Abstract
The complex repertoire of immune receptors generated by B and T cells enables recognition of diverse threats to the host organism. In this work, we show that massively parallel DNA sequencing of rearranged immune receptor loci can provide direct detection and tracking of immune diversity and expanded clonal lymphocyte populations in physiological and pathological contexts. DNA was isolated from blood and tissue samples, a series of redundant primers was used to amplify diverse DNA rearrangements, and the resulting mixtures of barcoded amplicons were sequenced using long-read ultra deep sequencing. Individual DNA molecules were then characterized on the basis of DNA segments that had been joined to make a functional (or nonfunctional) immune effector. Current experimental designs can accommodate up to 150 samples in a single sequence run, with the depth of sequencing sufficient to identify stable and dynamic aspects of the immune repertoire in both normal and diseased circumstances. These data provide a high-resolution picture of immune spectra in normal individuals and in patients with hematological malignancies, illuminating, in the latter case, both the initial behavior of clonal tumor populations and the later suppression or re-emergence of such populations after treatment.
Introduction
Antigen receptors with diverse binding activities are the hallmark of B and T cells of the adaptive immune system in jawed vertebrates and are generated by genomic rearrangement of variable (V), diversity (D), and joining (J) gene segments separated by highly variable junction regions (1). Initial calculations of the combinatorial and junctional possibilities that contribute to the human immune receptor repertoire greatly exceed the total number of peripheral T or B cells in an individual (2). One study in which small subsets of rearranged T cell receptor (TCR) subunit genes were extensively sequenced using a few segment-specific primers yielded extrapolations for the full TCR repertoire corresponding to 2.5 × 107 distinct TCRα-TCRβ pairs in the peripheral blood of an individual (3). Extensive repertoire analyses for the human B cell compartment have been more limited, although small-scale studies and focused analysis of immunoglobulin (Ig) class subsets such as IgE have been performed (4, 5). Advanced sequencing methods have recently been used to analyze B cell receptor diversity in the relatively simple model immune system in zebrafish (6). Against a background of continually generated novel DNA sequences, expanded clones of B cells with useful antigen specificities persist over time to enable rapid responses to antigens previously detected by the immune system. Systematic means for detection of such expanded clones in human beings would open much of our immunity to specific analysis and tracking, including measurement of clonal population sizes, anatomic distributions, and changes in response to immunological events (7).
In contrast to healthy immune systems, malignancies of B- or T-cell origin typically express a single dominant clonal Ig or TCR receptor. A variety of assays have been used to detect the presence of B cell clonality for diagnosis of lymphomas and leukemias, including analysis of Ig light chain gene restriction and Southern blotting or sizing of polymerase chain reaction (PCR) products from rearranged Ig or TCR loci (8, 9). While adequate for many applications, these strategies make limited use of the high information content inherent in rearranged immune receptor gene sequences and can give indeterminate results. A recent study using deep sequencing of clonal Ig heavy chain receptor genes (IgH) in chronic lymphocytic leukemia revealed unexpected intraclonal heterogeneity in a subset of cases, showing that fundamental features of leukemic cell populations have escaped notice using prior approaches (10). Detection of more subtle clonal populations (for example, to follow the response of lymphomas or leukemias to treatment) currently relies on time- and labor-intensive multiparameter flow cytometry or custom-designed patient- and clone-specific real-time PCR assays (11–13). Early diagnostic screening approaches may benefit from generalized and more efficient clonal detection. Indeed, a recent population-based epidemiological study showed that small amplified B-cell populations can be seen in almost all individuals who go on to develop chronic lymphocytic leukemia, further underscoring the importance of assessing lymphocyte clonality in human specimens (14).
Detection and analysis of clonality is also of fundamental interest in characterizing and tracking both normal and pathogenic immune reactions. For protective and healthy humoral immune responses, high-resolution analysis of immune receptor clonality and evolution offers the potential for definitive detection and monitoring of effective immune responses to vaccination and specific infections (15), while for some autoimmune disorders this type of analysis could facilitate diagnosis, long-term therapeutic monitoring strategies and, eventually, specific interventions (16).
Using a bar-coding strategy to allow pooling of multiple libraries of rearranged immunoglobulin heavy chain (IgH) V-D-J gene loci from many human blood samples, we have performed high-throughput pyrosequencing to characterize the B cell populations in a series of human clinical specimens (17). Deep sequencing of immune receptor gene populations offers specific and detailed molecular characterization as well as high sensitivity for detecting sequences of interest, and should help to transform our understanding of the human immune system, while aiding in diagnosis and tracking of lymphoid malignancies.
Results
Barcoded high-throughput pyrosequencing of rearranged IgH loci
We amplified rearranged IgH loci in human blood samples using BIOMED-2 nucleic acid primers adapted for high-throughput DNA pyrosequencing. A unique 6-, 7-, or 10-nucleotide sequence “barcode” in the primers used for a particular sample allowed pooling and bulk sequencing of many libraries together, and subsequent sorting of sequences from each sample (Fig. 1, Supplementary Table S1). Patient specimens in our initial 2 replicate experiments included peripheral blood of three healthy individuals, with experimental replicates of one individual’s blood sample at each of two different time points 14 months apart; tissue specimens from patients with lymphomas; and peripheral blood from patients with chronic lymphocytic leukemia. We also studied samples generated by serial 10-fold dilutions of a chronic lymphocytic leukemia peripheral blood specimen into a healthy control peripheral blood sample, to assess the sensitivity of the sequencing approach for detecting small numbers of clonal B cells among a background B cell population (Table 1, Supplementary Table S2). From all specimens pooled for Experiment 1, we obtained 299,846 different IgH rearrangement sequences, while Experiment 2 yielded 207,043 sequences. All sequence reads used for further analysis were full-length IgH amplicons extending from the V gene segment FR2 framework region primer to the J primer region.
