Abstract
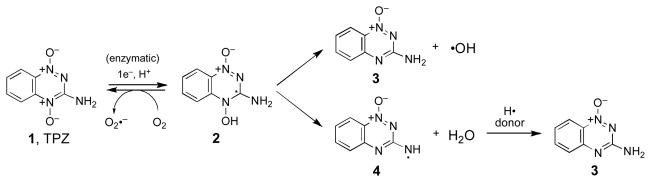
The antitumor agent 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine, TPZ, 1) gains medicinal activity through its ability to selectively damage DNA in the hypoxic cells found inside solid tumors. This occurs via one-electron enzymatic reduction of TPZ to yield an oxygen-sensitive drug radical (2) that leads to oxidatively generated DNA damage under hypoxic conditions. Two possible mechanisms have been considered to account for oxidatively generated DNA damage by TPZ. First, homolysis of the N–OH bond in 2 may yield the well known DNA-damaging agent, hydroxyl radical. Alternatively, it has been suggested that elimination of water from 2 generates a benzotriazinyl radical (4) as the ultimate DNA-damaging species. In the studies described here, the TPZ analogue 3-methyl-1,2,4-benzotriazine 1,4-dioxide (5) was employed as a tool to probe the mechanism of DNA damage within this new class of antitumor drugs. Initially, it was demonstrated that 5 causes redox-activated, hypoxia-selective oxidation of DNA and small organic substrates in a manner that is completely analogous to TPZ. This suggests that 5 and TPZ damage DNA by the same chemical mechanism. Importantly, the methyl substituent in 5 provides a means for assessing whether the putative benzotriazinyl intermediate 7 is generated following one-electron reduction. Two complementary isotopic labeling experiments provide evidence against the formation of the benzotriazinyl radical intermediate. Rather, a mechanism involving the release of hydroxyl radical from the activated drug radical intermediates can explain the DNA-cleaving properties of this class of antitumor drug candidates.
The compound 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine, TPZ, 1, Scheme 1) is currently undergoing a variety of phase I, II, and III clinical trials for the treatment of human cancers.1 TPZ gains medicinal activity from its ability to selectively damage DNA in the oxygen-poor (hypoxic) cells found inside solid tumors.2–8 This DNA-damage process begins with intracellular enzymatic reduction of TPZ to yield the drug radical intermediate (2, Scheme 1).9–12 In normally-oxygenated cells, 2 undergoes relatively harmless oxidation back to the parent drug (Scheme 1),9,10,13 while, under hypoxic conditions, the drug radical intermediate 2 leads to oxidatively generated DNA damage including hydroxylation of the nucleobases22,23 and strand breaks initiated by the abstraction of hydrogen atoms from the sugar-phosphate backbone of DNA.7–9,24–28
Scheme 1.
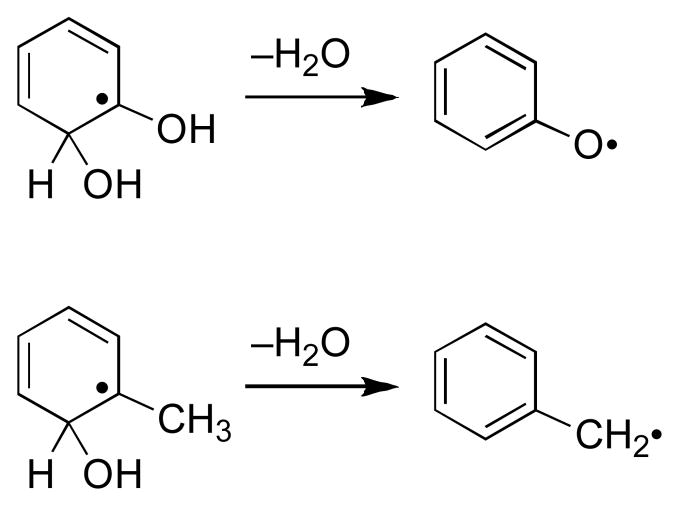
In the recent literature, two mechanisms have been considered to explain TPZ-mediated DNA damage. We have presented evidence22–24,26,29 supporting a mechanism involving homolysis of the N–OH bond in the neutral drug radical (2) to yield the mono-N-oxide metabolite 3 and the well known DNA-damaging agent hydroxyl radical (Scheme 1, upper branch).30 This radical fragmentation is calculated to be thermodynamically reasonable35 and finds a wealth of precedents in the organic literature. For example, the radical-induced fragmentation of Barton’s N-hydroxypyridinethione esters (Scheme 3) is directly analogous to that proposed for TPZ.36–42 In addition, several groups have studied similar photoinduced fragmentations of N-(alkoxy)pyridinium salts (Scheme 3).43–46 Alternatively, the results of pulse radiolysis experiments led to the suggestion that the activated intermediate 2 eliminates the elements of water from the exocyclic NH2 and the N–OH group to generate a benzotriazinyl radical (4) as the ultimate DNA-damaging species (Scheme 1, lower branch).47–50 The benzotriazinyl radical 4 has been generated via SO4•− oxidation of 3 and is, indeed, capable of oxidizing 2-deoxyribose and deoxyguanosine monophosphate;48 however, there is no direct evidence for the generation of this intermediate via bioreductive activation of TPZ. The dehydration process proposed for the formation of 4 finds some general precedent in radiation chemistry (Scheme 4). In particular, analogous eliminations of water from hydroxyl radical adducts of phenol and toluene have been reported,51,52 although to the best of our knowledge this type of transformation has not been observed in nitrogen-containing systems more closely related to the benzotriazine antitumor agents.
Scheme 3.
Scheme 4.
Development of novel 1,2,4-benzotriazine 1,4-dioxide drugs may be facilitated by improved understanding of the molecular mechanisms by which this new chemical class of antitumor agents damages DNA. In the studies described here, a TPZ analogue, 3-methyl-1,2,4-benzotriazine 1,4-dioxide (5), was employed as a tool to probe the mechanism of DNA damage by the 1,2,4-benzotriazine 1,4-dioxide antitumor drugs. First, we demonstrated that 5 causes redox-activated, hypoxia-selective oxidation of DNA and small organic substrates in a manner that parallels TPZ. Further, we found that one-electron reductive activation of 5 and TPZ under anoxic conditions generates analogous mono-N-oxide metabolites, 8 and 3, respectively.53 Having shown that the behavior of 5 mirrors TPZ, we recognized that the methyl group in this compound provides a “handle” for determining whether a benzotriazinyl radical intermediate (7) is generated following one-electron reduction of the heterocycle. Specifically, generation of the putative benzotriazinyl radical intermediate 7 in deuterated solvent mixtures would lead to formation of the deuterated, 3-(methyl-d)-1,2,4-benzotriazine 1-oxide (12) as a telltale metabolite. In the event, we find that bioreductive activation of 5 in solvent mixtures composed of CD3OD and D2O leads to the incorporation of less than 5% deuterium in the resulting mono-N-oxide metabolite. A complementary labeling experiment utilizing an analogue of 5 containing deuterium in the methyl group reveals no significant washout of label during bioreductive metabolism. Overall, the results provide evidence that benzotriazinyl radicals are not obligate intermediates in DNA damage by the 1,2,4-benzotriazine 1,4-dioxide family of antitumor agents. Rather, a mechanism involving the release of the well-known DNA-damaging agent hydroxyl radical from activated drug radical intermediates such as 2 and 6 may account for the DNA-cleaving properties of this family of drug candidates.
