Middle region of SET binds PIKE L. HEK293 cells were cotransfected with myc-PIKE-L and various GST-SET fragments. SET fragments were pulled down with glutathione beads, and the associated proteins were analyzed with myc antibody. (D). SET binds PIKE-L in vivo. Lysates of cortical neurons were immunoprecipitated with anti-SET antibody or Rabbit IgG. The associated proteins were analyzed with anti-PIKE antibody. (E). SET colocalizes with PIKE-L in the subnuclear spotty structures of the cotransfected nucleus. Transfected SET or PIKE-L alone mainly occurred in the nucleus (upper panel), whereas cotranfection of SET and PIKE-L provoked colocalization with spotty subnuclear structures (lower panel).
SET can be cleaved in acidic cytosolic extract
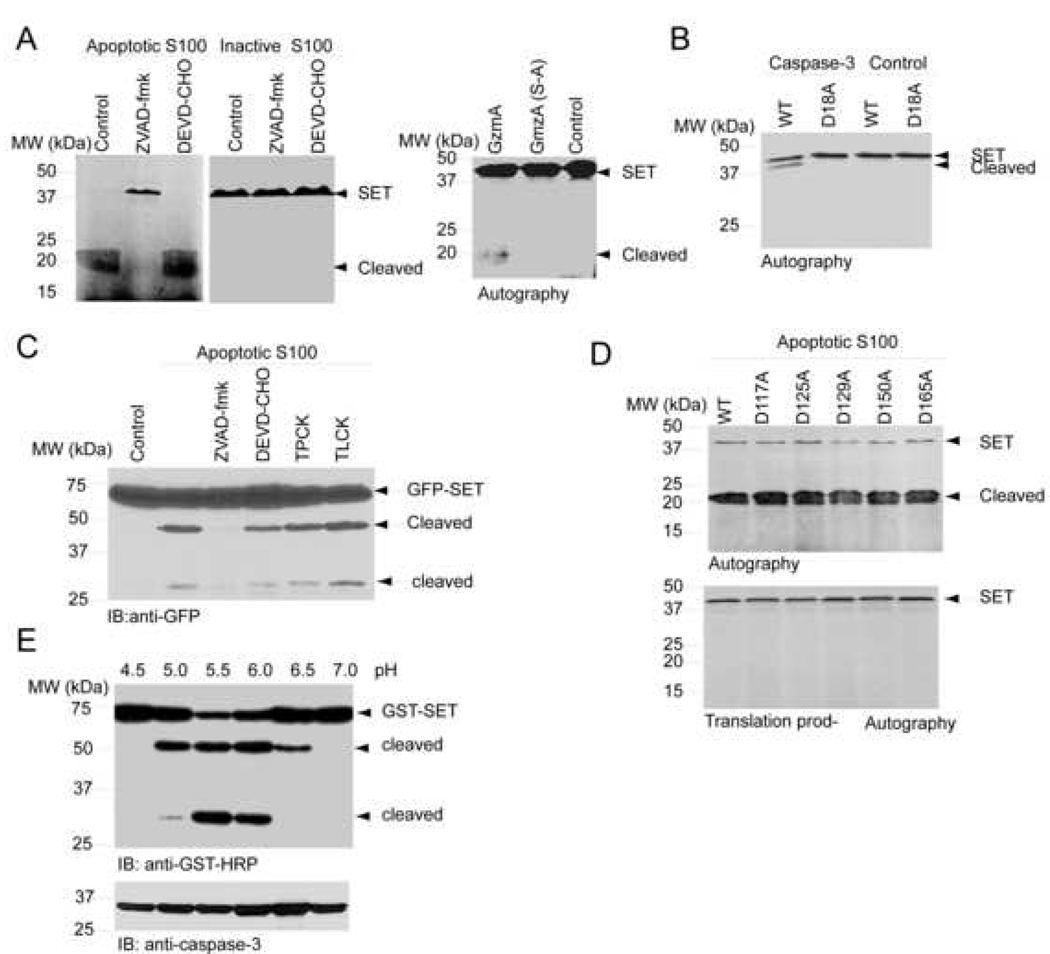
The cytotoxic T lymphocyte protease granzyme A (GzmA) initiates a novel caspase-independent cell death pathway characterized by single-stranded DNA nicking. GzmA cleaves the nucleosome assembly protein SET after Lys176 and disrupts its nucleosome assembly activity (Beresford et al., 2001). To explore whether cells other than T lymphocytes also contain protease activity against SET, we employed HEK293 cells. We conducted the cleavage assay with the cell-free apoptotic solution (S-100, pH at 7.5), supplemented with cytochrome c and dATP (Liu et al., 1996). The active apoptotic S-100 solution cleaved SET and yielded a smeared band at about 20 kDa. The pan-caspase inhibitor ZVAD-fmk but not the caspase-3 specific inhibitor DEVD-CHO completely blocked the proteolytic degradation of SET (Figure 2A, left panel). As a positive control, purified His-tagged GzmA but not protease inactive GzmA (S184A) recombinant protein robustly cleaved SET in buffer at pH 7.5 (Figure 2A, right panel). Thus, SET can be cleaved in HEK293 cells by proteases, related to ZVAD-fmk-sensitive caspases.
Figure 2. SET can be cleaved in acidic cytosolic solution.
(A). SET is cleaved in the cell-free apoptotic solution. The active or inactive cell-free apoptotic S-100 was respectively preincubated with 10 µM ZVAD-fmk and DEVD-CHO for 30 min, followed by addition of 35S-methionine-labeled SET. The reaction mixture was incubated at 37°C for another 2 h. The reactions were analyzed by SDS-PAGE and visualized by autography (left and middle panel). 35S-methionine-labeled SET was also incubated with 125 nM purified active GzmA and inactive GzmA(S184A) in 1 mM CaCl2, 1 mM MgCl2, and 50 mM Tris-HCl, pH 7.5 at 30 °C for 2 h (right panel). (B). D18 is the cleavage site of SET by caspase-3 in apoptotic solution. (C). Cleavage of SET was blocked by ZVAD-fmk in S-100. GFP-SET was incubated with the active apoptotic S100, which was preincubated with various inhibitors: ZVAD-fmk, DEVD-CHO, TPCK (0.1 mM) and TLCK (0.1 mM). The reactions were analyzed with anti-GFP antibody. (D). D into A mutation in the middle region fails to prevent SET from fragmentation in activated S-100. A variety of 35S-methionine-labeled SET mutants were incubated with activated S-100 at 37°C for 2 h. The reactions were analyzed by SDS-PAGE and visualized by autography. (E). SET is cleaved under acidic condition. GST-SET was incubated with S-100 at different pH values as indicated for 30 min at 37°C. The reactions were analyzed with GST-HRP. SET was markedly cleaved at pH 5.0 and 6.5 but not at pH 7.0 (upper panel). Caspase-3 was inactivated (lower panel).