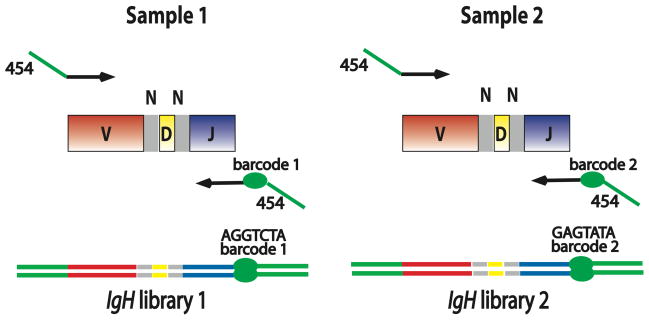
Fig. 1. Barcoded PCR amplicons for multiplexed IgH sequencing.
PCR primers used for preparing barcoded amplicons for high-throughput sequencing were designed using the FR2 IgH V gene segment family primers and the common IgH J segment primer from the BIOMED-2 consortium (19). Additional sequences required for emulsion PCR and pyrosequencing were added (indicated in green) at the 5′ end of the IgH-specific primers. In addition, a 6-, 7-, or 10-nucleotide sequence barcode was designed into the modified IgH J primer to identify the sample from which the PCR amplicons were derived. In the specimens analyzed using the 454 Titanium sequencer, an additional 10-nucleotide sample barcode was incorporated into the multiplexed IgH V gene segment primers used for amplification (Supplementary Table 1). Lines with arrowheads indicate PCR primers. Green segments: primer sequences needed for 454 sequencing protocol; red segments: V gene segment sequence; grey segments: non-templated N base sequences; yellow segments: D gene segment sequence; blue segments: J gene segment sequence; green ellipse: sample-specific barcode enabling pooling of IgH libraries for multiplexed sequencing. Sample 1 and Sample 2 could represent DNA template from any two clinical specimens, or independent DNA template aliquots from the same specimen.
Table 1.
Patient Specimens for IgH Sequencing.
| No. | Description | Sample Type | Clonality Assay Result |
|---|---|---|---|
| 1 | Healthy donor 1, time 0 | Blood | Negative |
| 2 | Healthy donor 1, time 0 | Blood | Negative |
| 3 | Healthy donor 1, time 14 months | Blood | Negative |
| 4 | Healthy donor 1, time 14 months | Blood | Negative |
| 5 | Patient 1; CLL/SLL time 0 | Blood | Positive |
| 6 | Patient 1; CLL/SLL time 3 months | Blood | Positive |
| 7 | Patient 2; FL | Lymph node | Positive |
| 8 | Patient 3; FL and SLL in Lymph node | Lymph node | Positive |
| 9 | Patient 4; CLL/SLL | Blood | Oligoclonal |
| 10 | Patient 5; PTLD, marrow infiltrate | Bone marrow | Positive |
| 11 | Patient 5; PTLD, liver DLBCL | Liver | Positive |
| 12 | Healthy donor 2 | Blood | Negative |
| 13 | Patient 6; CLL | Blood | Positive |
| 14 | Healthy donor 3 | Blood | Negative |
| 15 | Patient 6 CLL diluted 1:10 | Blood | Positive |
| 16 | Patient 6 CLL diluted 1:100 | Blood | Negative |
| 17 | Patient 6 CLL diluted 1:1000 | Blood | Negative |
| 18 | Patient 6 CLL diluted 1:10000 | Blood | Negative |
| 19 | Patient 6 CLL diluted 1:100000 | Blood | Negative |
The clonality assay results are those obtained using standard PCR amplification and capillary electrophoresis of product amplicons. Abbreviations: Blood: peripheral blood mononuclear cells; Lymph node: formalin-fixed paraffin-embedded lymph node tissue; Liver: formalin-fixed paraffin-embedded liver tissue; CLL/SLL chronic lymphocytic leukemia/small lymphocytic lymphoma; FL follicular lymphoma; PTLD post-transplant lymphoproliferative disease; DLBCL diffuse large B cell lymphoma.
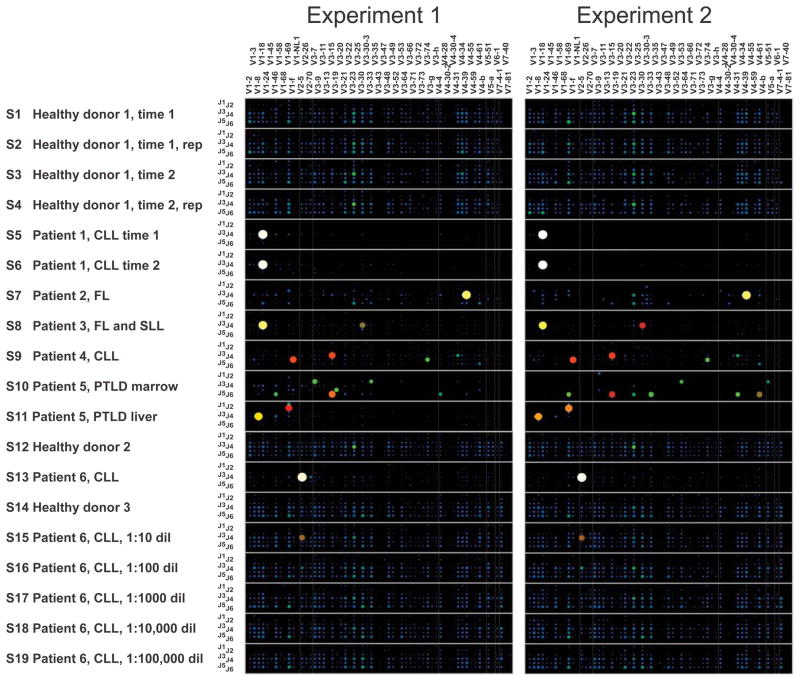
An overview of the IgH amplicon sequences in the data sets from Experiments 1 and 2 is shown in Fig. 2, with each point in the 2-dimensional grid for each sample indicating the V gene segment and the J gene segment used by a particular IgH V-D-J rearrangement. The size and color warmth of the circle at each point indicates what proportion of all sequences in the sample had the indicated V and J gene segment usage. Healthy peripheral blood lymphocyte populations showed a diverse use of different V and J gene segments, while samples that contained clonal IgH populations corresponding to lymphomas or chronic lymphocytic leukemia specimens were readily identified. Plots of the data showing the V, D and J segment usage are shown in Supplementary Figure S1.
