Abstract
Fluorescent RNA is an important analytical tool in medical diagnostics, RNA cytochemistry and RNA aptamer development. We have synthesized the fluorescent ribonucleotide analogue 1,3-diaza-2-oxophenothiazine-ribose-5′-triphosphate (tCTP) and tested it as substrate for T7 RNA polymerase in transcription reactions, a convenient route for generating RNA in vitro. When transcribing a guanine, T7 RNA polymerase incorporates tCTP with 2-fold higher catalytic efficiency than CTP and efficiently polymerizes additional NTPs onto the tC. Remarkably, T7 RNA polymerase does not incorporate tCTP with the same ambivalence opposite guanine and adenine with which DNA polymerases incorporate the analogous dtCTP. While several DNA polymerases discriminated against a d(tC-A) base pair only by factors < 10, T7 RNA polymerase discriminates against tC-A base pair formation by factors of 40 and 300 when operating in the elongation and initiation mode, respectively. These catalytic properties make T7 RNA polymerase an ideal tool for synthesizing large fluorescent RNA, as we demonstrated by generating a ~800 nucleotide RNA, in which every cytosine was replaced with tC.
Keywords: in vitro transcription, kinetics, mutagenesis, minor tautomeric forms, base analogue
INTRODUCTION
Several vibrant research fields are centered on the application of fluorescent RNA: fluorescent RNA cytochemistry, RNA aptamer development, and RNA dynamics. The discovery of the green fluorescent protein has enabled the localization and tracking of almost any given protein in a cell, and fluorescent RNA cytochemistry aims to accomplish the same within the RNA world. In RNA cytochemistry, fluorescent RNA is introduced into cells using microinjection or in vivo hybridization of fluorescent nucleic acids to endogenous RNAs and the transit path of the labeled RNA is visualized by fluorescent microscopy.1 Aptamers are short nucleic acids with extensive secondary structure that bind ligands with affinities and specificities comparable to that of antibodies.2 Similar to antibodies, labeled aptamers have been used in diagnostic assays, such as ELISA,3 Western blotting,4 microarrays,5 capillary electrophoresis6 and flow cytometry.7 Fluorescent RNA aptamers can be synthesized directly by solid-phase synthesis if the length does not exceed 80 nucleotides; the fluor is preferentially attached to a terminal position since internal labeling may have negative effects on the aptamer activity. To exclude the problem of dye interference, approaches have been developed to attach an additional RNA domain to the aptamer of interest, such as malachite green, which becomes fluorescent in response to ligand binding.8 Labeling of longer RNA can be accomplished enzymatically by transcription with T7 RNA polymerase (T7 RNAP)9–12 in the presence of fluorescent ribonucleotides or of so-called initiator nucleotides, such as guanosine monophosphothioate.13 The latter results in site-specific modification of the 5′-end, which can be rendered fluorescent via subsequent coupling to a fluorescent dye. Kimoto et al. has presented an elegant method for internal site-specific labeling of RNA that utilizes an unnatural base pair, the fluorescent base analogue 2-amino-6-(2-thienyl)purine, which T7 RNA polymerase incorporates opposite to pyrrole-2-carbaldehyde.14
In this study, we report the site-specific incorporation of 1,3-diaza-2-oxophenothiazine-ribose-5′-triphosphate (tCTP) opposite guanine in transcription reactions with T7 RNAP. Transcription of DNA by T7 RNAP takes place in two phases, initiation and elongation.15–18 During the initiation phase, T7 RNAP binds tightly to the promoter sequence via its N-terminal promoter binding domain, opens the DNA duplex and feeds the template into the active site.19–21 In this mode, RNA synthesis frequently results in abortive products that do not exceed 8–10 bases.18, 22 Subsequently, T7 RNAP releases the promoter sequence, which is achieved by a rigid body rotation of the promoter binding domain,23, 24 and enters the elongation mode.17, 25–27 Transcription becomes highly processive and efficiently produces full length transcripts.
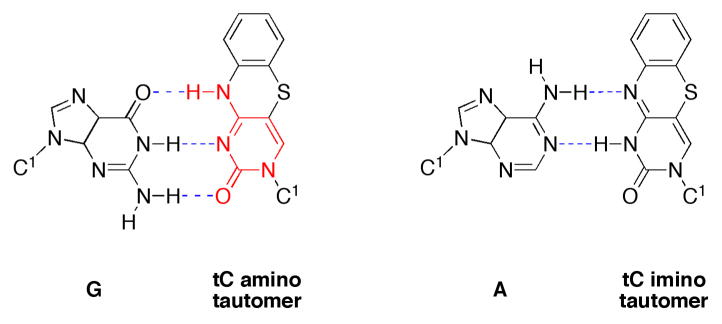
tC is a tricyclic cytosine analogue that absorbs light at 375 nm (ε375 = 4,000 M−1 cm−1) and fluoresces intensely in single- and double-stranded DNA at 505 nm (φF = 0.2).28, 29 Recently, we reported that dtCTP is a good substrate for several A and B family DNA polymerases.30–32 The analogue is incorporated with high catalytic efficiency opposite a template guanine, but also to a significant extent opposite a template adenine. The DNA polymerases efficiently extended a d(tC-G) base pair, whereas d(tC-A) base pairs primarily resulted in chain termination. We attributed the ambivalence of dtCTP incorporation to the propensity of N4-substituted cytosine analogues for forming the imino-tautomer, which is isosteric to thymine (Chart 1). Despite the somewhat mutagenic properties of dtCTP, we were able to produce large fluorescent DNA in polymerase chain reactions (PCR).31 However, the PCR was only efficient when < 70 % of the C’s were replaced with tC.
This article examines the potential of tCTP for labeling of RNA. We determined the catalytic efficiency of tCTP incorporation by T7 RNAP using synthetic DNA templates of defined sequence, which contained a unique G or A either within the initiation region or at a remote site. Both in the elongation and initiation mode, T7 RNAP polymerized tCTP with high catalytic efficiency across from a template G. Notably, T7 RNAP is much more discriminative against tC-A mismatches than DNA polymerases are against d(tC-A) mismatches. To demonstrate the merit of tCTP for RNA labeling, we produced large, 100% tC-labeled RNA by transcribing an 827 base pair DNA sequence, using tCTP in place of CTP.