Previous study shows that SET can be only cleaved at D18 in apoptotic cells (Morita et al., 2000). To confirm that D18 is the only apoptotic caspase cleavage site in SET, we radiolabeled wild-type and D18A SET with 35S-methionine and conducted in vitro cleavage assays. SET was actively cleaved by caspse-3, which was completely blocked by D18A mutation (Figure 2B), underscoring that D18 is the caspase cleavage site. Preincubation of the apoptotic S-100 solution with the pan-caspase inhibitor ZVAD-fmk completely blocked recombinant GFP-SET degradation; in contrast, the caspase-3 specific inhibitor DEVD-CHO or chymotrypsin inhibitors TPCK and TLCK failed to prevent SET proteolytic cleavage (Figure 2C). These results suggest that SET might be cut by proteinases other than chymotrypsin. To exclude the possibility that the 50 kDa GFP containing product is directly generated by caspases, we mutated individual aspartic acids in the middle region of SET into alanine. Wild-type and all mutants were robustly cleaved and yielded strong smear bands at 20–22 kDa (Figure 2D). Other point mutants with D into A outside the middle region exhibited similar results (data not shown), indicating that caspases are not implicated in this action, and unknown proteinases that are sensitive to ZVAD-fmk account for this activity. To explore whether the proteinases are regulated by pH, we prepared cytosolic solution from HEK293 cells at various pH values. Surprisingly, in the absence of cytochrome c or dATP, GST-SET recombinant protein displayed a pH-dependent cleavage with the strongest fragmentation occurred at pH 5.5. The cleavage activity decreased from pH 6.0 to 6.5. No degradation was detected at pH 7.0. The N-terminal tagged GST-SET yielded two fragments with molecular weights at 50 and 30 kDa, respectively (Figure 2E, upper panel). Interestingly, the 30 kDa fragment did not occur at pH 6.5, suggesting that the N-terminus of SET is not cleaved under these conditions. Caspase-3 was not processed to the mature form under different pH conditions (Figure 2E, lower panel). Other caspases including caspase-9 and 7 were not activated either (data not shown), suggesting that SET can be digested by proteinases other than caspases. The different pH buffers alone did not cleave SET (data not shown). Taken together, these data demonstrate that SET can be cut by proteinases, which are activated under acidic conditions.
N175 is the cleavage site in SET
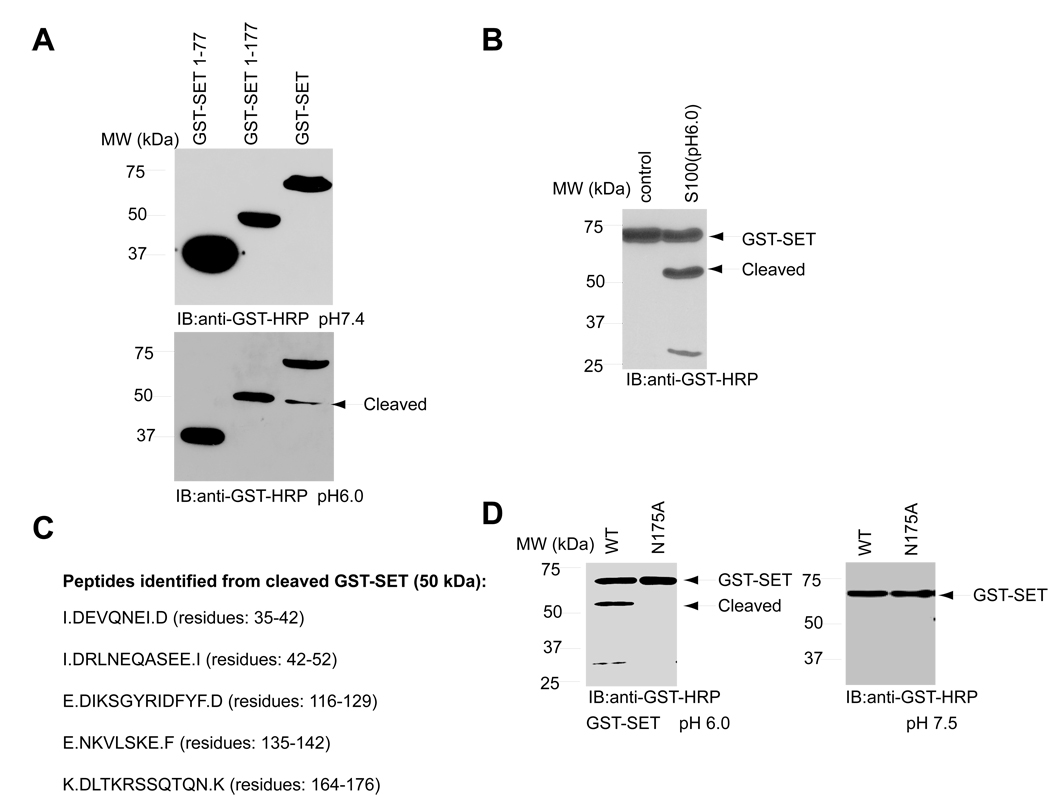
To locate the cleavage site in SET, we conducted a mapping assay with a variety of SET truncated recombinant proteins. Full-length SET produced a fragment with the same size as the 1–177 form (Figure 3A), indicating that SET is cut around position 177. Granzyme A, a specific tryptase, cleaves SET at K176 in K562 cells (Beresford et al., 2001). To explore whether any trypsin like enzymatic activity occurs in the acidic cytosolic solution, we mutated individual K or R residue around 177 region into A. Wild-type SET and its mutants were actively cleaved in the cytosolic solution at pH 6.0 but not at pH 7.5 (Supplemental Figure 1A), suggesting that none of the listed residues is the cleavage site. To search for the cleavage site in the middle region of SET, we purified a large amount of recombinant GST-SET protein and cut it with acidic cytosolic extract (pH at 6.0) (Figure 3B). The 50 kDa fragment was recovered and subjected to proteomic analysis. Numerous SET peptides, including a peptide with one Asn (N) cleavage end (K. DLTKRSSQTQN. K), which corresponds to amino acids 165–175, were identified as the potential C-terminal peptide (Figure 3C), with the MS/MS spectrum shown (Supplemental Figure 1B). To verify that N175 is the cleavage site, we mutated it into alanine. At pH 7.5, both wild-type SET and N175A mutant remained intact in the cytosolic extract. In contrast, at pH 6.0, wild-type SET was cleaved, whereas SET N175A remained uncleaved (Figure 3D), supporting that SET is selectively cut at N175 under acidic conditions.
Figure 3. Identification of N175 is cleavage site of SET in acidic condition.
(A). SET is cleaved around S177. GST-SET fragments were incubated with S-100 and analyzed by immunoblotting. (B). SET is cleaved in S-100 at pH 6.0. (C). Detection of the C-terminal peptide of cleaved SET protein by LC-MS/MS. The detected peptide sequences were listed. DLTKRSSQTQN was identified as the potential C-terminal peptide. (D). N175 mutant blocks SET cleavage. HEK293 cells were transfected with GST-SET wide-type or N175A mutant and lysed in pH 7.5 or 6.0 buffer and analyzed by immunoblotting with GST-HRP.