Results and Discussion
Hypoxia-Selective, Enzyme-Activated DNA Damage by 5
The hypoxia-selective cytotoxicity of 5 against human cancer cell lines is comparable to TPZ;54 however, the ability of 5 to damage DNA has not been examined previously. In the context of the mechanistic studies described here, it was first important to determine whether 5 generates redox-activated, hypoxia-selective DNA damage comparable to the lead compound TPZ.
Compound 5 and its expected bioreductive metabolites, 8 and 11, were synthesized by literature routes involving either BF3-catalyzed cyclization of a formazan precursor55 or PtO2-catalyzed cyclization of the 2-nitrophenylhydrazone of pyruvic acid,56 followed by N-oxidation using m-chloroperbenzoic acid.54 We employed either NADPH:cytochrome P450 reductase or xanthine/xanthine oxidase enzyme systems for the one-electron reductive activation of 5. These enzymes were chosen because NADPH:cytochrome P450 reductase, or a related enzyme,57 is thought to be responsible for in vivo activation of TPZ25,58,59 and xanthine/xanthine oxidase has been used successfully for the activation of TPZ in a variety of in vitro studies.9,23,26,60–62 For reactions carried out under anoxic conditions, molecular oxygen was removed from stock solutions by freeze-pump-thaw degassing and the assay mixtures prepared and incubated in an inert atmosphere glove bag.
We first explored the ability of 5 to generate strand breaks in double-stranded, supercoiled plasmid DNA. In this assay, single-strand cleavage converts supercoiled plasmid DNA (form I) to the open-circular form (form II).63–65 The two forms of plasmid DNA are then separated using agarose gel electrophoresis, the gel stained with a DNA-binding dye such as ethidium bromide, and the relative amounts of cut and uncut plasmid quantitatively determined by digital imaging analysis. The direct strand breaks (those not requiring thermal or basic workup) monitored in this type of experiment typically arise through abstraction of hydrogen atoms from the 2-deoxyribose backbone of DNA.32,33,66–69
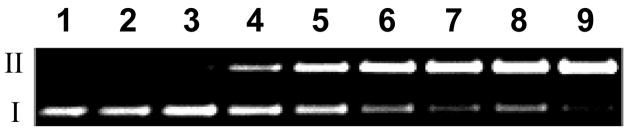
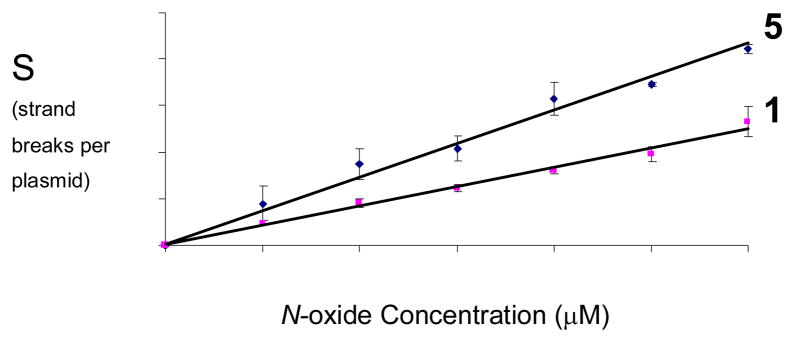
We find that compound 5 causes DNA strand cleavage when incubated with the NADPH:cytochrome P450 reductase enzyme system under anoxic conditions (Figure 1). The yields of DNA strand breaks generated by 5 are comparable to those produced by TPZ (Figures 1–3), with strand cleavage by 5 being somewhat superior to the parent drug TPZ. When the compound is incubated with the enzyme alone (no NADPH), NADPH alone (no enzyme), or with the complete enzymatic reducing system under aerobic conditions, no significant amounts DNA strand cleavage are observed (Figure 4). Compounds 8 and 11, the major metabolites expected53 to result from reductive metabolism of 5 (see below), do not cause DNA strand cleavage either alone or in the presence of the NADPH:cytochrome P450 reductase enzyme system under anoxic conditions (Figure 4). Comparable results were obtained using xanthine/xanthine oxidase as the enzymatic activating system (data not shown). Redox-activated DNA cleavage by 5 is substantially inhibited (60–90%) by classical radical-scavenging agents15 such as ethanol, methanol, t-butanol, DMSO, and mannitol (500 mM, Figure 4). Overall, the data indicate that one-electron reduction of 5 under anoxic conditions leads to direct DNA strand cleavage via radical mechanisms such as those characterized previously for TPZ.22–24,26,29
Figure 1.
DNA cleavage by various concentrations of reductively-activated 5 under anaerobic conditions. Supercoiled plasmid DNA (33 μg/mL, pGL-2 Basic) was incubated with 5 (25–150 μM), NADPH (500 μM), cytochrome P450 reductase (0.03 mU/μL), catalase (100 μg/mL), superoxide dismutase (10 μg/mL), sodium phosphate buffer (50 mM, pH 7.0), and desferal (1 mM) under anaerobic conditions at 25 °C for 4 h, followed by agarose gel electrophoretic analysis. Lane 1, DNA alone (S = 0.19 ± 0.02); lane 2, NADPH (500 μM) + reductase (0.03 mU/μL) (S = 0.2 ± 0.01); lane 3, 5 (150 μM) (S = 0.18 ± 0.03); lanes 4–9, NADPH (500 μM) + reductase (0.03 mU/μL) + 5 (25 μM, lane 4) (S = 0.46 ± 0.11); (50 μM, lane 5) (S = 0.71 ± 0.1); (75 μM, lane 6) (S = 0.81 ± 0.08); (100 μM, lane 7) (S = 1.13 ± 0.11); (125 μM, lane 8) (S = 1.22 ± 0.01); (150 μM, lane 9) (S = 1.46 ± 0.03). The values, S, represent the mean number of strand breaks per plasmid molecule and were calculated using the equation S = −ln fI, where fI is the fraction of plasmid present as in the supercoiled form I.
Figure 3.
Comparison of the efficiency of redox-activated DNA-cleavage by 5 and TPZ under anaerobic conditions. Supercoiled plasmid DNA (33 μg/mL, pGL-2 Basic) was incubated with TPZ or 5 (25–150 μM), NADPH (500 μM), cytochrome P450 reductase (0.03 mU/μL), catalase (100 μg/mL), superoxide dismutase (10 μg/mL), sodium phosphate buffer (50 mM, pH 7.0), and desferal (1 mM) under anaerobic conditions at 24 °C for 4 h, followed by agarose gel electrophoretic analysis. The values, S, are derived from agarose gel data such as that shown in Figures 1 and 2 and represent the mean number of strand breaks per plasmid molecule and were calculated using the equation S = −ln fI, where fI is the fraction of plasmid present as form I. Background cleavage in the untreated plasmid was subtracted to allow direct comparison of DNA strand cleavage yields between different experiments.
Figure 4.