Fig. 2. Immunoglobulin heavy chain V and J gene segment usage in healthy peripheral blood, oligoclonal or indeterminate specimens, and lymphoid malignancy specimens.
Barcoded IgH rearrangement libraries were PCR-amplified from genomic DNA of human specimens, pooled, and characterized by high-throughput pyrosequencing. Experiments 1 and 2 were independent experimental replicates beginning with different aliquots of the template DNA from each specimen. Each wide row represents the IgH sequences identified in a single sample. Samples (S1–S19) are labeled in the far-left column in the figure. The x-axis (across the top of the panels) indicates the V gene segment used in the receptor, and the y-axis (the column at the left of the panels) within each wide row represents the J gene segments used. The size and color of the circle at a given point indicates what proportion of all sequences in the sample used that particular combination of V and J gene segments. Sequences in which V, D or J segments or junctions could not be unambiguously assigned were filtered prior to generation of these plots. Rep: replicate sequence pool PCR amplified from an independent aliquot of template DNA; CLL: chronic lymphocytic leukemia; FL: follicular lymphoma; SLL: small lymphocytic lymphoma; PTLD: post-transplant lymphoproliferative disorder.
Evaluation of clonal malignancies
Human cancers are clonal proliferations of cells that have sustained mutational damage leading to dysregulated proliferation, survival, and response to the extracellular environment (18). Molecular clonality testing of IgH receptor and TCR gamma loci, accomplished with the use of PCR and capillary electrophoresis, is a helpful adjunct to morphological and immunophenotypic evaluation of suspected B or T cell malignancies (19). Blood or bone marrow samples from some patients give indeterminate or oligoclonal patterns of reactivity for a variety of reasons: Few lymphocytes may be present, there may be genuine oligoclonal lymphocyte populations, or clonal lymphocytes may have separately detected rearrangements from two chromosomes. We compared the results from DNA sequencing of the products of independent PCR replicates for such samples. One such difficult case is represented by the bone marrow and liver specimens from patient 5 in Table 1. The patient had undergone liver transplantation and subsequently developed a large B cell lymphoma in the liver as a manifestation of post-transplant lymphoproliferative disorder, a condition in which immunosuppression leads to B or T cell lymphomas that are typically associated with Epstein-Barr virus infection (Fig. 2). The patient’s bone marrow showed small lymphoid aggregates that were shown to contain B cells on morphological and immunohistochemical stain evaluation. Capillary electrophoresis sizing of VDJ rearrangements in the bone marrow sample gave support for a clonal population, but it was unclear whether this population represented involvement of the patient’s bone marrow by the lymphoma seen in the liver. The sequencing data resolved this uncertainty, showing no relationship between the liver lymphoma clone associated with IGHV1-8*01-IGHD2-8*01-IGHJ4*02 and the bone marrow B cells. Instead, a separate clonal B cell population that used gene segments IGHV3-15*04-IGHD3-9*01-IGHJ6*02 was present in the bone marrow. Patients with post-transplant lymphoproliferative disorder can develop multiple independent malignant clones, making the extra information provided by sequencing analysis of replicate PCR products particularly helpful. The other VDJ rearrangements detected in the patient’s bone marrow differed between the 2 replicate experiments, indicating the presence of small numbers of non-clonal B cells in the specimen. Another diagnostically challenging case, the chronic electrophoresis analysis. A consistent pattern was seen with deep sequencing of the sample. Finally, the two distinct V-D-J rearrangements in a lymph node from patient 3 indicated that there were two separate clonal B cell populations in the specimen, a conclusion supported by morphological and immunophenotypic evidence of two different B cell lymphomas (follicular lymphoma and small lymphocytic lymphoma) in the tissue.
Minimal residual disease testing by sequencing
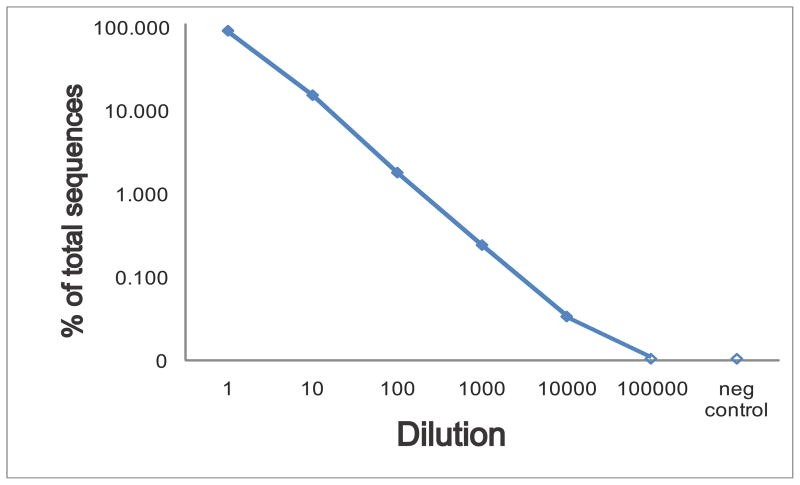
To evaluate the sensitivity of deep sequencing for detection of a clonal lymphoid population in a background of polyclonal cells, we performed serial 10-fold dilutions of a known clonal chronic lymphocytic leukemia blood sample into normal peripheral blood. The percentage of clonal sequences detected at each dilution is shown in Fig. 3 for Experiment 2, demonstrating detection down to a 1:10,000 dilution. This represents detection of 0.5 cells per microliter of blood when between 7,500 and 14,000 sequences are measured per sample of DNA template derived from approximately 10 microliters of blood.
