Abstract
Protein tyrosine phosphatases (PTPs) are cysteine-dependent enzymes that play a central role in cell signaling. Organic hydroperoxides cause thiol-reversible, oxidative inactivation of PTP1B in a manner that mirrors the endogenous signaling agent hydrogen peroxide.
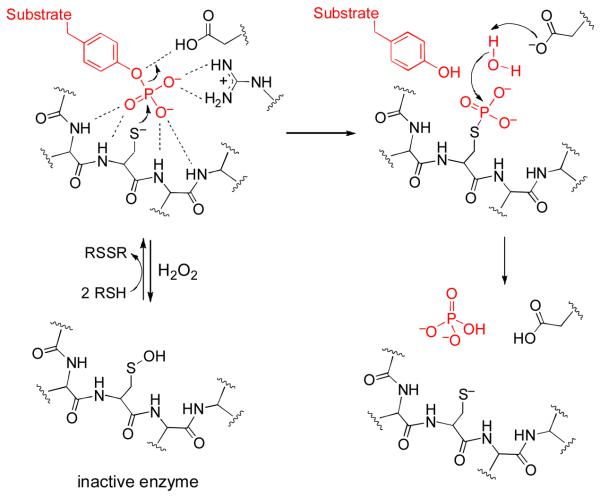
Protein tyrosine phosphatases (PTPs) are cysteine-dependent enzymes that catalyze the hydrolytic removal of phosphate groups from tyrosine residues in proteins (Scheme 1).1-3 Thus, PTPs, in concert with protein tyrosine kinases, play a central role in cell signaling by regulating the phosphorylation status and, in turn, the functional properties, of target proteins in various signal transduction pathways.4,5
Scheme 1.
The cellular activity of some PTPs is regulated by hydrogen peroxide (H2O2) that is produced as a second messenger in response to extracellular stimuli such as insulin, epidermal growth factor, and platelet derived growth factor.6-9 H2O2 inactivates PTPs via oxidation of the active site cysteine thiol residue to a sulfenic acid.6,10 In some cases, the cysteine sulfenic acid undergoes subsequent conversion to an active site sulfenyl amide linkage11-13 or disulfide.14,15 This type of oxidative inactivation of PTPs is slowly reversed upon reaction of the inactivated enzyme with biological thiols (Scheme 1).10-15
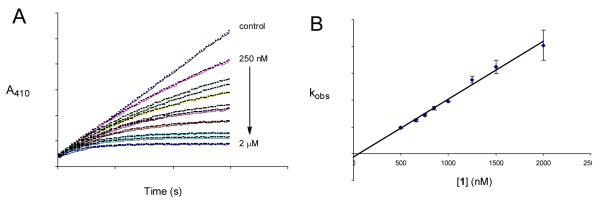
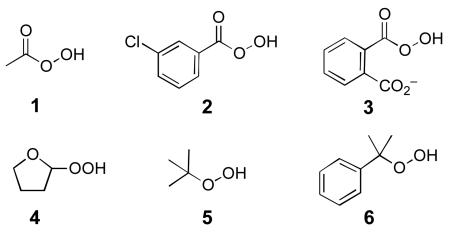
Given the biological importance of PTPs, it may be useful to identify organic reagents that regulate PTP activity through redox mechanisms.16-20 With this in mind, we examined the ability of several organic hydroperoxides (1-4) to effect thiol-reversible, oxidative inactivation of the archtypal member of the PTP family, PTP1B.21 We find that peracetic acid (1) is a potent, time-dependent inactivator of the catalytic subunit of PTP1B (a.a. 1-322) (Fig. 1A). The observation of time-dependent inactivation is consistent with a process involving covalent modification of the enzyme.22 A good linear fit is obtained in a plot of the apparent rates of inactivation versus concentration indicating that the inactivation reaction is a second-order process (Fig. 1B). The slope of the line reveals a rate constant of 2.1 ± 0.2 × 104 M−1 s−1 for the reaction of 1 with PTP1B. For comparison, hydrogen peroxide inactivates PTP1B with a rate constant of 10 M−1 s−1.10
Figure 1.