EXPERIMENTAL SECTION
Materials and Enzymes
Unlabeled NTPs were from Invitrogen and 32P-labeled NTPs from Perkin Elmer Life Sciences. T7 RNA polymerase and RNAse Out™ were from Invitrogen. Synthetic oligonucleotides were purchased from Integrated DNA Technology and the DNA sample used for transcription of the Borrelia miyamotoi gene (locus D3777, region 414–1241; Gen Bank 43777) was provided by the Center for Disease Control and Prevention, Fort Collins, CO, and PCR amplified in house to introduce the T7 promotor. The PCR amplification system we used to generate the E. coli vitamin B12 riboswitch33 (Gene Bank: M10112.1) was a generous gift from Dr. Robert Batey (University of Colorado at Boulder).
Synthesis of tCTP
We performed the synthesis of tCTP using using the known synthesis of the 1,3-diaza-2-oxophenothiazine nucleobase,34 Vörbruggen’s silyl-Hilbert-Johnson ribonucleoside synthesis,35 and the Ludwig method for triphosphate preparation.36 All reagents and dry solvents were purchased at high purity from commercial suppliers and used without further purification.
2,3,5-tri-O-acetyl-tC-ribonucleoside
The tC nucleobase (200 mg, 0.92 mmol) 34 was suspended in anhydrous CH3CN (10 ml), and N,O-bis(trimethylsilyl)acetamide (350 μl, 1.42 mmol) was added. The reaction mixture was heated at reflux under N2 for 20 min, then allowed to cool to room temperature. 1,2,3,5-tetra-O-acetyl-β-D-ribofuranose (300 mg, 0.94 mmol) and trimethylsilyl trifluoromethanesulfonate (210 μl, 1.17 mmol) were added. The reaction mixture was heated at reflux for 2.5 hours, the allowed to cool to room temperature. The reaction mixture was then poured into 5% NaHCO3 solution (100 ml), and extracted with CH2Cl2 (2 × 100 ml). After drying over anhydrous Na2SO4, the solvent was removed by rotary evaporation and the product was purified by flash chromatography (5% hexanes in EtOAc), yielding the product as a yellow oil (395 mg, 91%), which was found to consist of only the desired β anomer. 1H NMR (500 MHz, CDCl3) δ = 9.27 (br s, 1H, NH), 7.33 (s, 1H), 7.09 (m, 2H), 6.94 (m, 2H), 6.15 (d, J = 4.4 Hz, 1H), 5.35 (q, J = 4.5 Hz, 1H), 5.32 (m, 1H), 4.38 (d, J = 1.3 Hz, 3H), 2.22 (s, 3H, OAc), 2.10 (s, 3H, OAc), 2.09 (s, 3H, OAc) ppm. 13C NMR (100 MHz, CDCl3) δ = 170.4, 169.7, 169.2, 160.5, 154.5, 135.4, 133.1, 127.9, 126.1, 125.0, 118.5, 116.3, 98.2, 88.3, 79.8, 73.7, 69.9, 63.0, 21.1, 20.6, 20.6 ppm. HRMS for C21H22N3O8S [M + H] calcd: 476.1128, found: 47.1131 (error = +1.9 ppm).
tC-ribonucleoside
2,3,5-tri-O-acetyl-tC-ribonucleoside (340 mg, 0.715 mmol) was dissolved in dry MeOH (3 ml) under N2, and NaOMe (25 wt. % in MeOH, 25 μl) was added. After stirring for one hour at room temperature, acetic acid (100 μl) was added, and the solvent removed by rotary evaporation. The crude product was suspended in water (5 ml), filtered, and dried in a dessicator, yielding the pure product as a yellow solid (203 mg, 81%). 1H NMR (400 MHz, DMSO-d6) δ = 10.43 (br s, 1H, NH), 7.95 (s, 1H), 7.07 (m, 2H), 6.93 (m, 2H), 5.73 (d, J = 2.5 Hz, 1H), 5.36 (d, J = 3.2 Hz, 1H), 5.18 (t, J = 4.5 Hz, 1H), 5.01 (d, J = 2.5 Hz, 1H), 3.95 (s, 2H, OH), 3.84 (s, 1H, OH), 3.70 (m, 1H), 3.56 (m, 1H) ppm. 13C NMR (100 MHz, DMSO-d6) δ = 159.5, 154.0, 136.2, 135.7, 127.4, 126.0, 124.0, 116.9, 115.9, 94.3, 89.4, 84.1, 74.2, 68.9, 60.0 ppm. HRMS for C15H16N3O5S [M + Na] calcd: 372.0625, found: 372.0622 (error = −0.8 ppm).
tC-ribonucleoside triphosphate
tC (30 mg, 0.09 mmol) was dissolved in trimethyl phosphate (0.5 mL) under argon and cooled on ice. POCl3 (9 uL, 1.1 equivalent) in trimethyl phosphate (0.05 mL) was added dropwise and the mixture was stirred for 2 h while warming up to room temperature. Tributylammonium pyrophosphate (0.6 g, 1.1 mmol, 12 equivalents) in DMF (1 mL) was added followed by several droplets of tributyl amine. The mixture was stirred another 3 h at room temperature and then poured into 0.1 M triethylammonium bicarbonate (TEAB, 100 mL). The solvents (except DMF) were removed under vacuo, the product was redissolved in water (150 mL), purified on a TEAB equilibrated ion-exchange column (Sephadex-DEAE A-25, Sigma Aldrich); using a 0 to 1 M TEAB gradient for elution. Fractions were individually spotted on a MALDI plate with THAP as the matrix and triphosphate fractions were identified by their MALDI- peak (negative ion mode). This procedure was repeated until tetraphosphate (formed as a by-product) was fully removed. Fractions were collected and lyophilized, yielding 5.3 mg tCTP (yield = 13 %). MS (MALDI, neg.): 588 (M −1) calcd 588. 31P NMR (400 MHz, D2O): − 9.9 (bs, 2P, gamma-P and alpha-P), −22.5 (bm, 1P, beta-P).