Fig. 3. Titration of a chronic lymphocytic leukemia clonal sample into healthy peripheral blood.
Pooled barcoded IgH library sequencing was carried out on a series of 10-fold dilutions of a chronic lymphocytic leukemia blood sample (sample 13) into a healthy control blood sample (sample 14), to evaluate the sensitivity and linearity of high-throughput sequencing for detection of a known clonal sequence. The percentage of sequences matching the chronic lymphocytic leukemia clone in each diluted specimen is plotted on a log scale, with zero indicating that no sequences were detected. The counts of clonal sequences in each sample were as follows: CLL sample, 7805 clonal of 8612 total; healthy blood control, 0 clonal of 7518 total; 1:10 dilution, 2095 clonal of 13,717 total; 1:100 dilution, 156 clonal of 8674 total; 1:1000 dilution, 23 clonal of 9471 total; 1:10,000 dilution, 3 clonal of 8895 total; 1:100,000 dilution, 0 clonal of 6940 total. The negative control is the healthy donor blood sample used for diluting the clonal CLL sample. A second experiment measuring fewer sequences from independent PCR amplifications from the same samples detected the following number of clonal sequences in each sample: CLL sample, 422 clonal of 566 total; healthy blood control, 0 clonal of 270 total; 1:10 dilution, 189 clonal of 665 total; 1:100 dilution, 11 clonal of 230 total; 1:1000 dilution, 0 clonal of 344 total; 1:10,000 dilution, 0 clonal of 329 total; 1:100,000 dilution, 0 clonal of 208 total.
We next evaluated clinical specimens from patients with chronic lymphocytic leukemia who had undergone total lymphoid irradiation and anti-thymocyte globulin therapy followed by HLA-identical allogeneic peripheral blood progenitor cell transplantation (20–21), and compared the results of deep sequencing analysis to results from patient clone-specific real-time PCR assays (Table 2). In these experiments, the patients with chronic lymphocytic leukemia were different from the patients tested in our initial experiments described in Table 1, and the minimal residual disease sequencing was performed in a separate instrument run. Real-time PCR assay results were reported as confidently positive if at least 100 copies/μg of template DNA were detected. Table 2 demonstrates that all specimens showed agreement between the high-throughput sequencing data and real-time PCR assay, although for the lowest confidently positive real-time PCR result for chronic lymphocytic leukemia patient A, the clone was detected in only one of the two high-throughput sequencing sample replicates.
Table 2.
Comparison of High-Throughput Sequencing With Real-Time PCR Minimal Residual Disease Monitoring Assays.
| Patient | Specimen | Clone Copies1 | Total Sequences | % | Clone Copies2 | Total Sequences | % | RT-PCR (copies/μg) |
|---|---|---|---|---|---|---|---|---|
| CLL A sample 1 | Diagnostic Lymph Node | 7,227 | 11,190 | 64.6 | 5745 | 8935 | 64.3 | >100,000 |
| CLL A sample 2 | Blood | 0 | 341 | 0.0 | 0 | 670 | 0.0 | 10 |
| CLL A sample 3 | Blood | 38 | 1,477 | 2.6 | 60 | 3350 | 1.8 | 1,485 |
| CLL A sample 4 | Blood | 0 | 588 | 0.0 | 0 | 1657 | 0.0 | 91 |
| CLL A sample 5 | Blood | 0 | 430 | 0.0 | 0 | 491 | 0.0 | 37 |
| CLL A sample 6 | Bone Marrow | 0 | 1,471 | 0.0 | 21 | 2991 | 0.7 | 314 |
| CLL B sample 1 | Diagnostic Bone Marrow | 2,461 | 4,363 | 56.4 | 1964 | 3581 | 54.8 | >100,000 |
| CLL B sample 2 | BM | 1,080 | 1,974 | 54.7 | 1656 | 3002 | 55.2 | 5,496 |
| CLL B sample 3 | Blood | 0 | 162 | 0.0 | 0 | 208 | 0.0 | 24 |
| CLL B sample 4 | Blood | 0 | 114 | 0.0 | 0 | 117 | 0.0 | 10 |
| CLL B sample 5 | Bone Marrow | 188 | 493 | 38.1 | 343 | 1127 | 30.4 | 944 |
| Unrelated CLL | Blood | 0 | 5326 | 0.0 | 0 | 7673 | 0.0 | |
| Normal Control | Tonsil | 0 | 14,007 | 0.0 | 0 | 5167 | 0.0 |
For each patient specimen, IgH rearrangements were amplified from 200ng of genomic DNA of the indicated specimen types using barcoded primers adapted for 454 pyrosequencing. The IgH rearrangement libraries were pooled and sequenced. The number of clonal sequences (matching the initial diagnostic specimen clone) and the total number of sequences obtained are listed.
first replicate;
second replicate
Data from pyrosequencing were compared to the results of custom quantitative real-time PCR assays designed to amplify the patient’s malignant clonal sequence. The RT-PCR results were considered positive if >100 copies per microgram of template DNA were detected.