AEP cleaves SET in vitro and in vivo
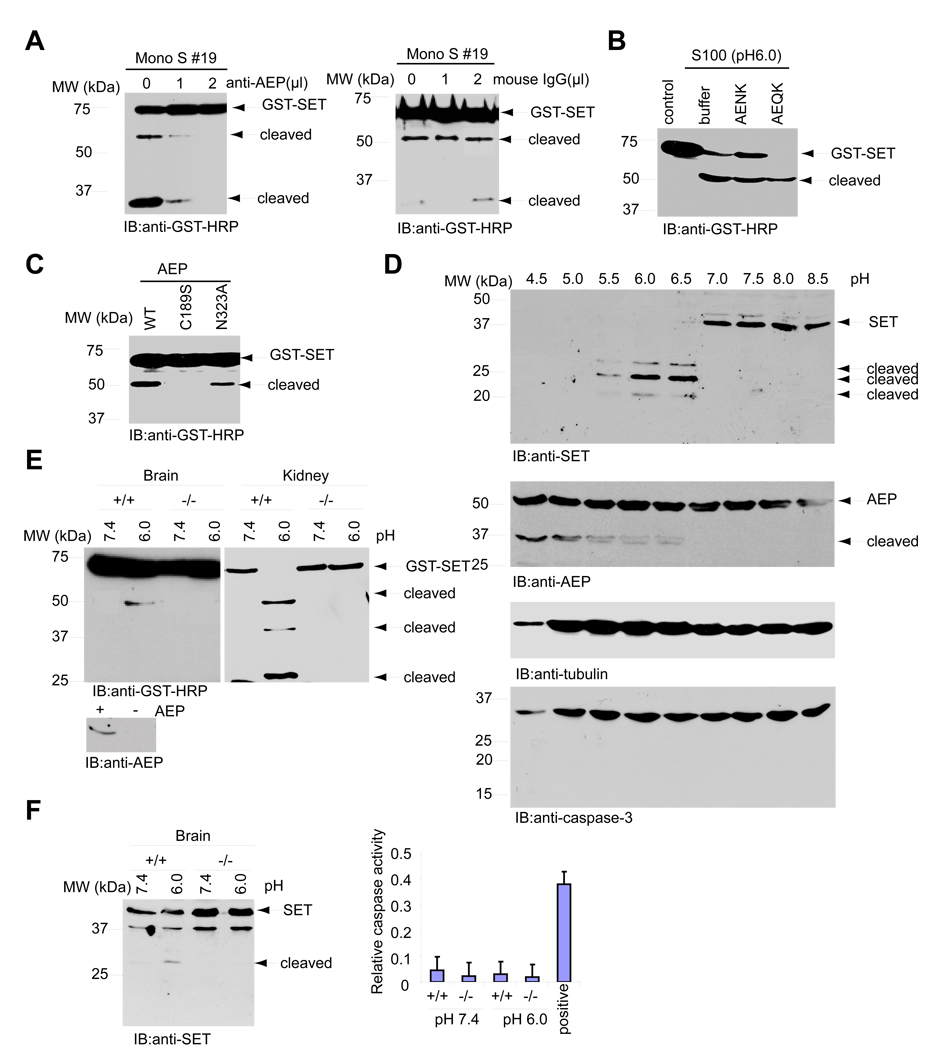
Biochemical purification revealed that AEP is the protease responsible for SET cleavage (Supplemental Figure 2). To confirm that AEP triggers SET proteolytic degradation, we conducted cleavage assay with the fraction 19 from Mono S column. Mouse monoclonal anti-AEP but not control mouse IgG selectively blocked the protease activity in a dose-dependent manner (Figure 4A). A previous study shows that AEP can be partially inhibited by AENK but not AEQK peptide (Loak et al., 2003). Cleavage of SET was selectively blocked by the competitive inhibitor AENK but not by the inactive control peptide AEQK (Figure 4B), suggesting that AEP is the protease exerting the enzymatic activity. The inactive 56-kDa human prolegumain is progressively converted to active 36-kDa AEP by sequential autocatalytic cleavage at N323 followed by D25, which is regulated by different pH thresholds. C189 is essential for its proteinase activity, and N323 cleavage also plays important role for the maturation of AEP (Li et al., 2003). As expected, wild-type AEP strongly cut SET and the activity was reduced with N323A mutant. The C189S mutant failed to degrade SET (Figure 4C), supporting the notion that SET is a substrate of AEP. To determine whether endogenous SET cleavage couples to AEP activation, we lysed HEK293 cells in buffers with different pH. At pH 4.5 and 5.0, SET was completely degraded. At pH 5.5 to 6.5, three cleaved SET fragments with different molecular weights occurred with 25 kDa fragment the strongest. Nevertheless, SET remained intact at pH 7.0 or above. Interestingly, caspase-3 was inactive when SET and AEP were actively cleaved, suggesting that SET and AEP cleavage is independent of caspase-3 activation. By contrast, AEP was strongly cleaved at low pH. Its activation decreased from pH 6.0 to 6.5. AEP remained inactive at pH 7.0 or above (Figure 4D). To further confirm that AEP is responsible for SET cleavage, we utilized the AEP-deficient mice (Shirahama-Noda et al., 2003). GST-SET remained intact in kidney lysates at pH 7.4 irrespective of wild-type or AEP knockout; by contrast, it was robustly cleaved into three fragments in wild-type but not AEP lacking lysates at pH 6.0. Similar cleavage activity occurred in brain lysates; however, only one cut fragment was detected (Figure 4E). The discrepancy might be due to the fact that kidney possesses much stronger AEP proteinase activity than brain (Chen et al., 1997). Endogenous SET displayed the similar cleavage pattern in brain, with degradation selectively associated with pH 6.0 condition. Caspase-3 was not activated under above conditions (Figure 4F). Collectively, these data demonstrate that SET is a physiological substrate of AEP in brain.
Figure 4. AEP cleaves SET in vitro and in vivo.
(A). AEP antibody diminishes the cleavage of SET. Fraction #19 (20 µl) was incubated with or without indicated amount of AEP antibody or mouse IgG at 4°C for 2 h. The reactions were centrifuged at 1 kg for 5 min at 4°C. The supernatants were incubated with GST-SET and analyzed with GST-HRP. (B). The proteolysis of SET was blocked by AENK peptide. GST-SET was incubated with S-100 at pH 6.0 with 0.1 mM AENK and AEQK peptides for 30 min at 37°C. Peptide AENK but not control peptide AEQK blocked SET cleavage. (C). C189S and N323A mutants of AEP diminish SET cleavage. GST-SET was incubated with purified AEP recombinant proteins in pH 6.0 buffer for 2 h at 37° and analyzed by immunoblotting. (D). SET cleavage couples to AEP activity. HEK293 cells were lysed in different pH buffers as indicated and immunoblotted against SET, AEP and tubulin antibodies. (E). AEP is the enzyme that cleaves SET. Kidney or brain from AEP +/+ and −/− mice were lysed in pH 7.4 or pH 6.0 buffer and incubated with GST-SET for 30 min at 37°C. The reactions were analyzed by immunoblotting. (F). SET is cleaved in pH 6.0 brain lysates from wild-type mice. The brain from AEP +/+ and −/− mice were lysed in pH 7.4 or 6.0 buffer and analyzed by immunoblotting. Endogenous SET was cleaved in pH 6.0 AEP +/+ but not −/− lysates (left panel). Caspase-3 was not activated (right panel).
PIKE-L prevents SET cleavage and DNA nicking
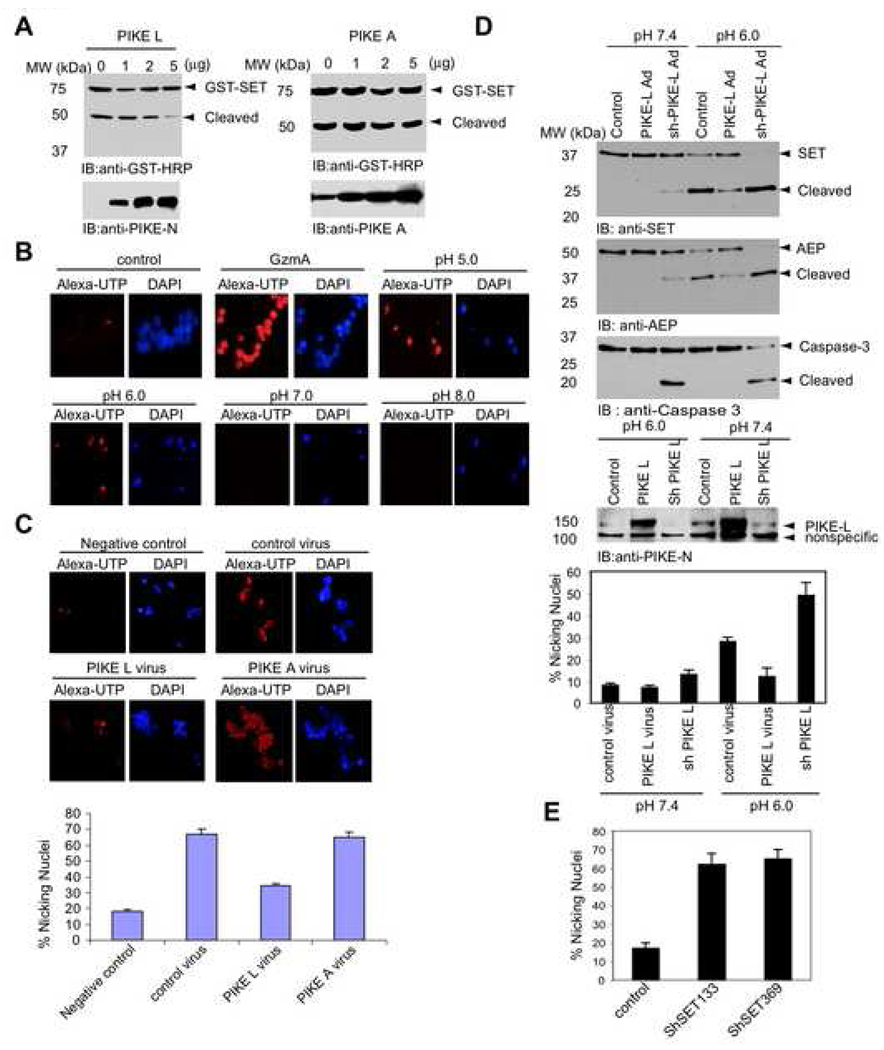
PIKE binds SET in vitro and in vivo. To examine whether PIKE-L protects SET from proteolytic degradation, we cotransfected GST-SET into HEK293 cells with different amount of PIKE-L. The transfected cells were lysed at pH 6.0. SET cleavage was gradually inhibited by the progressive increase of PIKE-L. In contrast, PIKE-L itself remained uncleaved. However, PIKE-A failed to protect SET, fitting with its inability to interact with SET (Figure 5A). Degradation of SET releases DNase NM23-H1, which triggers DNA nicking (Fan et al., 2003). To evaluate whether pH regulates DNA nicking through destruction of SET, we lysed HEK293 cells in buffers with different pH. The cellular extract was incubated with the isolated control nuclei in the presence of Texas-red-dATP and klenow DNA polymerase. As a negative control, the nuclei were barely labeled with control buffer (pH 7.4). Notably, purified granzyme A, the positive control, triggered promiscuous nicking of DNA. Marked DNA nicking also occurred to cellular extracts with low pH (5.0 and 6.0). By contrast, no DNA nicking was observed with pH 7.0 or 8.0 sample, fitting with the SET cleavage pattern (Figure 5B). These results demonstrated that SET cleavage correlated with DNA nicking mediated by NM23-H1. To explore whether PIKE-L blocks DNA nicking by protecting SET from degradation, we infected HeLa cells with various adenovirus expressing PIKE-L or PIKE-A, and prepared cellular extracts at pH 6.0. Approximately 70% nuclei were nicked in control virus-infected cellular extract. DNA nicking was evidently decreased when PIKE-L but not PIKE-A was overexpressed (Figure 5C), coupling with the observation that PIKE-L but not PIKE-A protected SET from proteolytic degradation. Quantitative data are summarized in the bottom panel of Figure 5C.