Rate of inactivation of PTP1B by compound 1. Panel A. Progress curves for the inactivation of PTP1B by 1. PTP1B was isolated as described previously19,20 and thiol-free samples of the enzyme were prepared by gel filtration of the protein through G25 Sephadex immediately prior to use according to reported procedures.19,20 Thiol-free PTP1B (25 nM final) was added to a solution of 1 (250 nM–2 μM) in 3,3-dimethyl glutarate buffer (50 mM, pH 7.0) containing the substrate p-nitrophenyl phosphate (p-NPP, 20 mM) at 23 °C. The apparent rate constant (kobs) for each concentration was calculated by the method of Voet and coworkers.23 Panel B. Determination of the second-order rate constant for the inactivation of PTP1B by 1. The apparent (pseudo-first-order) rate constants for inactivation of PTP1B by 1 were plotted versus concentration and the second-order rate constant for inactivation of PTP1B by 1 extracted from the slope. Inactivator concentrations >10-times the enzyme concentration were used in the plot. The concentration of 1 (and other peroxides) in stock solutions was determined by titration.24
The inactivation of PTP1B (250 nM) by 1 (1 μM, 3 min) is not reversed upon removal of excess inactivator by gel filtration of the enzyme through G25 Sephadex. The inactivation process is significantly slowed in the presence of phosphate ion, which is an active site-directed inhibitor of the enzyme (Ki = 17 mM). Specifically, treatment of the enzyme with 1 (15 μM) for 15 s inactivates 88% of the enzyme under standard conditions, while in the presence of phosphate ion (50 mM) only 49% inactivation is observed. Together, the results suggest that 1 inactivates PTP1B via covalent modification of the active site.
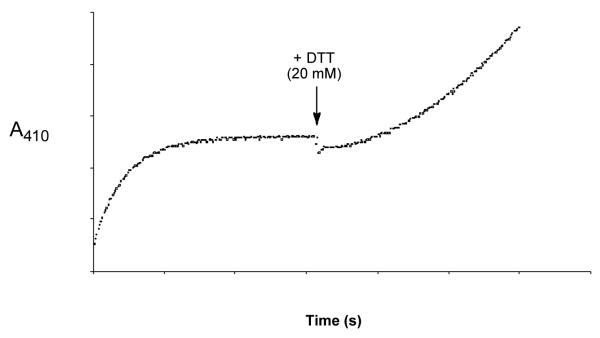
Inactivation of PTP1B by 1 can be reversed by treatment of the enzyme with thiols (Fig. 2). Specifically, inactivation of PTP1B (750 nM) with low concentrations of 1 (800 nM, 10 min, 23 °C), followed by treatment of the inactive enzyme with dithiothreitol (DTT, 100 mM final concentration), led to substantial return of enzymatic activity (73% of initial activity). When enzyme is not treated by 1, essentially all activity is retained under these conditions). This suggests that reaction of PTP1B with 1 predominantly converted the active site cysteine residue to the sulfenic acid oxidation state. In contrast, following treatment of the enzyme with higher concentrations of 1 (5 μM, 10 min, 23 °C), only 10% of the original activity returned upon treatment with DTT. Under these conditions, the active site cysteine presumably underwent significant amounts of “over-oxidation” to the sulfinic (RSO2H) or sulfonic acid (RSO3H) oxidation states that are not readily reduced to active enzyme by DTT. It is known that cysteine residues in redox regulated enzymes can be irreversibly “over-oxidized” to sulfinic and sulfonic acids by hydrogen peroxide under some conditions.12,14,25
Figure 2.
Inactivation of PTP1B by 1 can be reversed by addition of dithiothreitol (DTT). Thiol-free PTP1B (25 nM) was added to a solution of 1 (1 μM) in 3,3-dimethyl glutarate buffer (50 mM, pH 7.0) containing the substrate p-nitrophenyl phosphate (p-NPP, 20 mM) at 23 °C. When inactivation of the enzyme was nearly complete (300 s), a concentrated stock solution of DTT in water was added to give a final concentration of 20 mM. The experiment was conducted in a quartz cuvette and enzyme activity was measured by monitoring the enzyme-catalyzed release of p-nitrophenolate ion from the substrate p-NPP at 410 nm.