T7 transcription of synthetic oligonucleotides
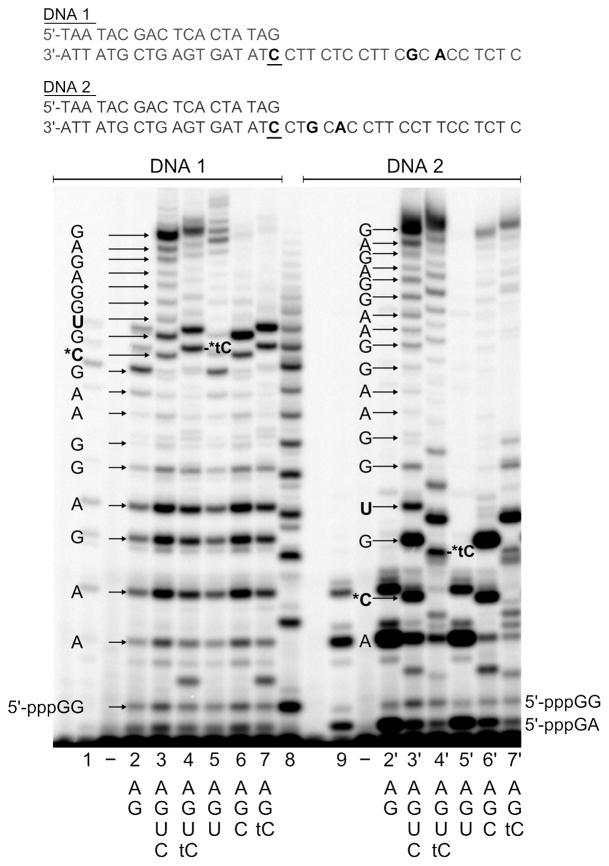
Standard 20 μL transcription reactions contained 1 μM DNA construct, 0.4 mM of each NTP, 5 mM DTT, [α-32P]GTP, commercial reaction buffer (40 mM Tris-HCl pH 8, 8 mM MgCl2, 2 mM spermidine, 25 mM NaCl) and 0.2 units/μL T7 RNA polymerase. The DNA constructs were prepared by hybridizing the T7 promotor (5′-TAA TAC GAC TCA CTA TAG-3′) to one of the following DNA templates: 3′-ATT ATG CTG AGT GAT ATC CTT CTC CTT CGC ACC TCT C-5′ (DNA1), 3′-ATT ATG CTG AGT GAT ATC CTG CAC CTT CCT TCC TCT C-5′ (DNA2), 3′-ATT ATG CTG AGT GAT ATC CTT CTC CTT CAC TCC TCT C-5′ (DNA3), 3′-ATT ATG CTG AGT GAT ATC CTA CTC CTT CCT TCC TCT C-5′ (DNA4) (see Figure 1 and 2). For single time points, the reaction was incubated for 30 or 60 min and stopped with a 2-fold excess of formamide. The RNA products were separated by denaturing gel electrophoresis (20 % polyacrylamide, 8 M urea gels) and visualized using phosphorimagery.
Figure 1.
Incorporation of tCTP by T7 RNA polymerase operating either in the initiation or elongation mode. Transcription starts at the underlined C and proceeds in the 3′-to-5′ direction. All reactions contained 0.2 units/μl T7 RNAP, 1 μM DNA and 0.4 mM of each of the indicated NTPs. All RNA products are visualized based on the incorporation of [α-32P]GTP, except for lanes 1 and 3, where [α-32P]ATP was used instead, together with ATP and GTP. Lane 2 displays a poly-rG ladder, which was generated as described in the experimental section.
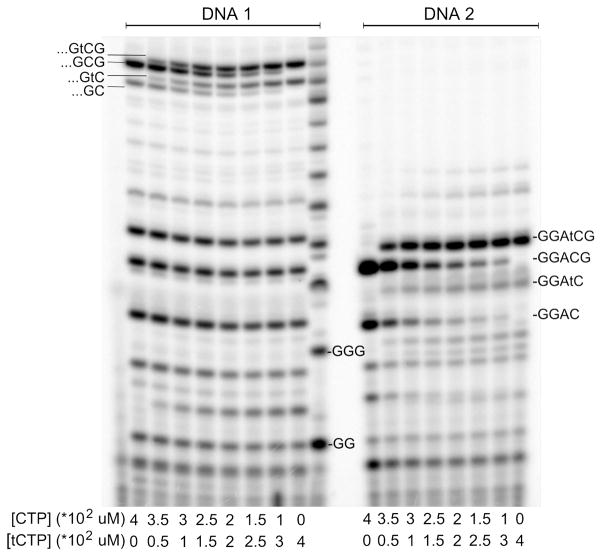
Figure 2.
Competitive incorporation of CTP and tCTP across from G. All reactions contained 0.2 units/μl T7 RNAP, 1 μM DNA and 0.4 mM [α-32P]GTP and 0.4 mM ATP. The [CTP]-to-[tCTP] ratio was systematically varied as indicated below the lanes and the reactions products were analyzed after incubating for 1h. The very left lane shows the no enzyme control, the poly-rG ladder is located in the middle between the two reactions series.
To produce a poly-G ribonucleotide ladder, a standard transcription reaction was carried out using only 0.4 mM GTP, [α-32P]GTP, 0.2 units/μL T7 RNA polymerase and 1 μM DNA template 3′-ATT ATG CTG AGT GAT ATC CCTT CTC CTT CGC ACC TCT C-5′.
The competitive incorporation of tCTP and CTP across from G was tested in standard assays (see above), this time containing 1 μM DNA1 or DNA2, 0.4 mM ATP, 0.4 mM GTP, [α-32P]GTP and one of the following: 400 μM CTP, no tCTP; 375 μM CTP, 25 μM tCTP; 350 μM CTP, 50 μM tCTP; 300 μM CTP, 100 μM tCTP; 250 μM CTP, 150 μM tCTP; 200 μM CTP, 200 μM tCTP; 150 μM CTP, 250 μM tCTP; 100 μM CTP, 300 μM tCTP; no CTP, 400 μM tCTP. The reactions were allowed to proceed for 1 hour and stopped and analyzed as described above. The relative catalytic activity of tCTP and CTP incorporation was determined using equation 1: , with fractionCTP being the amount of RNA extended by CTP, (kcat/KM)tCTP the catalytic efficiency for tCTP incorporation and (kcat/KM)CTP the catalytic efficiency for CTP incorporation.