Figure 5. PIKE-L prevents SET cleavage and DNA nicking.
(A). PIKE-L protects SET from cleavage. HEK293 cells were transfected with GST-SET and different amount of PIKE-L or PIKE-A construct as indicated. In 24 h, the cells were lysed in pH 6.0 buffer and analyzed with anti-GST-HRP. Overexpression of PIKE-L but not PIKE-A protected SET cleavage at pH 6.0. (B). In situ DNA nicking assay. DNA nicking occurred under acidic condition. (C). PIKE-L but not PIKE-A prevents DNA nicking. HeLa cells were infected with control adenovirus or adenovirus expressing PIKE-L or PIKE-A for 24 h and lysed in buffer (pH 6.0). The lysates were incubated with HeLa nuclei for 30 min at 37°C. The nuclei were labeled, fixed and analyzed quantitatively. (D). PIKE-L protects SET and AEP cleavage under pH 6.0 in cortical neurons. Primary cortical neurons were infected with PIKE-L and shRNA adenovirus or control adenovirus. In 24 h, the infected neurons were lysed in pH 7.4 or 6.0 lysis buffer. Quantitative DNA nicking analysis (mean ± SD)(bottom panels). (E). Knock-down of SET by its sh-RNAs provokes DNA nicking. Cortical neurons were infected with control adenovirus or adenovirus expressing sh-SET133 or sh-SET369 for 24 h. The isolated nuclei were labeled, fixed and analyzed quantitatively. The results were expressed as means ± S.D. calculated from three independent experiments.
To investigate whether PIKE-L protects SET in primary neuronal preparations, we infected cortical neurons with adenovirus expressing PIKE-L, PIKE-L shRNA or scrambled control shRNA. Compared to control shRNA, PIKE-L was selectively eliminated by its shRNA (Figure 5D, 4th panel). AEP, SET and caspase-3 remained intact in control shRNA infected neurons and PIKE-L overexpressed samples at pH 7.4. Surprisingly, ablation of PIKE-L elicited demonstrable AEP, SET and caspase-3 degradation, indicating that PIKE-L is essential for maintaining neuronal survival. At pH 6.0, both AEP and SET were robustly cleaved in control neurons. PIKE-L overexpression evidently decreased the proteolytic cleavage, suggesting that PIKE-L somehow protects AEP activation. Depletion of PIKE-L led to complete AEP and SET cleavage, underscoring that PIKE-L plays a critical role in suppressing AEP and SET cleavage. By contrast, caspase-3 exhibited the similar pattern at pH 7.4 and 6.0, further supporting that caspase-3 is not implicated in AEP or SET degradation (Figure 5D, top, 2nd and 3rd panels). DNA nicking tightly coupled to the SET cleavage (Figure 5D, bottom panel). Depletion of SET by its shRNA elicited marked DNA nicking compared to the control (Figure 5E), supporting the notion that SET is critical for preventing DNA nicking in neurons. Thus, PIKE-L selectively protects SET from proteolytic cleavage by AEP, thereby preventing nicking of DNA.
AEP is required for kainic acid-induced SET cleavage and DNA nicking
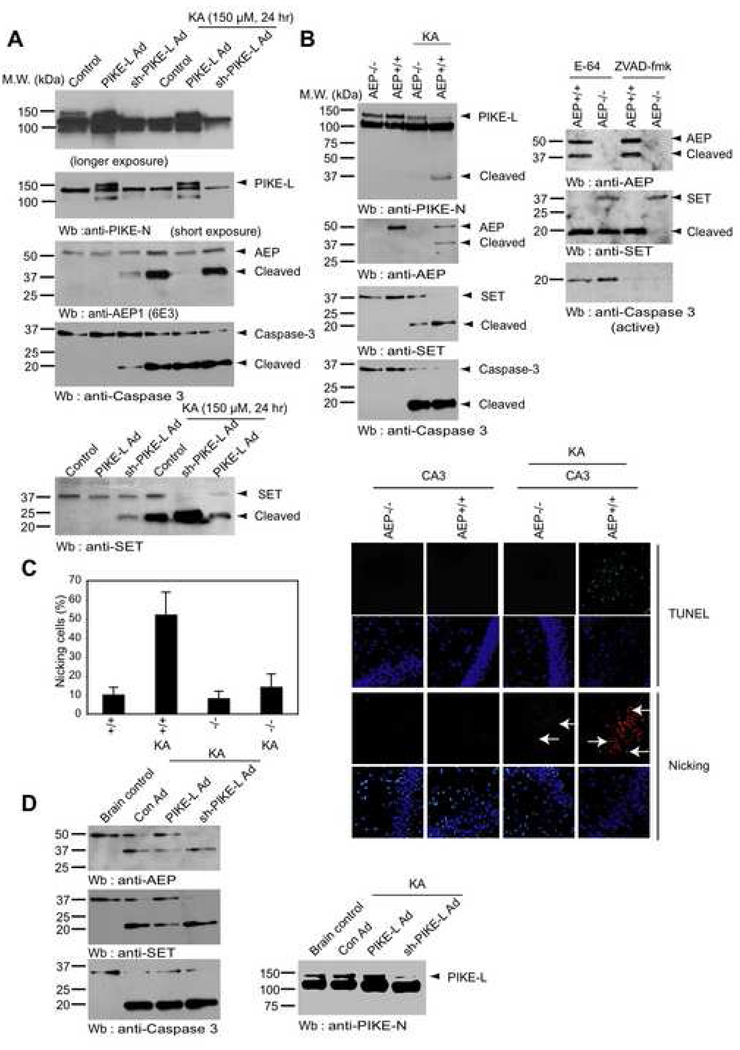
The interstitial milieu of the brain is buffered to pH of 7.3. Pathophysiological processes including kainic acid (KA)-triggered seizures produce global and prolonged acidification of interstitial pH (Ihle and Patneau, 2000). KA also induces substantial intracellular acidification in hippocampal neurons and glia cells (Deitmer and Schneider, 1997). To explore whether KA provokes AEP activation and leads to DNA nicking in neurons, we prepared primary neurons and infected with adenovirus expressing PIKE-L or its shRNA. The infected neurons were treated with 150 µM KA for 24 h. Compared to control shRNA, PIKE-L was selectively depleted by its shRNA (Figure 6A, top and 2nd panels). Ablation of PIKE-L elicited AEP activation and SET cleavage under control condition. KA triggered potent activation of AEP in control and PIKE-L-depleted neurons. By contrast, AEP activation was markedly blocked when PIKE-L was overexpressed. SET cleavage pattern tightly correlated with AEP activation. Notably, KA induced SET complete degradation in PIKE-L depleted neurons, which was substantially decreased in PIKE-L overexpressed samples, underscoring that KA-elicited SET cleavage can be inhibited by PIKE-L. Compared to control, KA also provoked caspase-3 activation in all samples. Interestingly, ablation of PIKE-L alone incurred caspase-3 activation as well (Figure 6A). It is worth noting that both AEP and SET fragments were substantially increased in control and PIKE-L-depleted samples after KA treatment. Conceivably, KA upregulated both AEP and SET protein synthesis, when PIKE-L protein level was low. To determine whether AEP is required for KA-induced SET cleavage, we intraperitoneally injected wild-type and AEP-null mice with 25 mg/kg KA and monitored SET degradation and nicking of DNA in 4 days. KA provoked robust AEP cleavage in wild-type mice; by contrast, it remained intact in control mice. PIKE-L exhibited the similar proteolytic pattern (Figure 6B, left top and 2nd panels). Surprisingly, SET was completely degraded in +/+ mice and weakly cleaved in AEP −/− mice, suggesting that KA triggers the activation of some unknown proteinases in AEP-null mice. The residual SET cleavage activity is neither pH sensitive nor DTT sensitive, indicating that cathepsins are not implicated in this action (data not shown). ZVAD-fmk but not E-64, a lysosomal cathepsin inhibitor, blocked SET cleavage in AEP −/− mice by KA (Figure 6B, right panels), supporting that caspase activation by KA is responsible for the residual SET degradation in AEP-null mice. Notably, KA triggered robust caspase-3 activation in both wild-type and AEP-null mice, which was blocked by ZVAD-fmk (Figure 6B). Consequently, prominent DNA nicking occurred in the hippocampus CA3 region of KA-treated +/+ mice, though modest DNA nicking was also detected in AEP −/− mice, suggesting that AEP plays a major role in KA-induced neuronal injury. TUNEL staining revealed that demonstrable apoptosis occurred in KA-treated wild-type AEP mice, which was not detected in AEP-null mice despite of potent caspase-3 activation (Figure 6C). To further explore whether PIKE-L protects AEP and SET from proteolytic degradation in vivo, we injected adenovirus expressing PIKE-L or its shRNA into the hippocampus of mouse brain, followed by KA treatment. Compared to control, KA provoked evident AEP and SET cleavage in control adenovirus infected mouse brains, which were markedly suppressed in PIKE-L adenovirus injected mouse brain. Infection of PIKE-L shRNA adenovirus substantially enhanced AEP and SET cleavage. Caspase-3 displayed the similar cleavage pattern as SET (Figure 6D), underscoring PIKE-L played an essential role in repressing AEP, SET and caspase-3 proteolytic degradation in response to KA.
Figure 6. AEP is required for kainic acid-induced SET cleavage and DNA nicking.
(A). PIKE-L prevents KA-elicited SET cleavage in cortical neurons. Primarily cultured cortical neurons were infected with control adenovirus and adenovirus expressing PIKE-L or its sh-RNA. In 24 h, the infected neurons were treated with 150 µM KA for 24 h. Verification of PIKE-L expression in infected cortical neurons (top and 2nd panels). Depletion of PIKE-L provoked AEP degradation, and KA treatment further increased AEP cleavage. Compared to control, KA treatment strongly provoked AEP cleavage (3rd panel). Ablation of PIKE-L also elicited caspase-3 and SET degradation even without KA treatment. KA induced demonstrable caspase-3 activation in all samples. SET cleavage in control neurons was decreased when PIKE-L was overexpressed. SET was completely degraded when PIKE-L was depleted (4th and bottom panel). (B & C). AEP is required for KA-triggered DNA damage in mouse brain. Wild-type and AEP −/− mice were injected with 25 mg/kg KA. In 4 days, brain lysates were monitored by immunoblotting and the brain slides were analyzed with in situ DNA nicking (white arrows) and TUNEL assays. Inhibition of caspases by ZVAD-fmk but not cathepsins by E-64 blocked the residual SET degradation by KA in AEP−/− mice (6B, right panels). The quantitative results were expressed as means ± S.D. calculated from three independent experiments. (D) Overexpressing PIKE-L in mouse brain suppresses AEP/SET cleavage, whereas depletion of PIKE-L increases AEP/SET cleavage. Mice brains were injected with adenovirus expressing PIKE-L or its shRNA. In 3 days, the mice were injected with 25 mg/kg KA. In 4 days, the mice brains were analyzed by immunoblotting.
AEP mediates stroke-provoked SET degradation and DNA damage
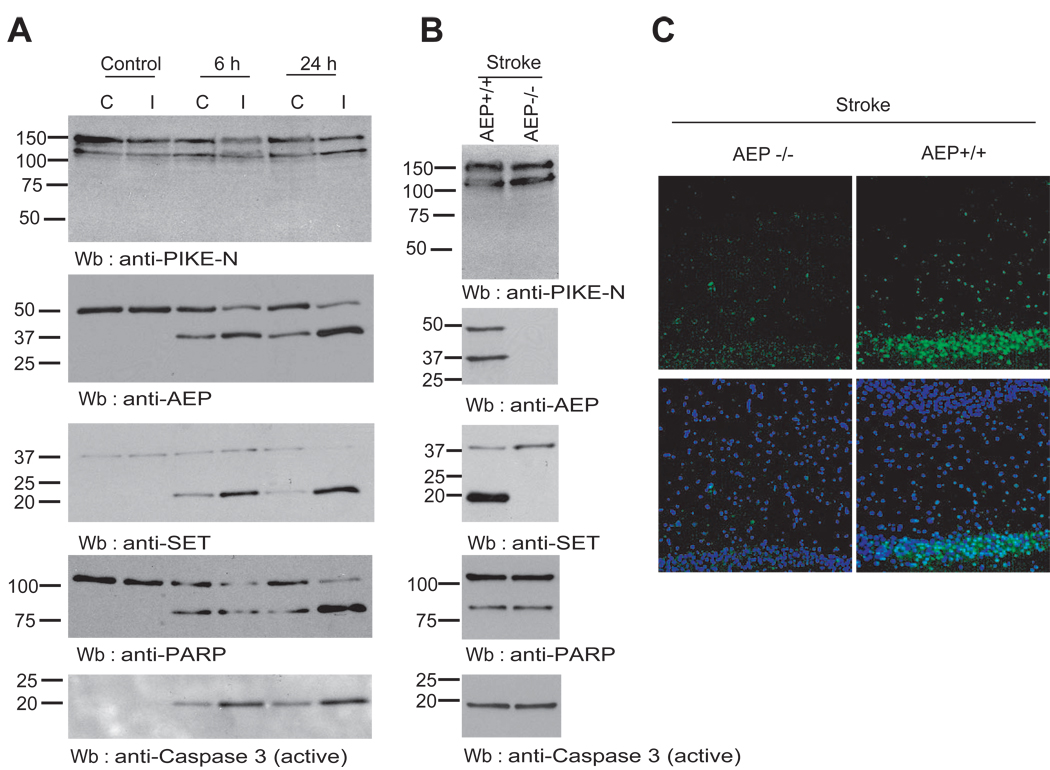
Stroke elicits acidosis in brain (Back et al., 2000). To investigate whether AEP is implicated in this process, we conducted stroke experiments using a transient middle cerebral artery occlusion (MCAO) model. In 48 h, we performed immunoblotting with mouse brain tissues and observed a time dependent AEP activation in ischemia affected regions but not in control tissues. Interestingly, the surrounding tissues next to ischemia core region also revealed weak AEP activation. By contrast, PIKE-L remained intact (Figure 7A, top and 2nd panels). As expected, SET degradation coupled to AEP cleavage activity. Both PARP and caspase-3 displayed much stronger fragmentation in ischemia-attacked lesion area than surrounding regions (3rd to bottom panels). Next, we performed stroke experiments on AEP wild-type and knockout mice and found that SET was selectively cleaved in wild-type but not AEP-null brain; by contrast PARP and caspase-3 were both robustly activated in both wild-type and AEP −/− animals (Figure 7B). TUNEL assay revealed that substantial DNA damage in wild-type but not AEP deficient mice (Figure 7C), underscoring AEP is indispensable for stroke-provoked SET proteolytic degradation and neuronal cell death. Taken together, our results support the hypothesis that PIKE-L binds SET and protects it from AEP, which is activated under acidic conditions, preventing neuronal cell death.