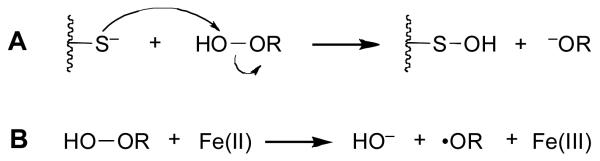
Two possible mechanisms can be considered for the inactivation of PTP1B by 1. Direct reaction of the active site cysteine residue with 1 would yield the inactive, sulfenic acid form of the enzyme (Scheme 2A).26-28 Alternatively, a metal-mediated Fenton-type reaction29 could generate diffusible oxygen radicals that might oxidize the active site cysteine residue.30 Two lines of experimentation argue against a Fenton-type process in the inactivation of PTP1B by 1. First, we find that addition of the classical oxygen radical scavenging agent mannitol29 has little effect on the inactivation of PTP1B by 1. Second, addition of FeSO4 (5 μM) inhibits, rather than facilitates, the inactivation process, presumably due the fact that Fe(II) mediates destruction of the peracid.29 With the Fenton-type mechanism excluded, it is reasonable to suspect26-28 that the inactivation of PTP1B by 1 proceeds via the mechanism shown in Scheme 2A.
Scheme 2.
Potential mechanisms for PTP inactivation by hydroperoxides.
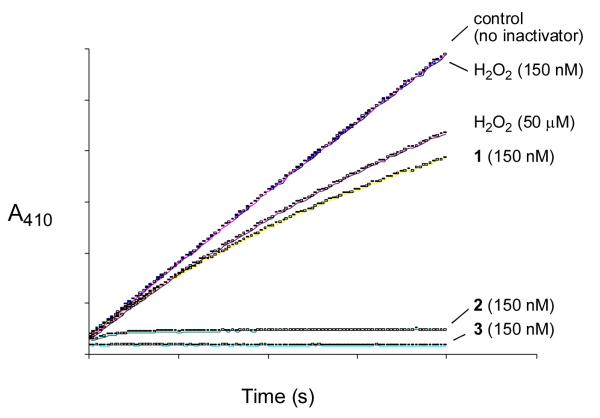
We find that aromatic peracids 2 and 3 inactivate PTP1B even more effectively than 1 (Fig. 3). For example, 2 and 3, at 150 nM concentrations, completely inactivate the enzyme within seconds. In contrast, when the enzyme is treated with the same concentration of 1, a significant amount of enzyme activity remains at 1 min (Fig. 3). For another point of comparison, 150 nM H2O2 has no measurable effect on enzyme activity under these conditions. Indeed concentrations of H2O2 approaching 100 μM are required to yield inactivation rates comparable to 150 nM 1 (Fig. 3). Finally, it is noteworthy that 1-3 can inactivate the enzyme in the presence of the biological thiol, glutathione.31 For example, treatment of PTP1B with 1 (5 μM, 15 s) causes a 29% loss of enzyme activity even in the presence of 1 mM glutathione.
Figure 3.
Comparision of the inactivation of PTP1B by compounds 1, 2, 3, and hydrogen peroxide. Thiol-free PTP1B (25 nM) was added to a cuvette containing 3,3-dimethyl glutarate buffer (50 mM, pH 7.0), p-NPP (10 mM), and the hydroperoxide of interest (1, 2, 3, or H2O2) at 24 °C. Enzyme inactivation progress curves showing the amount of PTP1B activity remaining as a function of time were obtained by monitoring the enzyme-catalyzed release of p-nitrophenolate ion from the substrate p-NPP at 410 nm. Note: curves for control “no inactivator” and 150 nM H2O2 are overlapping.
In addition, we examined the ability of 2-hydroperoxytetrahydrofuran (4) to inactivate PTP1B. This reagent was synthesized by the method of Gold and coworkers.32 We find that 4 inactivates PTP1B with a rate constant of 20.3 ± 0.8 M−1 s−1. This is comparable to the literature rate of 10 M−1 s−1 reported for hydrogen peroxide.10 Analogous to the inactivation of PTP1B by 1 and hydrogen peroxide, inactivation by 4 (50 uM, 10 min) is reversed upon reaction of the enzyme with DTT (54% return of activity, following treatment with 100 mM DTT for 1 h)..