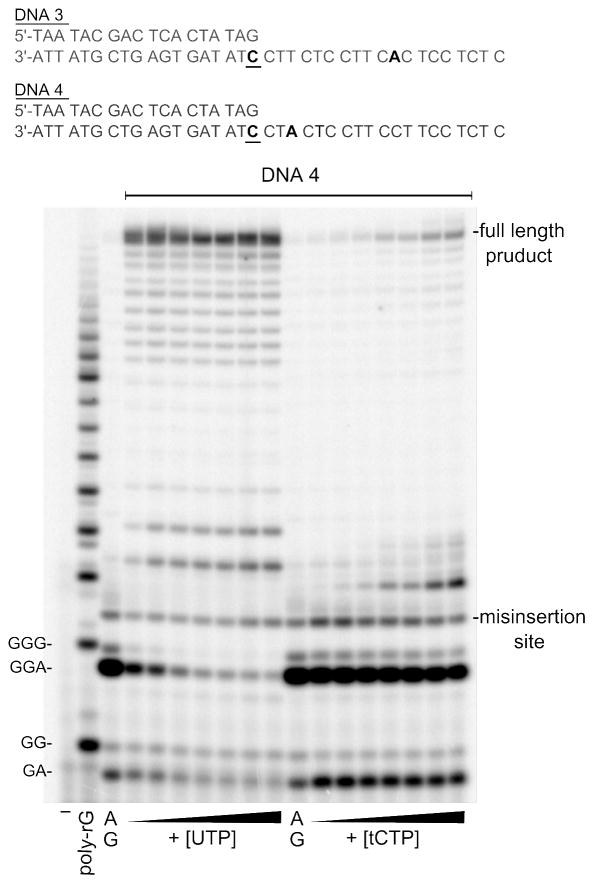
The kinetic parameters for tCTP incorporation across from A were measured under standard conditions. Assays were incubated at 37°C for 30 min and contained 1 μM DNA 3 or DNA 4, 0.4 mM ATP, 0.4 mM GTP, α-[32P]-GTP and either: 1, 5, 10, 25, 50, 100, 200 μM UTP or 1, 5, 10, 25, 50, 100, 200 μM tCTP. When DNA 3 was used, the percentage of RNA extended beyond 11 nucleotides was determined, in case of DNA 4, the percentage of RNA extended beyond 3 nucleotides was quantified. The percentage of extended RNA was plotted versus the UTP or tCTP concentration and the Michaelis-Menten parameters were derived by non-linear curve fitting.
Generation of long, fluorescent RNAs
Standard 20 μL transcription reactions contained 1 μL PCR amplified DNA template, 0.4 mM ATP, 0.4 mM GTP, 0.4 mM UTP, 5 mM DTT, 0.6 units/μL RNAse out, commercial reaction buffer (40 mM Tris-HCl pH 8, 8 mM MgCl2, 2 mM spermidine, 25 mM NaCl) and 2 units/μL T7 RNA polymerase. CTP and tCTP were added to match the following concentrations: 400 μM CTP, no tCTP; 375 μM CTP, 25 μM tCTP; 350 μM CTP, 50 μM tCTP; 300 μM CTP, 100 μM tCTP; 200 μM CTP, 200 μM tCTP; 100 μM CTP, 300 μM tCTP; no CTP, 400 μM tCTP. The samples were incubated for 1 hour, mixed with gel loading buffer (10 % ficoll 400, 10 % glycerol, 1 × TBE) and the RNA was separated using an 1.2 % agarose gel. The gels were exposed on an UV transilluminator prior to and after staining with ethidium bromide.
Absorbance and fluorescence emission spectra
To measure the absorbance and fluorescence emission spectra of tC labeled RNA, we synthesized large amounts of the B12 riboswitch by scaling-up the T7 transcription reaction described above to 400 μL and by prolonging the reaction time to 3 hours. We examined three different tC labeling densities: a) 25 μM tCTP and 375 μM CTP, b) 100 μM tCTP and 300 μM CTP, or c) 400 μM tCTP and no CTP. The RNA was precipitated with a 2.5 × volume of EtOH at −20 °C over night. The precipitate was dissolved in 90 % formamide and purified by denaturing gel electrophoresis (8 % polyacrylamide, 6 M urea). The excised bands were crushed and eluted into sterile 0.5X TE buffer. The RNA was concentrated and buffer exchanged 3 times with 0.5X TE using YM-10 centricon filters (Amicon). The fluorescence spectra were recorded in 0.5 × phosphate buffered saline (PBS) using a Quantamaster 40 fluorimeter (Photon Technology International).
The fluorescence quantum yield of tCTP was determined by recording the absorbance and fluorescence emission (λex=375 nm) spectra of tCTP solutions (in 0.5 × PBS buffer) using quinine sulfate in 0.1 M H2SO4 as a standard (quantum yield φF = 0.57). The quantum yield was calculated using the following expression: where the subscript S refers to the fluorescence standard, I is the integrated fluorescence intensity, E is the absorbance at the excitation wavelength and n is the refractive index.
Reverse transcription
Unlabeled and tC labeled RNA were reverse transcribed using SuperScript III reverse transcriptase (Invitrogen). The 20 μL assays contained 250 μM primer (5′-TGC CGC AGG TTT CAT CAA for B12 RNA and 5′-GCA TCT GAT GAT GCT GCT GG for Borrelia RNA), 500 μM of each dNTP, some [α-32P]TTP, 4 μL RNA template (T7 transcription reactions purified using the QIAquick PCR purification kit from Qiagen), 5 mM DTT, 40 units RNase OUT (Invitrogen), 50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2 and 100 units Super Script III reverse transcriptase. Reaction time was 50 min at 52 °C.