Figure 7. AEP mediates stroke-provoked SET degradation and DNA damage.
(A). Ischemia triggers AEP and SET cleavage and apoptotic activation. Wild-type mice were tested in a transient middle cerebral artery occlusion (MCAO) model. After 0, 6 and 24 h transient MCAO, the ischemia-attacked lesion area (Ipsilateral) and its surrounding regions (Contralateral) were collected and analyzed by immunoblotting. (B). SET is selectively cleaved in wild-type but not AEP–null mice. Immunoblotting analysis of stroke- attacked wild-type and AEP-null brain tissue. Both caspase-3 and PARP were activated by stroke (4th and bottom panels), whereas SET cleavage selectively occurred in wild-type but not AEP −/− mice (2nd and 3rd panels). (C) Stroke-triggered DNA damage is blocked in AEP knockout mice. TUNEL staining was conducted on both wild-type and AEP-null mice brains provoked by MCAO stroke model.
Discussion
In the present study, we identified AEP as a novel proteinase for SET in mouse brain. The inactive zymogen AEP was activated under acidic conditions, KA treatment or stroke. The mature AEP robustly cleaved SET after asparagine 175, resulting in activation and nuclear translocation of DNase NM23-H1, which triggers DNA nicking. Moreover, PIKE-L selectively bound SET and protected it from degradation by AEP, inhibiting DNA damage, fitting with the well-documented pro-survival functions of PIKE (Ahn et al., 2004a; Rong et al., 2003). In confirmation of AEP is the physiological proteinase cleaving SET in non-immune system, we provided compelling evidence that AEP played a major role in mediating SET fragmentation. Following SET proteolytic activity, we purified AEP from HEK293 cellular extract (pH 6.0) via a variety of biochemical columns (Supplemental Figure 2). Employing AEP deficient mice, we established that AEP is indispensable for SET cleavage under neuroexcitotoxicity or stroke.
In primary cortical neurons, KA induced AEP activation. Accordingly, SET exhibited the similar cleavage patterns. Interestingly, PIKE-L also mediated AEP proteolytic cleavage in addition to SET. The similar results were observed with PIKE-L adenovirus-injected mouse (Figure 6). It remains unknown exactly how PIKE-L protects AEP from maturation. It is also noteworthy that depletion of PIKE-L alone triggered caspase-3 activation, indicating that PIKE-L is essential for protecting neurons from apoptosis, fitting with our previous report (Rong et al., 2003). Moreover, we showed that both AEP and SET were selectively activated in KA-injected mouse brain. Unexpectedly, SET was also weakly cleaved in AEP −/− mice, suggesting that KA triggered unknown proteinases activation, which provokes SET cleavage that can be blocked by ZVAD-fmk (Figure 6B). In alignment with AEP/SET proteolytic degradation, KA elicited tremendous DNA nicking in +/+ mice; by contrast, DNA nicking was very weak in AEP −/− mice (Figure 6C). These data demonstrate that the intracellular acidosis might be attributed to ignite AEP maturation and consequent DNases NM23-H1 and TREX1 activation and DNA nicking. It remains unclear why TUNEL staining was negative in AEP −/− samples, although caspase-3 was robustly activated by KA in both wild-type and AEP-null brain. Probably, AEP is also required for the activity of the DNases responsible for DNA fragmentation. PIKE-L was selectively cleaved in +/+ but not −/− mice upon KA treatment, although caspase-3 was potently activated in both mice (Figure 6B). These data suggest that AEP somehow mediates PIKE-L proteolytic cleavage triggered by KA. Nevertheless, PIKE-L was intact in stroke-insulted mice, albeit AEP and caspase-3 were activated (Figure 7), suggesting that PIKE-L is not a substrate of either AEP or caspase-3. This finding that PIKE-L dictates AEP/SET cleavage suggests that PIKE-L might not only control apoptotic caspase activation through activating PI 3-kinase/Akt signaling but also monitor other proteinases activation, preventing cell death in apoptosis and necrosis.
AEP is a lysosomal cysteine proteinase belonging to the clan CD and homologous to plant legumain. Like papain-type cysteine proteinases, AEP is synthesized as an inactive zymogen, and processed into a mature form localized in lysosomes, which was confirmed by colocalizing with lysosome marker LAMP-1. Surprisingly, we found that KA substantially increased AEP expression in both the cell body and neurite processes (Supplemental Figure 3). Conceivably, KA-induced mature AEP cuts cytoplasmic and nuclear SET, which can be blocked by PIKE-L.
KA and stroke trigger necrosis and apoptosis in neurons in many regions of the brain via caspase-dependent and -independent mechanisms (Faherty et al., 1999; Glassford et al., 2002; Wang et al., 2005). Here, we showed that neural excitotoxic KA and stroke provoked neuronal damage through cysteine proteinase AEP cleavage of SET at N175, as well as caspases. In addition to AEP, low pH activates diverse proteases. For instance, under low pH, calpain proteases can be activated and cleave cytoskeleton proteins. We strongly believe that AEP is just one of the numerous proteases activated by KA or stroke. However, neuronal cell death was markedly blocked in AEP-deficient mice supports that AEP might be the major proteinase mediating this devastating process (Figure 6 & 7). Certainly, identification of these enzymes will further our understanding how neuroexcitotoxicity provokes neuronal injury. Taken together, our findings provide a molecular mechanism explaining how neural excitotoxic amino acid- or stroke-induced acidosis exerts its cytotoxic effect. Conceivably, inhibitors for AEP might be useful for therapeutic treatment of neurodegenerative diseases triggered by stroke or ischemia.