Peripheral blood B cell repertoire in healthy subjects
To identify potentially expanded B cell clones within healthy peripheral blood, we looked for independent occurrences of “coincident” IgH sequences (identical V, D, and J segments, and identical V-D and D-J junction sequences) in independent pools from the same individual. Such coincidences could have resulted from clonally related cells; indeed, clonal relationships are likely for a majority of these coincidences, given both the diversity of the potential repertoire of IgH rearrangements and the absence of rearrangements found in this individual from comparable sequence samples from different individuals. We note that any population with a limited IgH rearrangement repertoire would be expected to show large numbers of such coincidences. Instead, we observed only small numbers of coincident sequences in our data. From six independent amplification pools derived from the blood of a single individual at one time point, we observed only 19 potential coincidences from a total of 10,921 distinct IgH rearrangements sequenced. Seven independent amplification pools from a second time point (14 months later) gave comparable results (25 potential coincidences from a total of 7,450 distinct rearrangements sequenced) (Table 3).
Table 3.
Coincident Sequences in a Healthy Donor’s Peripheral Blood at 2 Time-points.
| T1r2 | T1r3 | T1r4 | T1r5 | T1r6 | T2r1 | T2r2 | T2r3 | T2r4 | T2r5 | T2r6 | T2r7 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1, replicate 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 0 | 2 | 2 | 1 |
| replicate 2 | 1 | 1 | 0 | 4 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | |
| replicate 3 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | ||
| replicate 4 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | 3 | 1 | |||
| replicate 5 | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 2 | ||||
| replicate 6 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | |||||
| T2, replicate 1 | 0 | 1 | 1 | 1 | 1 | 1 | ||||||
| replicate 2 | 0 | 2 | 2 | 0 | 1 | |||||||
| replicate 3 | 1 | 2 | 0 | 2 | ||||||||
| replicate 4 | 2 | 0 | 2 | |||||||||
| replicate 5 | 1 | 0 | ||||||||||
| replicate 6 | 5 |
IgH rearrangements from peripheral blood mononuclear cells of a healthy blood donor were PCR amplified in multiple independent replicate PCR reactions and sequenced. The table shows the number of identical sequences detected in more than one replicate (termed “coincident sequences”). Blood samples from two time-points separated by 14 months were analyzed. Sequences from different replicates were considered to be coincident sequences if they shared the same V, D, and J segment usage as well as the same V-D and D-J junctional nucleotide sequences. T1, initial timepoint; T2, second timepoint 14 months later; r1, replicate 1.
It is noteworthy that we see only slightly fewer coincidences when comparing aliquots between the two time points (0.76 coincidences per sample comparison versus 1.22 for comparisons within the same time point). Although the difference is statistically significant (P<0.05; Fischer’s exact 1-tailed), the modest ratio between intra-temporal and inter-temporal coincidence levels indicates a considerable degree of persistence in the clonal populations in this individual. The numbers of coincident sequences observed when comparing sequence data from any two aliquots provide strong evidence for substantial diversity in the IgH repertoire. Minimal estimates obtained using approaches similar to the “birthday problem” in probability theory (22) yield a lower bound of approximately 2 million different IgH rearrangements in these samples. The analysis leading to this lower bound estimate does not yield an upper bound on repertoire; in particular, it is not possible from these data to rule out a category of IgH rearrangements that are very diverse but present in single or low copy number in the approximately 2×109 B cells in peripheral blood. Thus the true complexity of the blood IgH repertoire could certainly be much greater than 2×106.
In addition to the total complexity of the IgH pool, it is of interest to evaluate the degree to which clonal cell populations above a certain size are present in normal peripheral blood. No sequence was identified in more that described above, we can derive an upper bound for the most abundant IgH rearrangements. For the healthy individual examined in these experiments, this analysis yields a maximum contribution to the sequence pool of 1/1000 for any individual clone (P<0.01) in this individual (23).
Within these experimental estimates of the lower bound of IgH repertoire size, and the upper bound of largest clone size, a variety of combinations of clonally expanded populations of different sizes could give rise to our observed data. Estimation of the upper limit of the IgH repertoire would require much more extensive sequencing to evaluate the extent of single-copy or very small clonal expansions of B cells, and would require characterization of a significant fraction of the blood volume of a healthy donor, which presents ethical concerns. It should be noted that this analysis of the blood does not exclude the possibility that other tissues may contain B cells that are clonally related to circulating cells, and does not address the exchange of B cells between the blood and other hematolymphoid compartments of the body. The sequences found in multiple replicates performed with blood from the healthy donor characterized in Table 3 are presented in Supplementary Table S4.
Diversity of clonal B cell expansions in healthy subjects of various ages
We extended our analysis of healthy human patients to an additional 23 subjects ranging in age from 19 to 79 years by sequencing sixfold replicate samples of peripheral blood IgHs from each individual. We detected considerable inter-individual variation in the number of expanded lymphocyte clones and expanded clone sizes (Table 4). Using an analysis similar to that performed for the healthy donor in Table 3, we calculated the minimum IgH repertoire size and the largest clone size for these additional subjects. Our data confirm that at least 15 of the 23 additional normal human samples had IgH pools of greater than 1,000,000 different rearrangements. Although the additional eight individuals may have comparable diversity, the lower bound estimates were somewhat lower, relative to the other 15 subjects, because of the greater numbers of weakly amplified clones detected and the lower total yield of sequences from these samples. For a majority of the healthy samples, no sequence appeared in more than two of six sequenced DNA aliquots; for these individuals, this places an upper limit of 0.1–0.3% of the measured B cell repertoire that could be dedicated to any single clone, similar to the results from the individual in Table 3.