RESULTS AND DISCUSSION
tCTP incorporation opposite a template guanine
Since T7 RNAP possesses different catalytic properties during the initiation and elongation phases of RNA synthesis, we examined tCTP incorporation during both phases by using two different synthetic DNAs. Both consisted of the 18 nucleotide (nt) T7 promotor hybridized to a complementary 37 nt DNA template (Figure 1). Transcription usually starts at the underlined C and proceeds in a 3′-to-5′ direction along the template. To study tCTP incorporation while T7 RNAP operates in the elongation mode, we employed DNA1, which exhibited a unique guanine 12 bases away from the start site of transcription. DNA2 featured the unique guanine at a position only four nucleotides from the start site, directing tCTP incorporation to the initiation region. It was necessary to start the templating region with CCT instead of CCC to avoid slippage of T7 RNAP during initiation, which resulted in non-sense poly-G ribonucleotides of different lengths.18
Figure 1 compares the lengths of the RNA products obtained in transcription reactions using DNA1 or DNA2, respectively, T7 RNAP, different combinations of NTPs and [α-32P]GTP. In the presence of only GTP and ATP (lanes 2 and 2′), transcription terminated as T7 RNAP reached the single template guanines of DNA1 and DNA2, respectively, and the length of the products verifies that transcription starts indeed at the underlined C. Oddly, T7 RNAP produces a large amount of abortive GA dinucleotide when transcribing DNA2, as revealed by RNA labeling with [α-32P]ATP in the presence of only ATP and GTP (Figure 1, lane 9). Reactions carried out in the presence of all four natural NTPs produced full length products (20 nt long) with both DNA constructs (lanes 3 and 3′), and so did the reactions in which CTP was fully substituted by tCTP (lanes 4 and 4′). The RNA transcripts containing tC are clearly upward shifted relative to the transcript exhibiting C at the same position, presumably due to the larger size of tC compared to C. To test if UTP can be misinserted opposite G, transcription of DNA1 and 2 was performed in the presence of GTP, ATP and UTP (lanes 5 and 5′). For DNA1, a significant amount of G-U mismatch was formed leading to full length product, whereas transcription terminated opposite the unique template G of DNA2. However, the RNA transcript that contains U instead of the correct C migrates distinctly different from the RNA exhibiting tC at this position. Omitting UTP from the reaction mixture in the presence of CTP or tCTP, respectively (lanes 6 and 7), lead to almost complete abortion of transcription at the unique A of DNA1. In both reactions only ~7 % of the corresponding abortive RNA transcripts were extended. Employing DNA2, the reactions containing ATP, GTP and CTP lead to 11 % extension past the unique template A (lane 6′), whereas it was 26 % for the reaction containing tCTP (lane 7′). Thus, T7 RNAP used neither ATP, GTP, CTP nor tCTP effectively to bypass the template adenine. This result suggests that T7 RNAP base pairs tCTP less ambivalently than we would have expected based on our previous study of dtCTP incorporation by DNA polymerases.30
Competition between tCTP and CTP
To measure the relative catalytic efficiency of tCTP incorporation versus the catalytic efficiency of CTP incorporation, (kcat/KM)tCTP/(kcat/KM)CTP, transcription reactions were carried out in the presence of fixed concentrations of GTP and ATP, and varying ratios of tCTP and CTP. The omission of UTP facilitated the quantification of the reaction products by stopping the polymerase at the unique template A, thus avoiding the challenge of electrophoretically separating a large number of longer RNA transcripts with differing migration characteristics. The slower migration of products containing tC allowed us to resolve the RNA strands that had been extended by either tC or C using gel electrophoresis (Figure 2). Using equation 1, we determined (kcat/KM)tCTP/(kcat/KM)CTP to be 2.3±0.1 and 2.5±0.1 for tCTP incorporation in the elongation and initiation modes, respectively. Other studies that used T7 RNAP for the incorporation of unnatural nucleotides reported drastic differences for the elongation and initiation mode. For instance, T7 RNAP cannot introduce the uridine analogue thieno[3, 4-d]pyrimidine nucleotide closer than 7 bases to the promoter.37
tCTP incorporation opposite a template adenine
Next, we examined the ability of T7 RNAP to discriminate against a tC-A mismatch more closely. Although we had not observed significant tC-A mismatch formation transcribing DNA1 and DNA2, we wanted to make sure this was not a result of the base sequence, where A succeeded G by only 1 nucleotide. Additionally, these reactions contained tCTP, and T7 RNAP polymerizes tCTP more efficiently than CTP. Thus, the lack of detectable incorporation of tC across from A might have resulted from tC incorporation at the immediately preceding position. Therefore we repeated the transcription reactions using DNA3 and DNA4 (Figure 3), which exhibit unique adenines at the same position where DNA1 and 2 exhibited guanine. Surprisingly, transcription reactions containing ATP, GTP, tCTP and DNA3 or DNA4, respectively, produced some full length RNA transcripts, whereas reactions containing only ATP and GTP did not. Thus, T7 RNAP synthesized tC-A mismatches in this base context, suggesting that the proximity of the tC-G base pair to the template A in DNA1 and 2 and/or sequence effects made the formation of the tC-A base pair less favorable with these DNAs. We determined the kinetic parameters for the formation of the U-A and tC-A base pairs by performing the transcription reactions with increasing UTP or tCTP concentrations, respectively, while ATP and GTP were kept at a constant high concentration (Figure 3). We did not use competitive assays to compare the catalytic efficiencies as shown in Figure 2 because the template base sequence did not offer the possibility to halt transcription shortly after the insertion of U or tC, respectively. Given that the U or tC containing RNA is extended by another 8 (DNA3) or 16 (DNA4) nucleotides, it would have been difficult to separate all RNA products containing U from those containing tC. The Michaelis Menten parameters were derived from plots of the percentage of RNA extended beyond 11 (DNA3) and beyond 3 nucleotides (DNA4), respectively, versus the UTP or tCTP concentration (Table 1). The experiments yielded a discrimination factor of 40 for the formation of a tC-A base pair when T7 RNAP is in the elongation mode and a factor of 300 when in the initiation mode. Thus, T7 RNAP discriminates significantly better against a tC-A mismatch than DNA polymerases do against a d(tC-A) mismatch.
Figure 3.
Incorporation of UTP and tCTP across from a template A. All reactions contained 0.2 units/μl T7 RNAP, 1 μM DNA4, 0.4 mM GTP, 0.4 mM ATP, some α-[32P]-GTP and increasing concentrations of UTP (left side) or tCTP (right side). Each reaction was stopped after 30 min. The UTP and tCTP concentrations, respectively, were as follows: 1, 5, 10, 25, 50, 100, 200 μM.
Table 1.