Experimental Procedures
Cells and Reagents
HEK293, HeLa cells were maintained in DMEM, supplemented with 10% fetal bovine serum (FBS), 2 mg/ml glutamine and 100 units penicillin-streptomycin at 37°C with 5% CO2 atmosphere in a humidified incubator. PC12 cells were maintained in DMEM, supplemented with 5% fetal bovine serum (FBS), 10% horse serum, 2 mg/ml glutamine and 100 units penicillin-streptomycin at 37°C with 5% CO2 atmosphere in a humidified incubator. The purified anti-PIKE-N antibody is against the N-terminus from residues 268 to 384 in PIKE-L. Anti-GFP and anti-SET antibodies were from Santa Cruz Biotech. Glutathione Sepharose 4B was supplied by Pharmacia Biotech. All the tyrosine kinase inhibitors and Anti-myc antibody were from Calbiochem. Protein A/G-conjugated agarose beads were from Sigma. All of beads used for purification were from Amersham Bioscience. C57BL/6J mice were obtained from The Jackson Laboratory. AEP-deficient mice have been described previously (Shirahama-Noda et al., 2003). Animals were bred in a pathogen-free environment in accordance with Emory Medical School guidelines. The sequences of two sh-RNA of human SET are: sh-RNA SET133 F: GATCCCG GGC CGA CGA GAC CTC AGA A GAAGCTTG T TCT GAG GTC TCG TCG GCC TTTTTT; sh-RNA SET369 F: GATCCCG GTC AAC CAT CCA CAA GTG TCT GAAGCTTG AGA CAC TTG TGG ATG GTT GAC TTTTTT
Protein-protein interaction assays
Ten-cm dishes of HEK293 cells were transfected with 10 µg DNA by the calcium phosphate precipitation method. In 48 h, the transfected cells were treated as indicated, collected and washed once in PBS, lysed in 1 ml lysis buffer (50 mM Tris, pH 7.4, 40 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1.5 mM Na3VO4, 50 mM NaF, 10 mM sodium pyrophosphate, 10 mM sodium b-glycerophosphate, 1 mM phenylmethylsulfonyl flouride (PMSF), 5 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A), and was centrifuged for 10 min at 14,000 × g at 4°C. Immune complexes were resolved by SDS-PAGE and analyzed by immunoblotting.
AEP Purification from HEK293 cytosolic fraction
HEK293 cells (20 liters) were cultured by National Cell Culture Center and lysated in buffer A (25 mM sodium citrate, pH 6.0, 25 mM KCl, 5 mM MgCl2, 1 mM mercaptoethanol, 1mM PMSF, plus protein inhibitor cocktail). The lysate (total protein 1.35g) was applied to Q sepharose (Amersham) equilibrated with buffer A and eluated with buffer A containing 0.2 M, 0.5 M or 1M NaCl. The SET cleavage fraction was applied to Heparin column (Amersham) equilibrated with buffer A and eluated with buffer A containing 0.2 M, 0.5 M or 1M NaCl. The SET cleavage fraction was applied to Mono Q column (pharmacia) equilibrated with buffer A and eluated with gradient from buffer A to buffer containing 1M NaCl in 15 ml. The SET cleavage fraction was applied to Mono S column (pharmacia) equilibrated with buffer A and eluated with gradient from buffer A to buffer containing 1M NaCl in 15 ml. The SET cleavage fraction was further purified by glycerol gradient from 30% to 15% glycerol in buffer A.
In Situ Nicking Assay
In situ nicking assay was processed as described (Fan et al., 2003) with minor change. Briefly, the isolated HeLa nuclei were treated for 30 min at 37°C with cell lysates with different pH or lysates from HEK293 cells, transfected with PIKE-L or infected with adenovirus expressing shRNA of SET. The reactions were washed with buffer B (l50mM Tris-HCl (pH 7.9); 50 µg/ml BSA; 5 mM MgCl2; 10 mM mercaptoethanol) and DNA nicking was labeled in 50 µl buffer B plus 2 U Klenow (New England); 100 µM dATP, dCTP, and dGTP; and 16 µM Alexa 488-12-UTP (Molecular Probes). Nuclei were then plated onto poly-Lys-coated slides and fixed and permeabilized using the Fix-and-Perm kit (Caltag Laboratories). After washing with PBS, slides were counterstained with 0.05 µg/ml DAPI (Sigma), washed again, and mounted with ProLong Antifade medium (Molec-ular Probes).
Cell-free apoptotic cleavage assay
The procedures are exactly as described (Liu et al., 1996). Briefly, the pellets of 293 cells were washed once with ice-cold PBS and resuspended in 5 vol of buffer C (20 mM Hepes–KOH [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, 1 mM dithiothreitol [DTT], and 0.1 mM PMSF, supplemented with protease inhibitors cocktail). After sitting on ice for 15 min, the cells were broken by passing 15 times through a G22 needle. After centrifugation in a microcentrifuge for 5 min at 4 °C, the supernatants were further centrifuged at 105 × g for 30 min in an ultracentrifuge (Beckman). The resulting supernatants were used for in vitro apoptosis assay. Cytochrome c and dATP were added into S-100 extract to initiate caspase cascade. After 30 min incubation at 37 °C, GST-SET or GFP-SET were introduced and incubated for 2 h. The reaction mixture was analyzed by immunoblotting analysis with anti-GST-HRP or GFP antibody, respectively. To evaluate AEP and SET cleavage under different pH conditions, we introduced HCl (1 M) into buffer C by dropwise and monitored pH values using pH Benchtop Meter (SymPhony, VWR).
TNT® Quick Coupled Transcription/ Translation Reaction of SET
The reaction components T7 Quick Master Mix (16 µl), [35S]methionine (1 µl), PCR enhancer (1 µl) and plasmid DNA template (1 µg) were assembled in a 1.5 ml microcentrifuge tube with a total volume of 20 µl. The reaction mixture was incubated at 25°C for 90 min. The translation products were determined by SDS-PAGE and autoradiograph. To conduct in vitro apoptotic cleavage, 2µl translation product was incubated with 200 µg (60 µl in volume) active S100 at 37°C for various time. The reaction mixture was then analyzed by SDS-PAGE, then the gel was dried and exposed to the film.
Adenovirus injection into mouse hippocampus
We determined the viral titer by measuring the number of infected HEK293 cells expressing GFP. We adjusted all viral stocks to 1 × 108 plaque-forming units/µl before their use. For injection of adenoviral vectors into the hypothalamus, we anesthetized male C57BL/6J mice aged 8–12 weeks (n = 6–14 per group) with 2.5% avertin. We performed standard surgical procedures to inject adenoviral vectors expressing either PIKE-specific siRNA with GFP or GFP alone using a stereotaxic table (David Kopf Instruments). The experiment was conducted as previously described (Sheng et al., 2006).
MCAO stroke on AEP wild-type and knockout mice
Six AEP-null mice and six wild-type littermates were anesthetized with 4% chloral hydrate. The rectal and masseter muscle temperatures were controlled at 37°C with a homeothermic blanket. Cerebral perfusion (CP) in the distribution of the middle cerebral artery was monitored throughout the surgical procedure with a laser Doppler (Perimed Inc., North Royalton, OH), and only animals with a >80% decrease in CP were included in this study. The middle cerebral artery was exposed and occluded with a 10-0 suture as described (Yepes et al., 2003). In 48 h, MCAO mice were sacrificed and brains were cut onto 5 µm sections and stained with TUNEL assay or DNA nicking assay.
Supplementary Material
Acknowledgement
This work is supported by grants from National Institute of Health RO1, NS045627 to K.Y. and NS-49478 to M.Y. The authors are thankful to the National Cell culture Center in Minneapolis for culturing large amount of HEK293 cells. We thank Dr. Daniel Kalman for critical reading of the manuscript.