Cleavage of supercoiled plasmid DNA by 5. All reactions contained DNA (33 μg/mL, pGL-2 Basic), sodium phosphate buffer (50 mM, pH 7.0), catalase (100 μg/ mL), superoxide dismutase (10 μg/mL), and desferal (1 mM) and were incubated under anaerobic conditions (except lane 9) at 25 °C for 3 h, followed by agarose gel electrophoretic analysis. Lane 1, DNA alone (S = 0.17 ± 0.03); lane 2, NADPH (500 μM) + NADPH:cytochrome P450 reductase (0.03 mU/μL) (S = 0.15 ± 0.01); lane 3, 5 (250 μM) + NADPH (500 μM) + reductase (0.03 mU/μL) (S = 1.01 ± 0.35); lanes 4–8, 5 (250 μM) + NADPH (500 μM) + reductase (0.03 mU/μL) + methanol (500 mM, lane 4) (S = 0.28 ± 0.04); ethanol (500 mM, lane 5) (S = 0.27 ± 0.03); tert-butyl alcohol (500 mM, lane 6) (S = 0.49 ± 0.03); DMSO (500 mM, lane 7) (S = 0.37 ± 0.13); mannitol (500 mM, lane 8) (S = 0.46 ± 0.06); lane 9, 5 (250 μM) + NADPH (500 μM) + reductase (0.03 mU/μL) + air (S = 0.17 ± 0.04); lane 10, mono-N-oxide 8 (250 μM) + NADPH (500 μM) + reductase (0.03 mU/μL) (S = 0.18 ± 0.06); lane 11, benzotriazine 11 (250 μM) + NADPH (500 μM) + reductase (0.03 mU/μL) (S = 0.18 ± 0.04); lanes 12–14, 5 alone (250 μM, lane 12) (S = 0.22 ± 0.07); 8 alone (250 μM, lane 13) (S = 0.16 ± 0.04); 11 alone (250 μM, lane 14) (S = 0.23 ± 0.08). The values, S, represent the mean number of strand breaks per plasmid molecule and were calculated using the equation S = −ln fI, where fI is the fraction of plasmid present as form I.
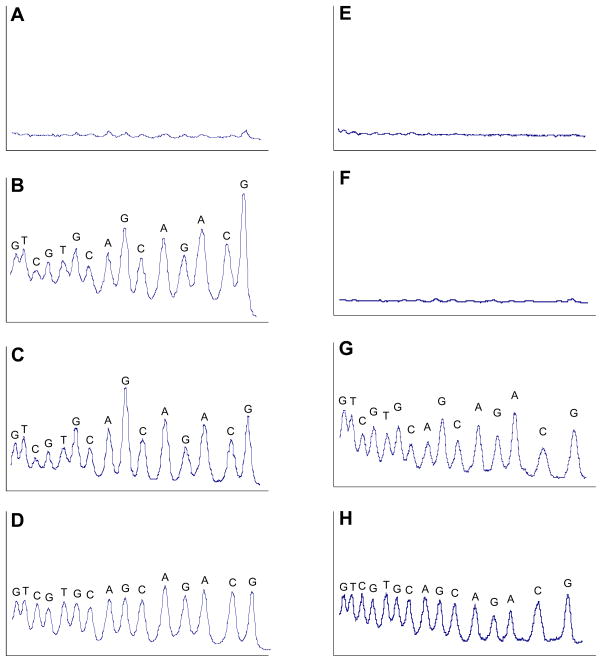
We also examined the sequence-specificity of strand cleavage by 5. In these experiments, a 30 base pair, 5′-32P-labeled oligodeoxynucleotide duplex was treated with 5 and the NADPH:cytochrome P450 reductase enzyme system under anoxic conditions. Following the DNA-damage reaction, the resulting 32P-labeled DNA fragments were resolved on a denaturing polyacrylamide sequencing gel and visualized by phosphorimager analysis. The relevant lanes from the gel are displayed as densitometry traces in which each cleavage band appears as a peak (Figure 5). We find that DNA strand cleavage by reductively-activated 5 occurs at every base pair in the duplex (Figure 5B). The pattern of sequence-independent DNA strand cleavage caused by 5 closely resembles that generated by TPZ (Figure 5C) and is generally characteristic of a highly oxidizing, small, diffusible species such as hydroxyl radical.30,31 We compared DNA strand cleavage by 5 and TPZ to that by an iron-EDTA system that generates hydroxyl radical (or a functionally equivalent species, Figure 5D).31 Comparison with the iron-EDTA cleavage lane reveals that both TPZ and 5 display some preference for cleavage at purine residues in the duplex. This result raises the possibility that iron-EDTA is not an appropriate control because system operates under aerobic conditions. Thus, we were driven to examine the sequence-specificity of DNA strand cleavage by hydroxyl radical. For this purpose, we employed a method developed by Macgregor involving the generation of hydroxyl radical via photolysis of hydrogen peroxide.97 This method is attractive because it can be carried out effectively under anaerobic conditions and there is no possibility for complications arising from any organic fragment that serves as a “delivery vehicle” for the HO• cleaving agent. In the event, we find that the anaerobic photolysis of H2O2 yields strand cleavage with almost no base specificity (Figure 5H). Indeed the result closely resembles DNA strand cleavage by the iron-EDTA system (compare Figure 5D and 5H). We carried out the photolysis reactions under the salt conditions (40 mM NaCl) employed by Macgregor.97 Therefore, it was necessary to examine strand cleavage by TPZ under identical buffer and salt conditions for comparison (Figure 5G). Control reactions under these conditions show little cleavage by photolysis in the absence of hydrogen peroxide (Figure 5F) or the enzyme system in the absence of TPZ (Figure 5E). It remains clear that the benzotriazine N-oxides cause DNA strand cleavage at every nucleotide, but with a subtle, yet significant, preference for cleavage at purine residues. In short, we believe that other experiments in our manuscript (vide infra and supra) seem best explained by the hypothesis that bioreductively-activated benzotriazine N-oxides decompose with release of hydroxyl radical. If so, why is the sequence specificity of strand cleavage by the N-oxides not identical to that of anaerobic hydroxyl radical?
Figure 5. Left side, A–D.
Comparison of DNA-cleavage patterns generated by enzymatically-activated TPZ, 5, and iron-EDTA. (A) Control; NADPH:cytochrome P450 reductase enzyme system (no 5), (B) TPZ activated by NADPH:cytochrome P450 reductase enzyme system under anaerobic conditions, (C) compound 5 activated by NADPH:cytochrome P450 reductase enzyme system under anaerobic conditions and, (D) the hydroxyl radical-generating Fe-EDTA system under aerobic conditions. DNA cleavage reactions were performed on a 30 base pair 5′-32P-labeled oligodeoxynucleotide duplex as described in the Experimental Section. Densitometer scans are from a portion of a 20% denaturing polyacrylamide gel and show the relative intensity of DNA cleavage at each base position. Lanes A, B and C were loaded with equal amounts (cpm) of labeled DNA and are plotted on the same vertical scale. Lane D, provided for comparison, is not plotted on the same vertical scale as A, B, and C. Right side, E–H. Comparison of DNA-cleavage patterns generated by enzymatically-activated TPZ and hydroxyl radical generated by the anaerobic photolysis of H2O2 in sodium phosphate buffer containing 40 mM NaCl. (E) Control, DNA treated with the NADPH:cytochrome P450 reductase enzyme system under anaerobic conditions in the absence of TPZ (F) Control, DNA subjected to photolysis under anaerobic conditions, (G) TPZ activated by NADPH:cytochrome P450 reductase enzyme system under anaerobic conditions in the same buffer and salt conditions employed for the peroxide photolysis, (H) DNA strand cleavage by hydroxyl radical generated via photolysis of H2O2. Lanes E and G and F and H were loaded with equal amounts (cpm) of labeled DNA and are plotted on the same vertical scale so they can be compared.