Kinetic parameters for the insertion of tCTP and UTP opposite adenine.
| NTP | DNA X | Vmax (SD) | KM (SD) [μM] | Vmax/KM | Discrimination* |
|---|---|---|---|---|---|
| tCTP | DNA3 | 2.66 (0.1) | 1.7 (0.5) | 1.6 | 41 |
| UTP | DNA3 | 3.29 (0.01) | 0.05 (0.01) | 66 | 1 |
| tCTP | DNA4 | 1.4 (0.4) | 108 (56) | 0.01 | 300 |
| UTP | DNA4 | 3 (0.1) | 1.0 (0.2) | 3 | 1 |
Discrimination is defined as Vmax/KM for the incorporation of UTP into DNAX, divided by Vmax/KM for the incorporation of tCTP into DNAX.
Previously, we measured discrimination factors of 7 and 6, respectively, for the formation of a d(tC-A) base pair by human DNA polymerase α and Klenow fragment.30 We rationalized this observation by proposing that tC is in rapid equilibrium between its amino and imino tautomer. The amino tautomer is competent to base pair with guanine, and the imino tautomer, being isosteric to thymine, would base pair with adenine (Chart 1). The observation that the structurally related pyrimidine analogue tetrafluorophenoxazine binds with equal affinity to G and A, as judged based on the melting temperatures of the corresponding DNA duplexes, supports the existence of tautomerism in this type of tricycle.38 If this tautomeric equilibrium operates and hydrogen bonding is a general determinant for nucleotide selection, DNA and RNA polymerases should polymerize such substrates with similar ambivalence.
Indeed, such a behavior has in fact been observed for the cytidine analogue P (3,4-dihydro-6H,8H-pyrimido[4,5-c][1,2]oxazin-2-one).39 For the P base, the ratio of the imino to amino form is 11:1.40 Strikingly, the relative catalytic efficiencies for the incorporation of dPTP opposite A versus G by Klenow fragment match this number, 11:1.40 Although the exact kinetic parameters are not available, Moriyama et al. reported that T7 RNAP preferentially inserts PTP opposite A, not G, suggesting that the tautomeric form also plays a role during nucleotide selection by T7 RNAP.41 How then do we explain the superior discrimination of T7 RNAP against tC-A base pairs? One possibility is that the occurrence of the tC imino tautomer is a templating effect, i.e. the alignment of tC with a templating adenine induces the imino tautomer to minimize the free energy of base pairing via hydrogen bonding. Such an inductive effect would be most powerful if the templating and the incoming base were aligned in one plane, as in duplex DNA. The X-ray structure of the open ternary T7 RNAP-DNA-NTP complex42 captures an interaction of the incoming nucleotide with the templating base at a putative preinsertion site that has not been observed in other open ternary polymerase-DNA-dNTP complexes so far, including BF polymerase,43 RB69,44 Taq polymerase45 and T7 DNA polymerase.46 In T7 RNAP, the incoming NTP binds to the fingers domain (which forms the roof of the active site in the closed ternary complex) and makes hydrogen bonding contacts with the templating base prior to entering the active site, albeit without the steric constraint of the active site and with a slight out of plane tilt. Thus, T7 RNAP likely has a mechanism to screen for base complementarity at the preinsertion site that probably contributes to selectivity. However, the open ternary complex of Taq polymerase, a structural homologue of Klenow fragment, captures the incoming nucleotide bound in the active site (the insertion site), while the templating base is occluded from interactions with the incoming nucleotide.45 Two studies confirmed that the closure of the fingers domain and the alignment of the templating base with the incoming base are both fast steps on the reaction coordinate of nucleotide polymerization by Klenow fragment.47, 48 This suggests that Klenow is in fast equilibrium between the open and closed conformation and that it screens for base complementarity at the insertion site after formation of Watson-Crick hydrogen bonds between the incoming dNTP and templating base. Taken together we suggest that T7 RNAP has a mechanism to reject the incoming tCTP at the preinsertion site prior to formation of Watson-Crick hydrogen bonds, whereas Klenow30 allows dtCTP access to the insertion site where hydrogen bonding with the templating base facilitates the formation of the imino tautomer, which is then accepted due to its resemblance to a T-A base pair.
tC labeling of large RNA
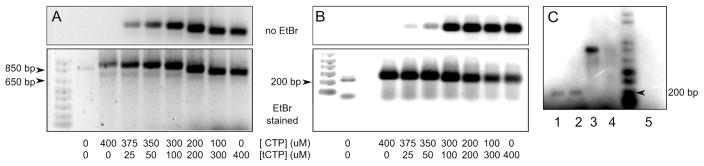
The generation of full length products on DNA1-4 in assays containing ATP, GTP, UTP, and tCTP suggested that it should be possible to use tCTP and T7 RNAP to generate highly fluorescent RNA. To present T7 RNAP with a more vigorous challenge and to demonstrate the merit of tCTP incorporation for fluorescent labeling of large RNA transcripts, we transcribed ~827 nucleotides of the 1398 bp Borrelia miyamotoi flagellin protein gene (locus D3777, region 414–1241; Gen Bank D43777) in the presence of tCTP. Formation of full length RNA requires T7 RNAP to incorporate 3 consecutive tCTPs at 6 different positions. To allow for T7 transcription, the particular gene fragment was PCR amplified using a primer that introduced the T7 promotor sequence into the final product. Figure 4A displays the RNA transcripts obtained at different mixing ratios of tCTP and CTP and separated by agarose gel electrophoresis. As we described recently for tCo labeled PCR products,31 tCo being a fluorescent oxo-ortholog of tC, it is possible to visualize the nucleic acids based on the tC fluorescence only, thus obviating the need for ethidium bromide staining. Fluorescent 827 nucleotide RNA was obtained at all tCTP/CTP ratios tested, from a ratio of 1/15 to the point of full substitution of CTP with tCTP. To demonstrate the generality of the labeling method, we used the same reaction conditions to transcribe a catalytic RNA, the 207 nucleotide long E. coli riboswitch for vitamin B12 (Figure 4B).33
Figure 4.