Reference
- Adachi Y, Pavlakis GN, Copeland TD. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J Biol Chem. 1994a;269:2258–2262. [PubMed] [Google Scholar]
- Adachi Y, Pavlakis GN, Copeland TD. Identification of in vivo phosphorylation sites of SET, a nuclear phosphoprotein encoded by the translocation breakpoint in acute undifferentiated leukemia. FEBS Lett. 1994b;340:231–235. doi: 10.1016/0014-5793(94)80144-4. [DOI] [PubMed] [Google Scholar]
- Ahn JY, Hu Y, Kroll TG, Allard P, Ye K. PIKE-A is amplified in human cancers and prevents apoptosis by up-regulating Akt. Proc Natl Acad Sci U S A. 2004a;101:6993–6998. doi: 10.1073/pnas.0400921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JY, Rong R, Kroll TG, Van Meir EG, Snyder SH, Ye K. PIKE (phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates Akt activity and mediates cellular invasion. J Biol Chem. 2004b;279:16441–16451. doi: 10.1074/jbc.M312175200. [DOI] [PubMed] [Google Scholar]
- Back T, Hoehn M, Mies G, Busch E, Schmitz B, Kohno K, Hossmann KA. Penumbral tissue alkalosis in focal cerebral ischemia: relationship to energy metabolism, blood flow, and steady potential. Ann Neurol. 2000;47:485–492. [PubMed] [Google Scholar]
- Beresford PJ, Zhang D, Oh DY, Fan Z, Greer EL, Russo ML, Jaju M, Lieberman J. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J Biol Chem. 2001;276:43285–43293. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- Brown CE, Lechner T, Howe L, Workman JL. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, Hewitt E, Watts C, Barrett AJ. Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J Biol Chem. 1997;272:8090–8098. doi: 10.1074/jbc.272.12.8090. [DOI] [PubMed] [Google Scholar]
- Chen JM, Dando PM, Stevens RA, Fortunato M, Barrett AJ. Cloning and expression of mouse legumain, a lysosomal endopeptidase. Biochem J. 1998;335(Pt 1):111–117. doi: 10.1042/bj3350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW. Glutamate receptors and the induction of excitotoxic neuronal death. Prog Brain Res. 1994;100:47–51. doi: 10.1016/s0079-6123(08)60767-0. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Deitmer JW, Schneider HP. Intracellular acidification of the leech giant glial cell evoked by glutamate and aspartate. Glia. 1997;19:111–122. [PubMed] [Google Scholar]
- Faherty CJ, Xanthoudakis S, Smeyne RJ. Caspase-3-dependent neuronal death in the hippocampus following kainic acid treatment. Brain Res Mol Brain Res. 1999;70:159–163. doi: 10.1016/s0169-328x(99)00143-6. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Zhang D, Lieberman J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol Cell Biol. 2002;22:2810–2820. doi: 10.1128/MCB.22.8.2810-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassford A, Lee JE, Xu L, Giffard RG. Caspase inhibitors reduce the apoptotic but not necrotic component of kainate injury in primary murine cortical neuronal cultures. Neurol Res. 2002;24:796–800. doi: 10.1179/016164102101200915. [DOI] [PubMed] [Google Scholar]
- Halfon S, Patel S, Vega F, Zurawski S, Zurawski G. Autocatalytic activation of human legumain at aspartic acid residues. FEBS Lett. 1998;438:114–118. doi: 10.1016/s0014-5793(98)01281-2. [DOI] [PubMed] [Google Scholar]
- Ihle EC, Patneau DK. Modulation of alpha-amino-3-hydroxy-5-methyl-4- isoxazolepropionic acid receptor desensitization by extracellular protons. Mol Pharmacol. 2000;58:1204–1212. doi: 10.1124/mol.58.6.1204. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Li DN, Matthews SP, Antoniou AN, Mazzeo D, Watts C. Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo. J Biol Chem. 2003;278:38980–38990. doi: 10.1074/jbc.M305930200. [DOI] [PubMed] [Google Scholar]
- Li M, Makkinje A, Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996;271:11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Loak K, Li DN, Manoury B, Billson J, Morton F, Hewitt E, Watts C. Novel cell-permeable acyloxymethylketone inhibitors of asparaginyl endopeptidase. Biol Chem. 2003;384:1239–1246. doi: 10.1515/BC.2003.136. [DOI] [PubMed] [Google Scholar]
- Madeira A, Pommet JM, Prochiantz A, Allinquant B. SET protein (TAF1beta, I2PP2A) is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain. Faseb J. 2005;19:1905–1907. doi: 10.1096/fj.05-3839fje. [DOI] [PubMed] [Google Scholar]
- Maehr R, Hang HC, Mintern JD, Kim YM, Cuvillier A, Nishimura M, Yamada K, Shirahama-Noda K, Hara-Nishimura I, Ploegh HL. Asparagine endopeptidase is not essential for class II MHC antigen presentation but is required for processing of cathepsin L in mice. J Immunol. 2005;174:7066–7074. doi: 10.4049/jimmunol.174.11.7066. [DOI] [PubMed] [Google Scholar]
- Manoury B, Hewitt EW, Morrice N, Dando PM, Barrett AJ, Watts C. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396:695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- Morita A, Suzuki N, Matsumoto Y, Hirano K, Enomoto A, Zhu J, Sakai K. p41 as a possible marker for cell death is generated by caspase cleavage of p42/SETbeta in irradiated MOLT-4 cells. Biochem Biophys Res Commun. 2000;278:627–632. doi: 10.1006/bbrc.2000.3860. [DOI] [PubMed] [Google Scholar]
- Moss CX, Matthews SP, Lamont DJ, Watts C. Asparagine deamidation perturbs antigen presentation on class II major histocompatibility complex molecules. J Biol Chem. 2005;280:18498–18503. doi: 10.1074/jbc.M501241200. [DOI] [PubMed] [Google Scholar]
- Neviani P, Santhanam R, Trotta R, Notari M, Blaser BW, Liu S, Mao H, Chang JS, Galietta A, Uttam A, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8:355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Rong R, Ahn JY, Huang H, Nagata E, Kalman D, Kapp JA, Tu J, Worley PF, Snyder SH, Ye K. PI3 kinase enhancer-Homer complex couples mGluRI to PI3 kinase, preventing neuronal apoptosis. Nat Neurosci. 2003;6:1153–1161. doi: 10.1038/nn1134. [DOI] [PubMed] [Google Scholar]
- Schauwecker PE, Steward O. Genetic determinants of susceptibility to excitotoxic cell death: implications for gene targeting approaches. Proc Natl Acad Sci U S A. 1997;94:4103–4108. doi: 10.1073/pnas.94.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Sheng G, Chang GQ, Lin JY, Yu ZX, Fang ZH, Rong J, Lipton SA, Li SH, Tong G, Leibowitz SF, Li XJ. Hypothalamic huntingtin-associated protein 1 as a mediator of feeding behavior. Nat Med. 2006;12:526–533. doi: 10.1038/nm1382. [DOI] [PubMed] [Google Scholar]
- Shirahama-Noda K, Yamamoto A, Sugihara K, Hashimoto N, Asano M, Nishimura M, Hara-Nishimura I. Biosynthetic processing of cathepsins and lysosomal degradation are abolished in asparaginyl endopeptidase-deficient mice. J Biol Chem. 2003;278:33194–33199. doi: 10.1074/jbc.M302742200. [DOI] [PubMed] [Google Scholar]
- von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3' half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GJ, Randall RD, Thayer SA. Glutamate-induced intracellular acidification of cultured hippocampal neurons demonstrates altered energy metabolism resulting from Ca2+ loads. J Neurophysiol. 1994;72:2563–2569. doi: 10.1152/jn.1994.72.6.2563. [DOI] [PubMed] [Google Scholar]
- Wang Q, Yu S, Simonyi A, Sun GY, Sun AY. Kainic acid-mediated excitotoxicity as a model for neurodegeneration. Mol Neurobiol. 2005;31:3–16. doi: 10.1385/MN:31:1-3:003. [DOI] [PubMed] [Google Scholar]
- Ye K, Aghdasi B, Luo HR, Moriarity JL, Wu FY, Hong JJ, Hurt KJ, Bae SS, Suh PG, Snyder SH. Phospholipase C gamma 1 is a physiological guanine nucleotide exchange factor for the nuclear GTPase PIKE. Nature. 2002;415:541–544. doi: 10.1038/415541a. [DOI] [PubMed] [Google Scholar]
- Ye K, Hurt KJ, Wu FY, Fang M, Luo HR, Hong JJ, Blackshaw S, Ferris CD, Snyder SH. Pike. A nuclear gtpase that enhances PI3kinase activity and is regulated by protein 4.1N. Cell. 2000;103:919–930. doi: 10.1016/s0092-8674(00)00195-1. [DOI] [PubMed] [Google Scholar]
- Yepes M, Sandkvist M, Moore EG, Bugge TH, Strickland DK, Lawrence DA. Tissue-type plasminogen activator induces opening of the blood-brain barrier via the LDL receptor-related protein. J Clin Invest. 2003;112:1533–1540. doi: 10.1172/JCI19212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.