We offer two potential explanations for the sequencing data. First, although the experiments presented in the manuscript indicate that generation of the benzotriazinyl radical 7 is not a major process (see below), it remains possible that small amounts of the 7 could contribute to the observed sequence specificity. Previous work has shown that the benzotriazinyl radical is capable of oxidizing guanine residues.48 Alternatively, intermediate hydroxyl radical adducts with deoxypurines may undergo secondary reactions with the parent di-N-oxides, the mono-N-oxide metabolites, or the radical anions and these reactions may lead to the generation of labile lesions (and strand cleavage in our experiments). There is precedent for this general type of process. For example, addition of hydroxyl radical to the 5,6-double bond of guanine, followed by loss of water, yields the so-called (G-H)• radical (see Scheme 2 in ref. 70b). Importantly, subsequent oxidation of (G-H) • by superoxide radical is thought to generate hydantoin or imidazolone products (Schemes 13, 18, and 21 in ref. 70b) that may be base labile.71 In the context of sugar damage, it has been shown that TPZ and its metabolites can oxidize radical intermediates.98,99 The possibility for interaction of the N-oxides with initially-generated hydroxyl radical adducts with the purine bases is unique to the benzotriazine N-oxide DNA-damaging system and, therefore, provides a plausible molecular mechanism for the observed differences in the sequence specificity of strand cleavage between TPZ and “authentic” hydroxyl radical. The nature of the ultimate nucleobase products generated by TPZ-mediated DNA damage deserves further study.
In Vitro Metabolism of 5 Generates the Mono-N-Oxide 8 As the Major Product
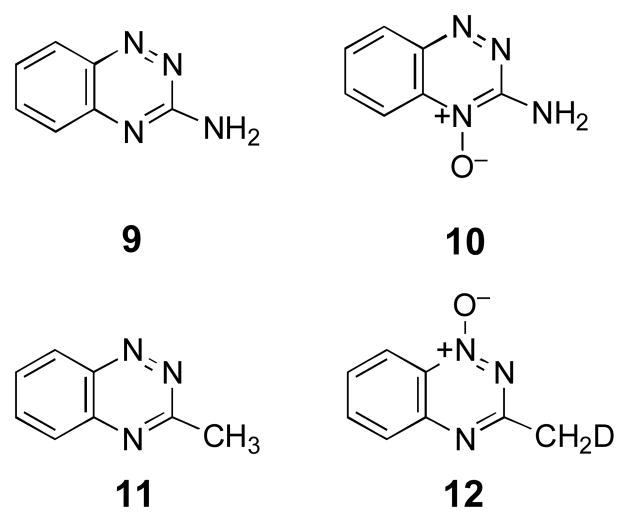
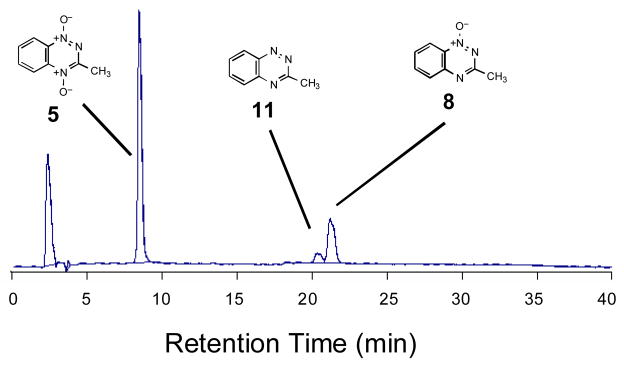
In vitro one-electron bioreductive activation of TPZ by enzyme systems such as NADPH:cytochrome P450 reductase or xanthine/xanthine oxidase produces the 3-amino-1,2,4-benzotriazine-1-oxide (3) as the major metabolite.9,53,72 Under some conditions, 3-amino-1,2,4-benzotriazine (9) is also observed along with small amounts of 3-amino-1,2,4-benzotriazine 4-oxide (10).9,53,72 A similar spectrum of metabolites is generated by cellular metabolism of TPZ.8,53,73 As part of the present investigation of the molecular mechanisms underlying strand cleavage by 5, we examined the products generated by one-electron reductive activation of this compound by the enzyme NADPH:cytochrome P450 reductase under anaerobic conditions. Reverse-phase HPLC analysis reveals two major products resulting from enzymatic activation of 5 (Figure 6). The products display the same retention times as authentic standards74 of 3-methyl-1,2,4-benzotriazine 1-oxide (8) and 3-methyl-1,2,4-benzotriazine (11). The identity of the metabolites was further confirmed by co-injection with authentic synthetic standards and LC/ESI-MS analysis operating in the positive ion mode. Thus, the major metabolites generated by one-electron bioreductive activation of 5 under anaerobic conditions are completely analogous to those produced by TPZ.
Figure 6.
HPLC analysis of products arising from in vitro metabolism of 5 by NADPH:cytochrome P450 reductase under anaerobic conditions. Compound 5 (500 μM) was incubated with NADPH:cytochrome P450 reductase (0.33 U/mL) and NADPH (500 μM) in sodium phosphate buffer (pH 7.0, 50 mM) containing desferal (1 mM) for 3 h, under anaerobic conditions. The resulting mixture was analyzed by reverse-phase HPLC, as described in the Experimental Section.
Isotopic labeling studies provide evidence that bioreductive activation of 5 does not generate the benzotriazinyl radical intermediate 7
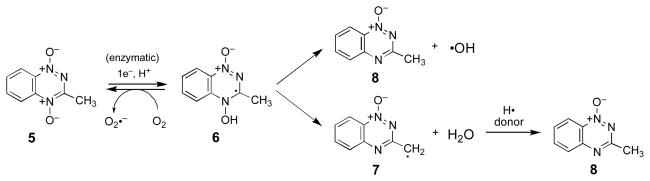
Overall, the results described above suggest that TPZ and 5 damage DNA by the same chemical mechanism. Thus, compound 5 may be employed as a tool to investigate the general mechanisms of DNA damage by the 1,2,4-benzotriazine 1,4-dioxide family of antitumor agents. In the context of 5, the two mechanisms under consideration here lead to the same metabolite 8 via chemically distinct pathways (Scheme 2, upper and lower routes). In the upper pathway, 8 is generated directly from 6 by homolytic fragmentation of the N–OH bond. On the other hand, in the lower pathway, compound 8 is formed only after the benzotriazinyl radical 7 abstracts a hydrogen atom from a donor molecule. The methyl group in 5 provides a means for detecting whether the benzotriazinyl radical intermediate (7) is generated following one-electron reduction of the heterocycle. Specifically, generation of 7 in deuterated solvent mixtures would lead to formation of the deuterated product 3-(methyl-d)-1,2,4-benzotriazine 1-oxide (12) as a telltale metabolite. For example, abstraction of a deuterium atom from a donor such as CD3OD would produce 12. Importantly, it is clear from the data shown in Figure 4 that methanol is able to quench the DNA-cleaving species generated upon one-electron reduction of 5. If hydrogen atom abstraction occurs at a position other than the methyl group in the resonance delocalized radical 7, tautomerization would intervene on the way to the observed metabolite 12 but, in CD3OD/D2O mixtures, deuterium incorporation would still be expected. Likewise, if 7 were to oxidize substrates via an electron transfer mechanism, incorporation of deuterium from CD3OD/D2O mixtures likely would result.
Scheme 2.