In summary, we find that peracetic acid (1) is a potent oxidative inactivator of PTP1B. At low concentrations of 1 (≤ 1 μM), enzyme activity was readily recovered by treatment of the inactivated enzyme with thiol. Overall, the results suggest that 1 inactivates the enzyme via oxidation of the active site cysteine thiol residue to the sulfenic acid oxidation state. Peracetic acid (1) is approximately 2000 times more potent than hydrogen peroxide, a known endogenous regulator of cellular PTP activity. The superior activity of 1 is likely due to the intrinsically higher reactivity of peracids compared to hydrogen peroxide.28 Similarly, the superior inactivating power of 2 and 3 compared to 1 likely reflects their higher intrinsic reactivity as oxidants.33 In short, the trends reported here provide the useful predictive observation that the PTP-inactivating power of sterically unhindered organic peroxides will track closely with the leaving group ability of the substituent RO− that is displaced by thiol attack on the peroxide (Scheme 2A). In addition, inactivation by peracids such as 2 and 3 may be fast because the aromatic rings in these compounds bind noncovalently to PTP1B prior to the inactivation reaction via interactions with phosphotyrosine-binding residues such as Tyr46 and Phe182.
Tanner and Denu previously reported that t-butyl hydroperoxide (5) and cumene hydroperoxide (6) do not inactivate PTPs.10 As part of this work, we confirmed their findings (data not shown). These hydroperoxides react with small-molecule thiols at rates comparable to hydrogen peroxide;34 therefore, it seems likely that their failure to inactivate PTPs stems, not from an inherent lack of reactivity, but from the fact that the substituents on these tertiary hydroperoxides form an “umbrella” of steric bulk that prevents the peroxyl residue from penetrating the deep active site cavity of PTP1B to reach the catalytic cysteine residue. Indeed, as part of this study, we showed that the less hindered hydroperoxide 4 inactivates PTP1B with potency comparable to hydrogen peroxide. Evidently, the planar peracids 1-3 clearly are not sterically precluded from entering the enzyme's active site.
Our observation that organic peroxides have the ability to cause thiol-reversible, oxidative inactivation of PTPs has some significant implications. Inactivation of PTPs could contribute to the biological activity of some hydroperoxide-containing natural products.35 Furthermore, a variety of organic hydroperoxides including lipid and amino acid hydroperoxides are generated in cells.36,37 Indeed, while our work was in progress, Davies and coworkers reported the inactivation of PTP1B by amino acid hydroperoxides whose chemical structures, as yet, remain undefined.38 Our work provides quantitative measurement of the PTP-inactivating power of several structurally well defined organic hydroperoxides. In addition, it is interesting to note that 1 and other peracids are produced by a number of bacterial and eukaryotic enzymes.39-43 This offers the possibility that peracids could be involved in the physiological or pathophysiological regulation of PTPs and other cysteine-dependent enzymes. Overall our work provides new evidence that endogenous and exogenous organic hydroperoxides have the potential to regulate or dysregulate cellular signaling pathways. Continued investigation of PTP inactivation by this class of molecules is warranted.
Acknowledgements
We thank the National Institutes of Health for financial support of this work (CA 83925 and CA 119131) and the reviewers for helpful comments.
References and Notes
- 1.Zhang Z-Y. Acc. Chem. Res. 2003;36:385–392. doi: 10.1021/ar020122r. [DOI] [PubMed] [Google Scholar]
- 2.Neel BG, Tonks NK. Curr. Opin. Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 3.Stone RL, Dixon JE. J. Biol. Chem. 1994;269:31323–31326. [PubMed] [Google Scholar]
- 4.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 5.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Ostermann A, Godzik A, Hunter T, Dixon JE, Mustelin T. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Tonks NK. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Rhee SG. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 8.Mahedev K, Zilbering A, Zhu L, Goldstein BJ. J. Biol. Chem. 2001;276:21938–21942. doi: 10.1074/jbc.C100109200. [DOI] [PubMed] [Google Scholar]