Table 4.
Coincident IgH Sequences in Peripheral Blood of Healthy Donors of Various Ages.
| Age | Total Sequences | Coincidences | Minimum Diversity | ||||
|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | |||
| 19 | 19368 | 22 | 0 | 0 | 0 | 0 | 2,136,616 |
| 20 | 12598 | 61 | 0 | 2 | 0 | 0 | 704,883 |
| 23 | 6964 | 11 | 0 | 0 | 0 | 0 | 1,133,759 |
| 25 | 6522 | 10 | 0 | 0 | 0 | 0 | 1,328,380 |
| 31 | 4086 | 10 | 1 | 1 | 0 | 0 | 474,366 |
| 32 | 6112 | 9 | 0 | 0 | 0 | 0 | 1,328,380 |
| 35 | 5358 | 4 | 0 | 0 | 0 | 0 | 1,860,053 |
| 37 | 5253 | 4 | 1 | 1 | 0 | 0 | 1,973,903 |
| 38 | 2173 | 18 | 2 | 1 | 0 | 0 | 70,876 |
| 42 | 4094 | 11 | 0 | 0 | 0 | 0 | 381,515 |
| 44 | 2249 | 3 | 0 | 0 | 0 | 0 | 438,241 |
| 45 | 6781 | 65 | 2 | 2 | 0 | 0 | 325,619 |
| 45 | 7697 | 12 | 0 | 0 | 0 | 0 | 1,409,687 |
| 50 | 6841 | 6 | 1 | 0 | 0 | 0 | 1,718,401 |
| 54 | 10822 | 13 | 1 | 0 | 0 | 1 | 3,369,228 |
| 55 | 3426 | 7 | 0 | 0 | 0 | 0 | 513,469 |
| 60 | 5173 | 8 | 3 | 0 | 0 | 0 | 704,883 |
| 61 | 5092 | 1 | 0 | 0 | 0 | 0 | 6,349,446 |
| 68 | 7028 | 11 | 1 | 2 | 0 | 1 | 1,897,254 |
| 70 | 5552 | 10 | 0 | 0 | 0 | 0 | 1,276,797 |
| 75 | 7064 | 5 | 0 | 1 | 0 | 0 | 3,303,164 |
| 78 | 5895 | 4 | 0 | 0 | 0 | 0 | 3,051,613 |
| 79 | 7127 | 11 | 0 | 0 | 0 | 0 | 1,587,537 |
Peripheral blood samples from 23 healthy donors of ages ranging from 19 to 79 years old were analyzed by deep-sequencing IgH rearrangements in 6 replicates from each sample. The number of distinct sequences detected in more than one replicate (termed “coincident sequences”) from each individual is tabulated below. Sequences from different replicates were considered to be coincident sequences if they shared the same V, D, and J segment usage as well as the same V-D and D-J junctional nucleotide sequences. Calculation of the minimum IgH repertoire diversity in each patient as indicated by the number of coincident sequences detected is described in the Methods section.
Two of the apparently healthy blood donors in our sample set had expanded B cell clones that were large enough to be detected in all 6 sequencing replicates. The size of these larger clones can be estimated by the expanded clonal sequence’s proportion of total sequences obtained from these patients: For the 54-year-old patient this value was 0.15%, while for the 68-year-old patient the value was 1.5% of the total sequences.
These data demonstrate detection of clonal populations that make up greater than 0.1% of the total B cell population is readily possible with the small blood samples used for this work (less than 0.1 ml of blood was sufficient for the multiple replicates from these specimens). Further, these results suggest that searches for persistent pre-malignant or pathological clonal populations at the 0.1% level might be facilitated in certain cases by the limited set of amplified candidates in the normal repertoire.
Deep sequencing data sets of this kind should enable explicit detection of preferentially rearranged or selected combinations of V, D or J segments in IgHs in specific populations. Using the healthy control specimens in our current data sets, we have seen evidence of preferential pairwise segment associations for at least 3 combinations (D2-2 with J6, D3-22 with J3, and D3-3 with J6) across the group of individuals. Overrepresentation of these D/J combinations (i.e. a frequency of the DJ combination that is greater than the products of the D and J frequencies) was observed in 122/138, 113/138, and 119/138 sequenced aliquots, respectively. With a false discovery rate of less than 10−7 (no examples of overrepresentation in this number of aliquots were found in 107 randomly shuffled datasets), these were the most consistent non-random associations seen with the dataset (certainly other associations might emerge from a larger dataset). We interpret these results as reflecting non-random character in rearrangement or selection in this specific population of individuals (Stanford’s blood donor pool in a fixed time frame). One could certainly expect (and it would be of great interest to follow) different specific nonrandom characters in other populations with distinct histories of community immune response and genetic compositions.