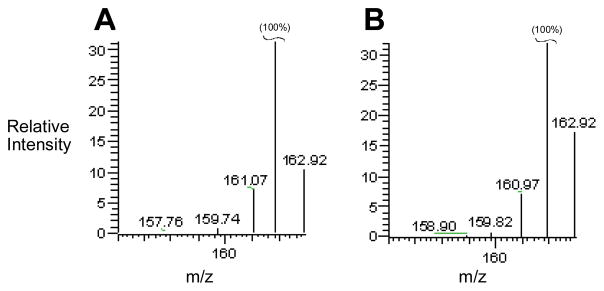
In accord with the experimental design described above, we carried out NADPH:cytochrome P450 reductase-mediated activation of 5 in deuterated sodium phosphate buffer containing the deuterium atom donor CD3OD (2 M) and analyzed the isotopic content of the mono-N-oxide product (8/12) by LC/ESI-MS operated in the positive ion mode. We find that bioreductive activation of 5 in solvent mixtures composed of CD3OD and D2O leads to the incorporation of less than 5% deuterium in the metabolite 8/12 (Figure 7).
Figure 7.
In vitro metabolism of 5 in deuterated buffer and in the presence of deuterated solvents CD3OD and D2O does not lead to incorporation of deuterium in the metabolite 8. Panel A shows mass spectrometric data obtained by ESI-LC/MS analysis of authentic (protio) 8 in the positive ion mode. The base peak at m/z 162 is the (M+H)+ ion of 8. The peak at m/z 162.9 is due to natural abundance of 13C, 15N, and 2D in the molecule and is close to the expected value of 10% of the base peak. Panel B shows mass spectrometric data obtained by LC/MS analysis of the mono-N-oxide metabolite generated by incubation of 5 (800 μM) with NADPH:cytochrome P450 reductase (0.6 units/mL) and NADPH (3 mM) in deuterated sodium phosphate buffer (50 mM, pD 6.6) and deuterated methanol (CD3OD, 2 M) under anaerobic conditions for 12 h. The small increase in the peak at m/z 163 may indicate that approximately 2–5% deuterated metabolite (12) was produced during one-electron reduction of 5.
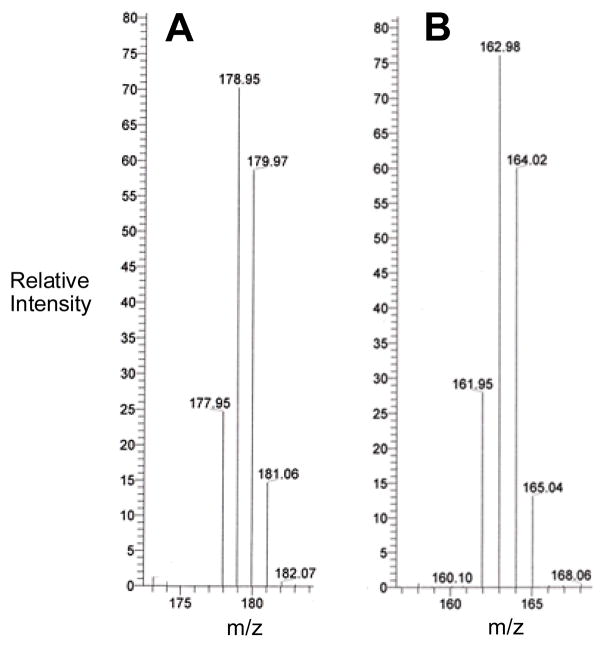
In addition, we performed a complementary experiment with an analogue of 5 containing deuterium in the methyl group. Using this compound, we investigated whether deuterium in the methyl group of 5 “washes out” during redox-activated conversion of the drug to its mono-N-oxide metabolite. The analogue of 5 containing deuterium in the methyl group was prepared by incubation of 5 in slightly basic D2O (pD = 7.6) at 24 °C for 25 days. Slow exchange produced a mixture of non-, mono-, di-, and tri-deuterated isomers of 5. Mass spectrometric analysis of this material revealed a set of peaks with relative intensities of 15:41:35:9 for these isotopologues (Figure 8). In vitro metabolism of this material by NADPH:cytochrome P450 reductase under anoxic conditions yields a mixture of mono-N-oxide metabolites in which the ratio of non-, mono-, di-, and tri-deuterated isomers is not significantly different from that seen in the mixture of starting di-N-oxides (Figure 8).
Figure 8.
In vitro metabolism of deuterium labeled 5 in sodium phosphate-methanol results in no washout of label. Panel A shows the family of M+H ions corresponding to the mixture of non-, mono-, di-, and tri-deuterated isomers of 5, at m/z 178, 179, 180, and 181 respectively, obtained using LC-ESI/MS operating in the positive ion mode (plotted at 7× actual abundance). Panel B shows the mass spectrum of the mono-N-oxide metabolites generated by in vitro metabolism of deuterated 5 by NADPH:cytochrome P450 reductase (0.6 units/mL), NADPH (3 mM), in phosphate buffer 50 mM, pH 7, containing methanol (2 M), under anoxic conditions, obtained using LC-ESI/MS operating in the positive ion mode (plotted at 2× actual abundance). The relative intensity of M+H ions for the resulting mixture of non-, mono-, di-, and tri-deuterated isomers of 8, at m/z 162, 163, 164, and 165 respectively, is not significantly altered from that observed in the starting mixture of deuterated di-N-oxides 5.
Overall, we find that significant amounts of hydrogen or deuterium atoms do not exchange in or out of the methyl group in 5 during bioreductive metabolism. Taken together, the results indicate that the benzotriazinyl radical 7 is not a precursor to the mono-N-oxide metabolite (8). These results led us to further examine the alternative hypothesis involving the release of hydroxyl radical (HO•) from the reductively-activated drug 7.
Generation of characteristic hydroxyl radical-derived products by redox-activated 5 under anoxic conditions
In light of our hypothesis that reductive activation of 1,2,4-benzotriazine 1,4-dioxides under hypoxic conditions leads to the release of hydroxyl radical, we felt it would be interesting to examine the enzymatic activation of 5 in the presence of reagents such as DMSO and salicylic acid that yield characteristic products upon reaction with HO•.
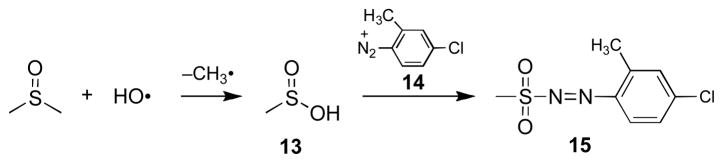
The reaction of hydroxyl radical with DMSO yields methanesulfinic acid (13) as a characteristic product.75–77 Methanesulfinic acid, in turn, can be quantitatively detected as the methane diazosulfone (15) resulting from derivatization with the diazonium salt 14 (Scheme 5).75,77,78 We find that enzymatic activation of 5 (500 μM) with NADPH:cytochrome P450 reductase under anaerobic conditions in the presence of DMSO (1 M), followed by treatment with 14, produces a 41% yield of the methanesulfinic acid derivative 15 (based on 5). By way of comparison, TPZ produces an 89% yield of the methanesulfinic acid derivative under these conditions. The yields are corrected for the small amounts of methanesulfinic acid generated in the incubation of DMSO with 5 alone, or with the NADPH:cytochrome P450 reductase enzyme system. These results are consistent with the notion that TPZ and 5 produce substantial yields of hydroxyl radical following bioreductive activation.
Scheme 5.