Large fluorescent RNA generated by transcription in the presence of CTP and tCTP at different concentrations. The top row of A and B shows the 1.2 % agarose gel exposed by UV light prior to staining with ethidium bromide (EtBr). The CTP and tCTP concentrations are provided below the images. The reaction in the lane next to the marker contained DNA template, primers, enzyme but no CTP or tCTP. A) 827 nucleotide RNA obtained from transcription of the Borrelia miyamotoi flagellin protein gene. B) 207 nucleotide long B12 riboswitch of E. coli. C) Reverse transcription of unlabeled or tC labeled RNA. The products of the reverse transcription reactions were analyzed on a 1.4 % agarose gel and visualized based on the incorporation of [α-32P]dTTP. Unlabeled B12 riboswitch (lane 1); 100 % tC labeled B12 riboswitch (lane 2); unlabeled Borrelia RNA (lane 3); 100 % tC labeled Borrelia RNA (lane 4); reaction as in lane 1 but without enzyme (lane 5).
For the Borrelia and the B12 RNA transcripts, increasing the tC content led to slightly faster migration of the product RNA, and the overall yield was somewhat diminished. The altered migration of tC labeled RNA could indicate premature termination of T7 transcription at a specific site, or more likely, the RNA adopts a different secondary structure due to the increased hydrophobicity of tC as compared to C. To distinguish between these two scenarios, we tried to verify the length of the RNA by reverse transcription. Using SuperScript III reverse transcriptase we synthesized DNA from RNA templates in which either none or all C’s we substituted by tC. If both types of RNA consist of the same number of nucleotides, reverse transcriptase reactions will generate DNA products of identical length. For the B12 riboswitch, the reverse transcription of the unlabeled and the 100 % tC labeled version of the same RNA produced DNA strands of identical length (Figure 4c). However, we were not able to obtain transcription products using 100 % tC labeled Borrelia RNA as template, which we attribute to the length of the template and the larger number of tC repeats. Judged based on the results obtained for the B12 RNA it is possible to replace all C’s of an arbitrary RNA sequence by tC using T7 RNAP. Importantly, the experiments also demonstrate that SuperScript III reverse transcriptase is capable of transcribing a template containing tC into DNA, however, the precise templating properties of tC remain to be characterized.
Fluorescence properties of tCTP
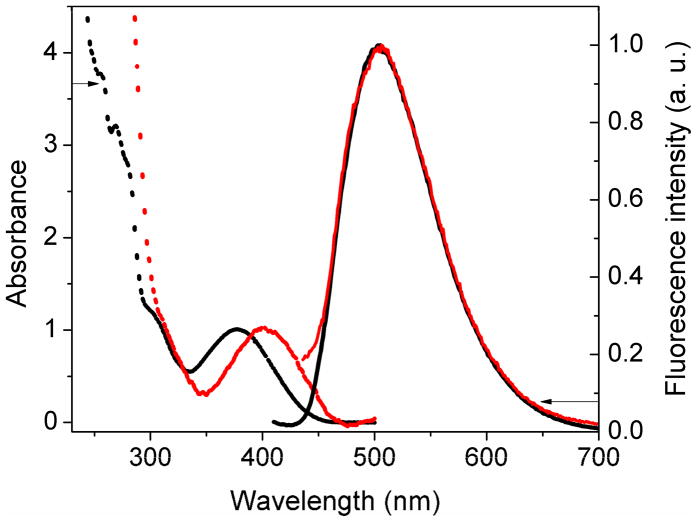
Besides the enzymatic efficiency of tCTP incorporation, the fluorescent properties of tC in the context of RNA are crucial to its potential use for labeling RNA. As expected based on the small difference in chemical structure, the fluorescent properties of free tCTP are virtually identical to those of dtCTP.29 The lowest energy absorption band of tCTP lies at 377 nm at pH 7.4 (Figure 5). The corresponding extinction coefficients are ε260=24,000 and ε377=4,800 M−1 cm−1, respectively. The fluorescence emission of tCTP peaks at 504 nm, the fluorescence quantum yield is φF=0.18 (λex=375 nm). We synthesized large amounts of the B12 riboswitch using T7 transcription in the presence of different tCTP/CTP ratios and purified the RNA and recorded their absorbance and emission spectra. The tCTP/CTP ratios examined were 1:15, 1:4 and 1:0. For all three RNA samples, the absorbance peak red-shifted by ~23 nm, whereas the fluorescence emission of tC labeled RNA perfectly overlapped with the emission of free tCTP (Figure 5). Based on the correlation between absorbance and integrated fluorescence intensity, we estimate the fluorescence quantum yield of tC in the context of RNA to be φF ~ 0.1, indicating only a small amount of quenching especially as compared to other fluorescent base analogues.
Figure 5.
Absorbance and fluorescence emission spectra of tCTP (black) and the purified B12 riboswitch (red) the C’s of which have been fully replaced by tC. The absorbance and fluorescence emission peaks are normalized to 1 and do not reflect relative intensities.
CONCLUSIONS
We have presented a method for the synthesis of fluorescently labeled RNA that is based on the incorporation of the fluorescent cytidine analogue tCTP by T7 RNA polymerase in transcription reactions. Compared to the insertion of dtCTP by DNA polymerases, tCTP insertion by T7 RNAP is more precise; the enzyme discriminates sufficiently well between tC-G and tC-A base pairs to avoid U → C mutations during transcription. The base analogue tC combines several properties that make it an attractive candidate for the design of regulatory and catalytic RNAs: as with other N4-substituted cytosine analogues it likely engages in variable hydrogen bonding patterns depending on its tautomerization state, it stabilizes DNA-RNA and DNA-DNA duplexes,28, 49 and it is fluorescent and hence traceable within the limits of its fluorescence quantum yield. Since the hydrophobicity, rigidity, size, and hydrogen bonding characteristics of tC’s tricyclic structure will alter the folding pattern and flexibility of catalytic RNAs significantly, it may be possible to artificially evolve new catalytic function. While tC was originally introduced as a probe for anti-sense nucleic acids, it will be exciting to explore the base analogue’s merit for producing functionally expanded RNA libraries using T7 transcription combined with SELEX approaches.50
Figure 6.
Chart 1: Base pairing of the tC amino and the tC imino tautomer.
Acknowledgments
This work was supported by NIH grants GM54194 and AI59764. B.W.P. thanks the University of Denver for financial support. We also thank James E. Johnson of the Batey Lab (University of Colorado at Boulder) for help with the B12 riboswitch.