Discussion
Modern DNA sequencing methods open a new window of investigation into the complex gene rearrangements necessary for human lymphocyte function. Our results using multiplexed barcoded IgH sequencing of multiple replicate samples of blood from 24 healthy subjects represent the most extensive characterization to date of human B cell populations. For a majority of the healthy individuals, our results were sufficient to place a lower limit of 1,000,000 on the number of distinct IgH rearrangements in circulating lymphocytes, and an upper bound of 0.1–0.3% of total B cells on the representation of any single clone within the repertoire. A small number of individual amplified clones with greater representation were observed in healthy individuals in our sample set, with the largest clonal populations (seen in patients aged 54- and 68-years-old) accounting for 0.15–1.5% of total sequences of the observed sequence space from circulating B cells. These larger expanded clones may be the result of physiological responses to environmental antigens or pathogens; alternatively, these could represent the precursors to lymphoid malignancies, such as chronic lymphocytic leukemia, that have a strong association with advanced patient age. Recent and older literature describing monoclonal B cell lymphocytosis (MBL) using multi-parameter flow cytometry assays to detect B cells with aberrant surface protein expression has indicated that between 5–12% of adults have these atypical B cell populations, and essentially all patients who develop chronic lymphocytic leukemia can be shown to have had preceding MBL (14, 24, 25). An important caveat is that most patients who show MBL do not go on to develop chronic lymphocytic leukemia (24, 26). High-throughput immune receptor sequencing provides an unprecedented degree of sensitivity and specificity in tracking monoclonal B cell expansions and enables detection of clonal B cell populations that do not show aberrant cell surface marker expression; it remains to be seen whether this augmented form of tracking will be of use in dissecting the additional clinical and molecular variables that lead some clonal expansions to progress to frank leukemias.
Deep sequencing of IgH rearrangements simplifies the assessment of overt populations of suspected malignant B cells in clinical samples and shows preliminary success in minimal residual disease testing after treatment of leukemia patients. A substantial advantage of the minimal residual disease detection approach used here is that all patient samples can be analyzed with a single uniform assay, rather than having to tailor individual real-time PCR assays to each patient’s clonal malignant sequence and to validate these assays individually as unique clinical tests, an expensive and laborious process likely to limit the accessibility of minimal residual disease (MRD) testing. Having a sequence-based assay that can detect variants from the original malignant clonal sequences present at diagnosis may be an advantage in screening for disease relapse. Recent microarray-based data from studies of acute lymphoblastic leukemias suggest that genomic copy number changes may occur relatively frequently at immune receptor loci between initial diagnostic specimens and relapse specimens (27). For the most sensitive detection of residual disease and clonal variants in a variety of B cell neoplasms, particularly those such as follicular lymphoma that have ongoing hypermutation of rearranged IgH gene loci, it will likely be advisable to use several different primer sets (for example, making use of all three framework regions of the IgH V genes) to avoid false-negative results that arise from mutations at primer-binding sites.
In a broader perspective, the deep-sequencing approach to lymphocyte population analysis may provide insights into autoimmune and infectious diseases, medical manipulations of the immune system such as vaccination, and harmful outcomes of current therapies, such as graft-versus-host disease after stem cell transplantation. We expect that immune receptor sequencing in medical scenarios that involve lymphoid malignancies or immune-mediated diseases will be broadly useful for gathering diagnostic, prognostic, and disease-monitoring information.
Supplementary Material
Fig. S1. V-D-J plots of healthy peripheral blood and lymphoid malignancies.
Fig. S2. Sequence complexity of healthy-donor blood specimens.
Table S1. Sequencing primers. (A) Primers used in Experiments 1 and 2. (B) Primers used in Experiment 3 (C) Primers used in healthy subjects of various ages.
Table S2. Patient specimens used in Experiment 2
Table S3. Number of sequences determined per specimen
Tables S4. Sequences found in more than one replicate from healthy-donor blood samples
Acknowledgments
We thank Kenneth Weinberg, Dan Arber, Poornima Parameswaran, Gil Chu, Perry Blackshear, Gerald Marti, Alex Lucas, Mark Davis, Bob Ohgami, Ron Levy, Shoshona Levy, Sandy Feng, Claus Niemann, David Lewis, Ash Alizadeh, Steve Galli, Emmanuel Mignot, Magali Fontaine, Bill Robinson, May Han, Yaso Natkunam, Andrew Collins, and members of our laboratories for helpful discussions, Chaya Krishna for excellent technical support. We are grateful to the Lucille-Packard Child Health Research Program for support of research materials, and for individual support from the Walter V. and Idun Berry Fellowship program (SDB), NCI (P01CA049605 [JLZ]), NIGMS (T32GM07365-34 [ELM]), the Stanford VPUE Research Program (LNZ), and the Stanford Immunity, Transplantation, and Infectious Disease Institute (KDN, KCN).
Footnotes
This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencetranslationalmedicine.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
A complete electronic version of this article and other services, including high-resolution figures, can be found at: http://stm.sciencemag.org/content/1/12/12ra23.full.html
Information about obtaining reprints of this article or about obtaining permission to reproduce this article in whole or in part can be found at: http://www.sciencemag.org/about/permissions.dtl
Conflicts of Interest
Boyd, Marshall, Merker, Maniar, Zhang, Sahaf, Jones, Nadeau, Nguyen, Miklos, Zehnder, Fire: None Simen, Hanczaruk, and Egholm are employees of 454 Life Sciences, A Roche Company
Authorship Contributions
Designed research: Boyd, Fire, Marshall, Merker, Miklos, Sahaf, Zehnder, Jones