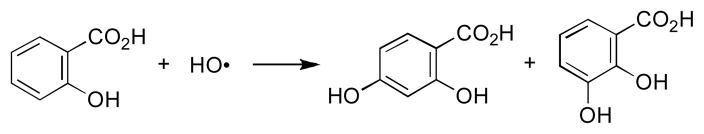
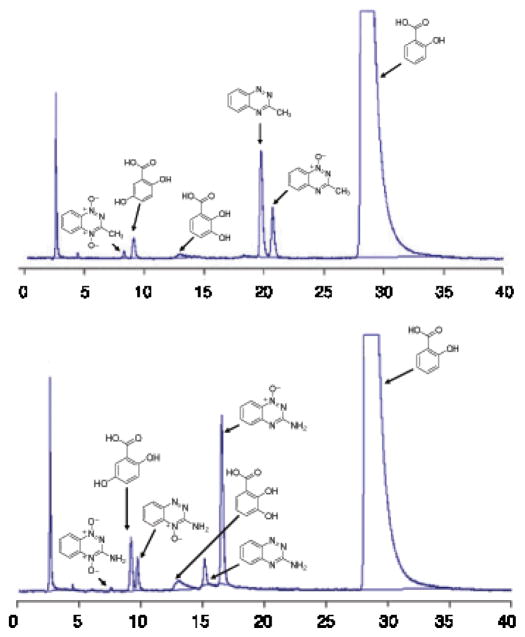
Salicylic acid has long been employed as a hydroxyl radical trapping agent.15,79,80 The reaction of hydroxyl radical with salicylic acid generates hydroxylated aromatic products including 2,3-dihydroxybenzoic acid and 2,5-dihydroxybenzoic acid (Scheme 6).81 We find that enzymatic activation of 5 (250 μM) under anaerobic conditions in the presence of salicylic acid (10 mM) produces the expected hydroxylated metabolites, 2,3-dihydroxybenzoic acid and 2,5-dihydroxybenzoic acid (Figure 9). Reductive activation of TPZ also yields 2,3-dihydroxybenzoic acid and 2,5-dihydroxybenzoic acid under these conditions. The identity of these hydroxylated aromatic species was confirmed by co-injection with authentic standards and LC/ESI-MS operated in the negative ion mode. Control reactions show that incubation of salicylic acid with either the NADPH:cytochrome P450 reductase or xanthine/xanthine oxidase enzyme systems in the absence of 5 does not produce significant amounts of these products. Further control experiments show that incubation of TPZ or 5 with salicylic acid in the absence of the enzyme does not generate significant amounts of the hydroxylated metabolites. Again, these results are consistent with the hypothesis that one-electron reduction of TPZ and 5 under anoxic conditions leads to the generation of hydroxyl radical.
Scheme 6.
Figure 9.
HPLC analysis reveals that anoxic metabolism of 5 (above) and TPZ (below) in the presence of salicylic acid generates the characteristic hydroxyl radical-derived products 2,3-dihydroxybenzoic acid and 2,5-dihydroxybenzoic acid. In vitro metabolism reactions were carried out as described in the Experimental Section.
Conclusions
The results described here provide useful information regarding the structure-activity relationships and the mechanism of DNA damage by 1,2,4-benzotriazine 1,4-dioxide antitumor agents. Many analogues in this structural series have been prepared and characterized54,61,62,82,83,84,85–90 but the 3-alkyl-1,2,4-benzotriazine 1,4-dioxides are of particular interest. These analogues display hypoxia-selective activity against human cancer cell lines that are comparable to TPZ54 and extravascular transport properties that may be superior to the parent drug.91,92 Accordingly, the 3-alkyl analogues have been described as potential clinical “back-ups” for TPZ.54 Despite their promising preclinical properties, the redox-activated DNA-damaging properties of 3-alkyl-1,2,4-benzotriazine 1,4-dioxides, such as 5, have not been examined previously. We find that 5 causes hypoxia-selective, oxidatively generated DNA damage in a manner completely analogous to TPZ. These findings support the expectation that, like TPZ, the bioactivity of 3-alkyl-1,2,4-benzotriazine 1,4-dioxides stems from their DNA-damaging properties.2–8
Having shown that the DNA-damaging activity of 5 mirrors that of TPZ, we recognized that the methyl group in this compound provides a “handle” for examining the mechanism of DNA strand cleavage. The results of two complementary isotopic labeling experiments provided evidence that 5 does not generate substantial amounts of the benzotriazinyl radical 7 upon one-electron reduction. Rather, we suggest that bioreductive activation of 5 under anoxic conditions leads to release of hydroxyl radical from the intermediate drug radical 6. Consistent with this hypothesis, we find that enzymatic one-electron reduction of both 5 and TPZ in the presence of DMSO or salicylic acid leads to the generation of characteristic hydroxyl radical-derived products. Indeed, a mechanism involving release of the well-known DNA-damaging agent hydroxyl radical from the reductively-activated drug radical intermediates (e.g. 2 and 6) can explain the DNA-damaging properties of the entire 1,2,4-benzotriazine 1,4-dioxide family of antitumor agents.
Experimental Section
Materials
Materials were of the highest purity available and were obtained from following sources: sodium phosphate, mannitol, xanthine, DMSO, and TLC plates from Aldrich Chemical Co. (Milwaukee, WI); NADPH, desferal, cytochrome P450 reductase, catalase, calf thymus DNA, and superoxide dismutase (SOD) from Sigma Chemical Co. (St. Louis, MO); xylene cyanol, bromophenol blue, formamide, and urea from United States Biochemical; T4 polynucleotide kinase from New England Biolabs; [γ-32P]-dATP from Perkin-Elmer Life Sciences; oligonucleotides were purchased from Integrated DNA Technologies; acrylamide and bisacrylamide from Roche Diagnostics; xanthine oxidase from Roche Diagnostics; agarose from Seakem; HPLC grade solvents (acetonitrile, methanol, ethanol, tert-butyl alcohol, ethyl acetate, hexane, and acetic acid) from Fischer (Pittsburg, PA); ethidium bromide from Roche Molecular Biochemicals (Indianapolis, IN); Silica gel (0.04–0.063 mm pore size) for column chromatography from Merck. The plasmid pGL2BASIC was prepared using standard protocols.93 Tirapazamine (1, TPZ), 3, and 9 were synthesized according to literature methods.53 High resolution mass spectroscopy was performed at the University of Illinois Urbana-Champaign Mass Spectroscopy facility and low resolution mass spectroscopy were performed at the University of Missouri-Columbia.