References
- 1.Pederson T. Nucleic Acids Res. 2001;29:1013–1016. doi: 10.1093/nar/29.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer G. Angew Chem Int Ed Eng. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 3.Drolet DW, MoonMcDermott L, Romig TS. Nat Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- 4.Murphy MB, Fuller ST, Richardson PM, Doyle SA. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gng110. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brody EN, Willis MC, Smith JD, Jayasena S, Zichi D, Gold L. Mol Diagn. 1999;4:381–388. doi: 10.1016/s1084-8592(99)80014-9. [DOI] [PubMed] [Google Scholar]
- 6.Berezovski M, Drabovich A, Krylova SM, Musheev M, Okhonin V, Petrov A, Krylov SN. J Am Chem Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- 7.Huang YF, Chang HT, Tan WH. Anal Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 8.Stojanovic MN, Kolpashchikov DM. J Am Chem Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 9.Fiammengo R, Musilek K, Jaschke A. J Am Chem Soc. 2005;127:9271–9276. doi: 10.1021/ja051179m. [DOI] [PubMed] [Google Scholar]
- 10.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao L. J Am Chem Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 11.Moriyama K, Kimoto M, Mitsui T, Yokoyama S, Hirao I. Nucleic Acids Res. 2005:33. doi: 10.1093/nar/gni128. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milligan JF, Uhlenbeck OC. Meth Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 13.Seelig B, Jaschke A. Bioconjug Chem. 1999;10:371–378. doi: 10.1021/bc980085h. [DOI] [PubMed] [Google Scholar]
- 14.Kimoto M, Mitsui T, Harada Y, Sato A. Nucleic Acids Res. 2007;35:5360–5369. doi: 10.1093/nar/gkm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durniak KJ, Bailey S, Steitz TA. Science. 2008;322:553–557. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma KY, Temiakov D, Jiang ML, Anikin M, McAllister WT. J Biol Chem. 2002;277:43206–43215. doi: 10.1074/jbc.M206658200. [DOI] [PubMed] [Google Scholar]
- 17.Jia YP, Patel SS. Biochemistry. 1997;36:4223–4232. doi: 10.1021/bi9630467. [DOI] [PubMed] [Google Scholar]
- 18.Martin CT, Muller DK, Coleman JE. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee S, Brieba LG, Sousa R. Cell. 2002;110:81–91. doi: 10.1016/s0092-8674(02)00815-2. [DOI] [PubMed] [Google Scholar]
- 20.Brieba LG, Sousa R. Embo J. 2001;20:6826–6835. doi: 10.1093/emboj/20.23.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brieba LG, Sousa R. Biochemistry. 2001;40:3882–3890. doi: 10.1021/bi002716c. [DOI] [PubMed] [Google Scholar]
- 22.Moroney SE, Piccirilli JA. Biochemistry. 1991;30:10343–10349. doi: 10.1021/bi00106a036. [DOI] [PubMed] [Google Scholar]
- 23.Yin YW, Steitz AA. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 24.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 25.Temiakov D, Mentesana PE, Ma KY, Mustaev A, Borukhov S, McAllister WT. Proc Natl Acad Sci USA. 2000;97:14109–14114. doi: 10.1073/pnas.250473197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang JB, Sousa R. J Mol Biol. 2000;303:347–358. doi: 10.1006/jmbi.2000.4150. [DOI] [PubMed] [Google Scholar]
- 27.Anand VS, Patel SS. J Biol Chem. 2006;281:35677–35685. doi: 10.1074/jbc.M608180200. [DOI] [PubMed] [Google Scholar]
- 28.Lin KY, Jones RJ, Matteucci M. J Am Chem Soc. 1995;117:3873–3874. [Google Scholar]
- 29.Sandin P, Wilhelmsson LM, Lincoln P, Powers VEC, Brown T, Albinsson B. Nucleic Acids Res. 2005;33:5019–5025. doi: 10.1093/nar/gki790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandin P, Stengel G, Ljungdahl T, Borjesson K, Macao B, Wilhelmsson LM. Nucleic Acids Res. 2009;37:3924–3933. doi: 10.1093/nar/gkp266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stengel G, Urban M, Purse BW, Kuchta RD. Anal Chem. 2009;81:9079–85. doi: 10.1021/ac9017555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stengel G, Purse BW, Wilhelmsson LM, Urban M, Kuchta RD. Biochemistry. 2009;48:7547–7555. doi: 10.1021/bi9006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. RNA. 2003;9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandin P, Lincoln P, Brown T, Wilhelmsson LM. Nat Prot. 2007;2:615–623. doi: 10.1038/nprot.2007.80. [DOI] [PubMed] [Google Scholar]
- 35.Vorbruggen H, Bennua B. Cem Ber. 1981;114:1279–1286. [Google Scholar]
- 36.Ludwig J. Acta Biochim Biophys Hung. 1981;16:131–133. [PubMed] [Google Scholar]
- 37.Srivatsan SG, Weizman H, Tor Y. Org Biomol Chem. 2008;6:1334–1338. doi: 10.1039/b801054d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang JY, Lin KY, Matteucci MD. Tetrahedron Lett. 1998;39:8385–8388. [Google Scholar]
- 39.Lin PKT, Brown DM. Nucleic Acids Res. 1989;17:10373–10383. doi: 10.1093/nar/17.24.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris VH, Smith CL, Cummins WJ, Hamilton AL, Adams H, Dickman M, Hornby DP, Williams DM. J Mol Biol. 2003;326:1389–1401. doi: 10.1016/s0022-2836(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 41.Moriyama K, Negishi K, Briggs MSJ, Smith CL, Hill F, Churcher MJ, Brown DM, Loakes D. Nucleic Acids Res. 1998;26:2105–2111. doi: 10.1093/nar/26.9.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 43.Johnson SJ, Taylor JS, Beese LS. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franklin MC, Wang JM, Steitz TA. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Korolev S, Waksman G. Embo J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 47.Joyce CM, Potapova O, DeLucia AM, Huang XW, Basu VP, Grindley NDF. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 48.Rothwell PJ, Mitaksov V, Waksman G. Mol Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 49.Engman KC, Sandin P, Osborne S, Brown T, Billeter M, Lincoln P, Norden B, Albinsson B, Wilhelmsson LM. Nucleic Acids Res. 2004;32:5087–5095. doi: 10.1093/nar/gkh844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keefe AD, Cload ST. Curr Opin Chem Biol. 2008;12:448–456. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]