Performed research: Boyd, Marshall, Merker, Zhang, Sahaf, Jones, Nguyen, Simen, Hanczaruk
Contributed analytical tools and reagents: Fire, Boyd, Marshall, Maniar, Zhang, Egholm, Simen, Hanczaruk, Zehnder, Nadeau, Miklos
Analyzed data: Boyd, Fire, Marshall, Maniar, Zhang, Simen, Sahaf, Miklos, Zehnder, Jones
Wrote manuscript: Boyd, Marshall, Fire
Read and approved manuscript: all authors
References and Notes
- 1.Schatz DG. Antigen receptor genes and the evolution of a recombinase. Semin Immunol. 2004;16:245–256. doi: 10.1016/j.smim.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 3.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 4.Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 5.Lim A, Luderschmidt S, Weidinger A, Schnopp C, Ring J, Hein R, Ollert M, Mempel M. The IgE repertoire in PBMCs of atopic patients is characterized by individual rearrangements without variable region of the heavy immunoglobulin chain bias. J Allergy Clin Immunol. 2007;120:696–706. doi: 10.1016/j.jaci.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson SM, Wilson PC, James JA, Capra JD. Human B cell subsets. Adv Immunol. 2008;98:151–224. doi: 10.1016/S0065-2776(08)00405-7. [DOI] [PubMed] [Google Scholar]
- 8.Rezuke WN, Abernathy EC, Tsongalis GJ. Molecular diagnosis of B- and T-cell lymphomas: fundamental principles and clinical applications. Clin Chem. 1997;43:1814–1823. [PubMed] [Google Scholar]
- 9.Arber DA. Molecular diagnostic approach to non-Hodgkin’s lymphoma. J Mol Diagn. 2000;2:178–190. doi: 10.1016/S1525-1578(10)60636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayala HA, Rawstron AC, Hillmen P. Minimal residual disease assessment in chronic lymphocytic leukaemia. Best Pract Res Clin Haematol. 2007;20:499–512. doi: 10.1016/j.beha.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Ladetto M, Donovan JW, Harig S, Trojan A, Poor C, Schlossnan R, Anderson KC, Gribben JG. Real-Time polymerase chain reaction of immunoglobulin rearrangements for quantitative evaluation of minimal residual disease in multiple myeloma. Biol Blood Marrow Transplant. 2000;6:241–253. doi: 10.1016/s1083-8791(00)70006-1. [DOI] [PubMed] [Google Scholar]
- 13.Rawstron AC, Villamor N, Ritgen M, Bottcher S, Ghia P, Zehnder JL, Lozanski G, Colomer D, Moreno C, Geuna M, Evans PA, Natkunam Y, Coutre SE, Avery ED, Rassenti LZ, Kipps TJ, Caligaris-Cappio F, Kneba M, Byrd JC, Hallek MJ, Montserrat E, Hillmen P. International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia. Leukemia. 2007;21:956–964. doi: 10.1038/sj.leu.2404584. [DOI] [PubMed] [Google Scholar]
- 14.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, Marti GE, Caporaso NE. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–667. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinna D, Corti D, Jarrossay D, Sallusto F, Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur J Immunol. 2009;39:1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wardemann H, Nussenzweig MC. B-cell self-tolerance in humans. Adv Immunol. 2007;95:83–110. doi: 10.1016/S0065-2776(07)95003-8. [DOI] [PubMed] [Google Scholar]
- 17.Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, Fire AZ. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 19.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 20.Kohrt HE, Turnbull BB, Heydari K, Shizuru JA, Laport GG, Miklos DB, Johnston LJ, Arai S, Weng W-K, Hoppe RT, Lavori PW, Blume KG, Negrin RS, Strober S, Lowsky R. TLI and ATG conditioning with low risk of graft-versus-host disease retains antitumor reactions after allogeneic hematopoietic cell transplantation from related and unrelated donors. Blood. 2009;114:1099–1109. doi: 10.1182/blood-2009-03-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowsky R, Takahashi T, Liu YP, Dejbakhsh-Jones S, Grumet FC, Shizuru JA, Laport GG, Stockerl-Goldstein KE, Johnston LJ, Hoppe RT, Bloch DA, Blume KG, Negrin RS, Strober S. Protective conditioning for acute graft-versus-host disease. N Engl J Med. 2005;353:1321–1331. doi: 10.1056/NEJMoa050642. [DOI] [PubMed] [Google Scholar]
- 22.The “Birthday Problem” refers to the calculation of probability that at least two individuals will share a single birthday in a group of n people. The large number of possible pairwise combinations from a group of n [the number is n*(n−1)/2] makes this probability surprisingly high, even with a value of n that is much fewer than the number of days in a year. This type of calculation is readily expanded to coincidences between groups of individuals, and to “value” spaces other than “days of the year”. Such calculations have been used extensively to evaluate minimum diversity in populations (e.g., Schnabel, Z.E. 1938. The estimation of the total fish population of a lake. American Mathematician Monthly 45:349-352.)
- 23.Materials, experimental procedures, and computational methods are described in detail in the online supplement section.
- 24.Dagklis A, Fazi C, Sala C, Cantarelli V, Scielzo C, Massacane R, Toniolo D, Caligaris-Cappio F, Stamatopoulos K, Ghia P. The immunoglobulin gene repertoire of low-count chronic lymphocytic leukemia (CLL)-like monoclonal B lymphocytosis is different from CLL: diagnostic implications for clinical monitoring. Blood. 2009;114:26–32. doi: 10.1182/blood-2008-09-176933. [DOI] [PubMed] [Google Scholar]
- 25.Nieto WG, Almeida J, Romero A, Teodosio C, Lopez A, Henriques AF, Sanchez ML, Jara-Acevedo M, Rasillo A, Gonzalez M, Fernandez-Navarro P, Vega T, Orfao A. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009;114:33–37. doi: 10.1182/blood-2009-01-197368. [DOI] [PubMed] [Google Scholar]
- 26.Rawstron AC, Bennett FL, O’Connor SJ, Kwok M, Fenton JA, Plummer M, de Tute R, Owen RG, Richards SJ, Jack AS, Hillmen P. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359:575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 27.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. V-D-J plots of healthy peripheral blood and lymphoid malignancies.
Fig. S2. Sequence complexity of healthy-donor blood specimens.
Table S1. Sequencing primers. (A) Primers used in Experiments 1 and 2. (B) Primers used in Experiment 3 (C) Primers used in healthy subjects of various ages.
Table S2. Patient specimens used in Experiment 2
Table S3. Number of sequences determined per specimen
Tables S4. Sequences found in more than one replicate from healthy-donor blood samples