Cleavage of plasmid DNA
Individual components of the DNA-cleavage assays, except DNA, NADPH, and enzymes were deoxygenated using three cycles of freeze-pump-thaw degassing in Pyrex tubes and then torch-sealed under high vacuum. Sealed tubes were scored, opened in an argon-filled glove bag, and used to prepare individual reactions. The enzymes, NADPH, and DNA were diluted with degassed water in the glove bag to prepare stock solutions. The DNA-cleavage assays contained supercoiled plasmid DNA (33 μg/mL), NADPH (500 μM), cytochrome P450 reductase (0.03 units/mL), catalase (100 μg/mL), superoxide dismutase (10 μg/mL), sodium phosphate buffer (50 mM, pH 7.0), and desferal (1 mM) in a final volume of 30 μL and were incubated with different concentrations of 5 or 1 (25 μM-150 μM) under anaerobic conditions at 25 °C for 4 h. To suppress possible background DNA damage resulting from enzymatic reduction of traces of molecular oxygen to superoxide, we employed desferal to sequester adventitious trace metals in a non-redox active form, thus preventing the conversion of superoxide-derived hydrogen peroxide to hydroxyl radical.15 In addition, the assays contained superoxide dismutase and catalase to decompose any superoxide radical and hydrogen peroxide that was produced.15 Following incubation, the reactions were quenched by addition of 5 μL of 50% glycerol loading buffer and were loaded onto a 0.9% agarose gel. The gel was electrophoresed for approximately 2.5 h at 82 V in 1x TAE buffer before staining in a solution of aqueous ethidium bromide (0.3 μg/mL) for 3 h. DNA in the gel was visualized by UV-transillumination, and the amount of DNA in each band was quantified using an Alpha Innotech IS-1000 digital imaging system. The values reported are not corrected for differential staining of form I and form II DNA by ethidium bromide.94 DNA-cleavage assays containing radical scavengers were carried out as described above, with the exception that degassed solutions of radical scavengers such as a methanol, ethanol, tert-butyl alcohol, DMSO, or mannitol (500 mM) were added to the reactions prior to the addition of cytochrome P450 reductase.
Sequence specificity of DNA strand cleavage by 5
A 30 base 2′-deoxyoligonucleotide 5′-GTCACGTGCTGCAGACGACGTGCTGAGCCT-3′ was 5′-end labeled with 32P using [γ-32P] dATP and T4 polynucleotide kinase and was purified on a 20% denaturing polyacrylamide gel.93 The labeled single strand oligonucleotide was then annealed with its complimentary strand by heating the mixture to 90 °C sodium phosphate (pH 7, 20 mM) followed by slow cooling to room temperature overnight. DNA cleavage by 1 and 5 was performed in a dialysis chamber placed in deoxygenated aqueous buffer (three freeze-pump-thaw cycles) inside an inert atmosphere glove bag as described previously.23 The degassed reaction mixture containing duplex oligonucleotide (250,500 cpm), 1 (250 μM) or 5 (250 μM), sodium phosphate buffer (10 mM, pH 7.0), desferal (1 mM), catalase (100 μg/mL), superoxide dismutase (10 μg/mL), NADPH (1 mM) and cytochrome P450 reductase (0.05 U/mL) in a total volume of 100 μL was pipetted into a Slide-A-Lyzer minimal unit (Pierce, 3000 MW cutoff). The dialysis unit was then placed with a floater into 2 mL of solution containing 1 (250 μM) or 5 (250 μM), NADPH (1 mM), sodium phosphate buffer (10 mM, pH 7.0), and desferal (1 mM) and gently stirred for 16 h in a glove bag filled with argon, followed by the removal of DNA-containing solution from inside the dialysis unit. The assay shown in Figure 5H was carried out in an identical manner, except the assay contained 40 mM NaCl. The oligonucleotide was precipitated using 0.3 M sodium acetate, 70% ethanol (final concentration), and 5 μg of carrier DNA (herring sperm), and then washed three times with 80% ethanol-water. The resulting pellet was air dried, the DNA fragments dissolved in formamide loading buffer, heated at 90 °C for 5 min, then immersed in ice water. An equal number of counts were loaded in each lane of 20% denaturing polyacrylamide gel. The gel was electrophoresed for 3 h at 1200 V in 1x TBE buffer. The resolved fragments of DNA in the gel were visualized by phosphorimager analysis (Molecular Imager® FX, Imaging Screen-K, cat 170–7841, Bio-Rad, using Quantity One® Version 4.2.3, Bio-Rad). Maxam-Gilbert A + G and G-reactions shown in Figure 5 were carried out according to the literature methods.95 DNA cleavage by iron EDTA/H2O2/ascorbate was performed according to the protocol of Tullius and co-workers.31
DNA strand cleavage by hydroxyl radical generated by anaerobic photolysis of hydrogen peroxide
The DNA duplex described above was dissolved in a solution containing 10 mM sodium phosphate (pH 7), 40 mM NaCl, 500 mM hydrogen peroxide and subjected to freeze-pump-thaw degassing to remove dissolved gasses as described above. The mixture was subjected irradiation with UV light (Spectroline MODEL ENF-240C, 115 Volts, 60 Hz, 0.20 amps, long wavelength 365 nm) in Pyrex tubes (6mm i.d.) for 10–15 min at room temperature (24 °C).
The oligonucleotide was precipitated using 0.3 M sodium acetate, 70% ethanol (final concentration), and 5 μg of carrier DNA (herring sperm), and then washed three times with 80% ethanol-water. The resulting pellet was air dried, the DNA fragments resuspended in formamide loading buffer, heated at 90 °C for 5 min, then immersed in ice water. An equal number of counts were loaded in each lane of 20% denaturing polyacrylamide gel. The gel was electrophoresed for 3 h at 1200 V in 10x TBE buffer. The resolved fragments of DNA in the gel were visualized by phosphorimager analysis (Molecular Imager® FX, Imaging Screen-K, cat 170-7841, Bio-Rad, using Quantity One® Version 4.2.3, Bio-Rad). Sequencing lanes were prepared as described above.
Characterization of products arising from in vitro metabolism of 5
A solution containing 5 (500 μM) and desferal (1 mM) in sodium phosphate (pH 7, 50 mM) was deoxygenated by three freeze-pump-thaw cycles and then torch-sealed under vacuum. The sealed tube was scored before being transferred to an argon-filled glove bag. The tube was then opened and the degassed solution was transferred to a microcentrifuge tube. To this solution, NADPH (1 mM), and cytochrome P450 reductase (0.33 U/mL), were added and the resulting samples incubated in an argon-filled glove bag at 25 °C for 3 h. The protein was removed by centrifugation through an Amicon Microcon (YM3) filter. The filtrate was analyzed by HPLC employing a C18 reverse phase Rainin Microsorb-MV column (5 μm particle size, 100 Å pore size, 25 cm length, 4.6 mm i.d.) eluted with gradient starting with 80% A (0.5% acetic acid in water) and 20% B (2:1 methanol/acetonitrile) followed by linear increase to 30% B from 0 min to 20 min. The column was eluted with 30% B for 10 min, then decreased to 20% B over the next 10 min. A flow rate of 0.9 mL/min was used and the products were monitored by their UV-absorbance at 240 nm. In vitro metabolism of 5 by NADPH:cytochrome P450 reductase yields two major products in the HPLC chromatogram. The major product eluting at ~19 min was identified as 8 and the minor product eluting at ~18 min as 11 by co-injection with authentic synthetic standards and by LC/ESI-MS operated in the positive ion mode. In LC/ESI-MS analysis, metabolites were extracted into ethyl acetate and dried, and redissolved in 50:50 methanol:water. LC/ESI-MS experiments were carried out on a Finnigan TSQ 7000 triple quadrupole instrument interfaced to a ThermoSeparations liquid chromatograph (TSP4000). Positive ion electrospray was used as the means of ionization. The heated inlet capillary temperature was 250 °C and electrospray needle voltage was 4.5 kV. Nitrogen sheath gas was supplied at 80 psi.