- 9.D'Authrëaux B, Toledano MB. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 10.Denu JM, Tanner KG. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 11.Sivaramakrishnan S, Keerthi K, Gates KS. J. Am. Chem. Soc. 2005;127:10830–10831. doi: 10.1021/ja052599e. [DOI] [PubMed] [Google Scholar]
- 12.Salmeen A, Anderson JN, Myers MP, Meng T-C, Hinks JA, Tonks NK, Barford D. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 13.van Montfort RLM, Congreeve M, Tisi D, Carr R, Jhoti H. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 14.Sohn J, Rudolph J. Biochemistry. 2003;42:10060–10070. doi: 10.1021/bi0345081. [DOI] [PubMed] [Google Scholar]
- 15.Lee S-R, Yang K-S, Kwon J, Lee C, Jeong W, Rhee SG. J. Biol. Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 16.Bialy L, Waldmann H. Angew. Chem. Int. Ed. Eng. 2005;44:3814–3839. doi: 10.1002/anie.200461517. [DOI] [PubMed] [Google Scholar]
- 17.Johnson TO, Ermolieff J, Jirousek MR. Nature Rev. Drug Discov. 2002;1:696–709. doi: 10.1038/nrd895. [DOI] [PubMed] [Google Scholar]
- 18.Hooft van Huijsduijnen R, Sauer WHB, Bombrun A, Swinnen D. J. Med. Chem. 2004;47:4142–4146. doi: 10.1021/jm030629n. [DOI] [PubMed] [Google Scholar]
- 19.LaButti JN, Chowdhury G, Reilly TJ, Gates KS. J. Am. Chem. Soc. 2007;129:5320–5321. doi: 10.1021/ja070194j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seiner DR, Gates KS. Chem. Res. Toxicol. 2007;20:1315–1320. doi: 10.1021/tx700213s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z-Y. Ann. Rev. Pharmacol. Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 22.Silverman RB. The Organic Chemistry of Enzyme-Catalyzed Reactions. Academic Press; San Diego: 2000. [Google Scholar]
- 23.Kraut D, Goff H, Pai RK, Hosea NA, Silman I, Sussman JL, Taylor P, Voet JG. Mol. Pharmacol. 2000;57:1243–1248. [PubMed] [Google Scholar]
- 24.Sully BD, Williams PL. Analyst. 1962;87:653–657. [Google Scholar]
- 25.Jacob C, Holme AL, Fry FH. Org. Biomol. Chem. 2004;2:1953–1956. doi: 10.1039/b406180b. [DOI] [PubMed] [Google Scholar]
- 26.Ishii A, Komiya K, Nakayama J. J. Am. Chem. Soc. 1996;118:12836–12837. [Google Scholar]
- 27.Forlando P, Olabe JA, Magallanes JF, Blesa MA. Can. J. Chem. 1997;75:9–13. [Google Scholar]
- 28.Bruice TC. JCS Chem. Comm. 1983:14–15. [Google Scholar]
- 29.Halliwell B, Gutteridge JMC. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Zhang N, Schuchmann H-P, von Sonntag C. J. Phys. Chem. 1994;98:6541–6547. [Google Scholar]
- 31.Meister A, Anderson ME. Ann. Rev. Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 32.LIang G, Gannett P, Shi X, Zhang Y, Chen F-X, Gold B. J. Am. Chem. Soc. 1994;116:1131–1132. [Google Scholar]
- 33.Davies DM, Jones RM. J. Chem. Soc. Perkin 2. 1989:1323–1326. [Google Scholar]
- 34.Bell IM, Hilvert D. Biochemistry. 1993;32:13969–13973. doi: 10.1021/bi00213a029. [DOI] [PubMed] [Google Scholar]
- 35.Casteel DA. Nat. Prod. Rep. 1999;16:55–73. [Google Scholar]
- 36.Porter NA. Acc. Chem. Res. 1986;19:262–268. [Google Scholar]
- 37.Winterbourn CC, Parsons-Mair HN, Gebicki S, Janusz M, Davies MJ. Biochem. J. 2004;381:241–248. doi: 10.1042/BJ20040259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gracanin M, Davies MJ. Free Rad. Biol. Med. 2007;42:1543–1551. doi: 10.1016/j.freeradbiomed.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Bunik VI, Schloss JV, Pinto JT, Gibson GE, Cooper AJL. Neurochem. Res. 2007;32:871–891. doi: 10.1007/s11064-006-9239-z. [DOI] [PubMed] [Google Scholar]
- 40.Bernhardt P, Hult K, Kazlauskas RJ. Angew. Chem. Int. Ed. Eng. 2005;44:2742–2746. doi: 10.1002/anie.200463006. [DOI] [PubMed] [Google Scholar]
- 41.Bugg TDH. Bioorganic Chem. 2004;32:367–375. doi: 10.1016/j.bioorg.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Picard M, Gross J, Lübbart E, Tölzer S, Krauss S, van Pée K-H, Berkessel A. Angew. Chem. Int. Ed. Eng. 1997;36:1196–1199. [Google Scholar]
- 43.Hoffmann B, Tölzer S, Pelletier I, Altenbuchner J, van Pée KH, Hecht HJ. J. Mol. Biol. 1998;279:889–900. doi: 10.1006/jmbi.1998.1802. [DOI] [PubMed] [Google Scholar]