Isotopic labeling experiments: in vitro metabolism of 5 in deuterated solvents
A solution of 5 (800 μM) was prepared in deuterated sodium phosphate buffer (pD 6.6, 50 mM). Deuterated buffer was prepared by dissolving Na2HPO4 (20.5 mg, 28.9 mmol) and NaH2PO4 (12.7 mg, 21.2 mmol) in D2O (5 mL). The resulting solution of 5 was deoxygenated using three cycles of freeze-pump-thaw in Pyrex tubes, followed by torch sealing. Pyrex tubes were scored, opened and the contents transferred to microcentrifuge tubes in an argon-filled glove bag. To these mixtures, NADPH:cytochrome P450 reductase (0.6 U/mL) and NADPH (3 mM) were added either in the presence or absence of CD3OD (2 M). Incubations were carried out in an argon atmosphere at 24 °C for 16 h. The metabolites generated from these reactions were extracted into ethyl acetate (2 mL) and the resulting solutions dried and redissolved in 1:1 methanol:water. Then the products were analyzed using LC/ESI-MS in the positive ion mode as described above. Analysis of the control reactions revealed less than 2% deuterium incorporation into 5 above background. The assays containing NADPH:cytochrome P450 reductase (0.6 U/mL), NADPH (3 mM), and CD3OD (2 M) contained less than 5% deuterium above background. The assay containing NADPH:cytochrome P450 reductase (0.6 U/mL), NADPH (3 mM), and no CD3OD contained less than 2% deuterium above background. Identical results were obtained when reactions were conducted in either the presence or absence of desferal.
Preparation and in vitro metabolism of deuterated 5
Compound 5 (1 mg, 5.7 mmol) was dissolved in 2 mL of deuterated sodium phosphate buffer (pD 7.6, 100 mM) and the resulting solution incubated at 25 °C for 25 days. Deuterated buffer (pD 7.6, 100 mM) was prepared by dissolving Na2HPO4 (66.2 mg, 93.3 mmol) and NaH2PO4 (4.1 mg, 6.7 mmol) in D2O (5 mL). Deuterium incorporation into the methyl group of 5 was detected by using 1H-NMR and LC/ESI-MS, as described above. The 1H-NMR shows no changes in the relative intensities of the aromatic protons but a 44% decrease in the intensity of peak corresponding to the methyl hydrogens. Mass spectrometric analysis reveals that the exchange reaction yields a mixture of non-, mono-, di-, and tri-deuterated 5, as shown in Figure 8.
In the experiment, a solution of deuterated 5, synthesized as described above, was prepared in sodium phosphate buffer (pH 7, 50 mM). This solution was deoxygenated using three cycles of freeze-pump-thaw in Pyrex tubes, followed by torch sealing. The Pyrex tubes were scored, opened and contents transferred into a microcentrifuge tube in an argon-filled glove bag. In vitro metabolism reactions were carried out by addition of NADPH (3 mM) and NADPH/cytochrome P450 reductase (0.6 U/mL), both in the presence and absence of CH3OH (2 M) and were incubated under an argon atmosphere at 24 °C for 16 h. Control samples were prepared without addition of enzyme, NADPH, or methanol. The metabolites generated from these reactions were extracted into ethyl acetate (2 mL) and the resulting solutions dried under a stream of air. The products were analyzed by using LC/ESI-MS in a positive ion mode as described above.
Oxidation of DMSO to methanesulfinic acid during in vitro metabolism of 1 and 5
Methanesulfinic acid produced by the oxidation of DMSO during in vitro metabolism of 1 and 5 was quantitatively measured using a modified version of the protocol reported by Fukui et al.77 In a typical assay, individual components of the reactions were deoxygenated as described above. To a degassed solution containing sodium phosphate (50 mM, pH 7.0), 1 or 5 (500 μM) and desferal (1 mM), deoxygenated DMSO (1 M), NADPH (2 mM) and cytochrome P450 reductase (0.29 U/mL) were added. The reaction (1 mL final volume) was capped, mixed, and allowed to incubate under an argon atmosphere at 24 °C for 4 h. Then sodium phosphate (0.5 mL, 500 mM, pH 4.0) was added to the reaction, followed by Fast Red TR diazonium salt (14, 0.5 mL of a 10 mg/mL solution water) and the mixture allowed to stand at room temperature for 10 min. During this time an orange color developed and the resulting solution was extracted with ethyl acetate (2 × 1 mL) and exactly 1.2 mL of the upper ethyl acetate layer removed by pipet. A portion of this ethyl acetate solution (20 μL) containing the methane diazosulfone was then analyzed by HPLC. The diazosulfone conjugate 15 was observed by its absorbance at 310 nm at a retention time of approximately 9 min on a Rainin Microsorb-MV propylamine column eluted with hexane-2-propanol (100:3) at a flow rate of 1 mL/min. Authentic 15 for comparison was prepared as previously described.96 The yield of methanesulfinic acid reported here for 1 are higher than that reported previously.26 The assays described here utilized a higher concentration of the hydroxyl radical trapping agent DMSO and the yields are corrected for unmetabolized starting drug. For the construction of calibration curves, known amounts of methanesulfinic acid was dissolved in sodium phosphate buffer (50 mM, pH 7.0) to make 1 mL solutions containing different concentrations of methane sulfinic acid (100–500 μM). To these solutions, sodium phosphate buffer (0.5 mL, 500 mM, pH 4.0) was added, followed by Fast Red TR diazonium salt (14, 0.5 mL of 10 mg/mL). The reactions were allowed to stand at room temperature for 10 min and the resulting orange solutions extracted with ethyl acetate (2 × 1 mL) and exactly 1.2 mL of the upper ethyl acetate layer removed by pipet. A portion of this ethyl acetate solution (20 μL) containing the methane diazosulfone was then analyzed and quantified by HPLC. Injection-to-injection variation in this system is less than ± 5%. Parallel control reactions lacking either NADPH or enzyme were performed. The final yields of 15 reported here are corrected for background levels measured in these control reactions and for unmetabolized di-N-oxide.
Hydroxyl radical trapping by salicylic acid
Assay solutions were degassed in Pyrex tubes using three cycles of freeze-pump-thaw, followed by torch sealing. Enzyme solutions were prepared in degassed water. The Pyrex tubes containing degassed solutions were scored, opened in an argon-filled glove bag, and the assays prepared in microcentrifuge tubes. Typical assays (300 μL final volume) contained 1 or 5 (250 μM, final concentrations), salicylic acid (10 mM), catalase (50 μg/mL), desferal (1 mM), sodium phosphate buffer (50 mM, pH 7), and NADPH (1 mM), NADPH:cytochrome P450 reductase (0.05 U/mL in the case of 5), or xanthine (750 μM), xanthine oxidase (0.6 U/mL, in the case of 1). Control reactions were prepared in an identical manner except without di-N-oxide or without the enzymatic reducing system. The reaction mixtures were incubated for 16–18 h in an argon-filled glove bag. Following incubation, enzymes were removed by Microcon filtration and the reaction products analyzed by their absorbance at 310 and 278 nm using reverse-phase HPLC with a C18 column eluted with a gradient solvent system starting with 95% A (2% acetic acid in water) and 5% B (methanol) for 2 min, followed by an increase to 25% B from 2–10 min, an increase to 40% B from 10–20 min, an increase to 50% B from 20–30 min and, finally an increase to 80% B from 30–45 min at a flow rate of 0.9 mL/min. The identity of hydroxylated products was confirmed by co-injection with authentic standards and by LC/ESI/MS operating in the negative ion mode, employing conditions analogous to those described above.
Figure 2.

DNA cleavage by various concentrations of reductively-activated TPZ (1) under anaerobic conditions. Supercoiled plasmid DNA (33 μg/mL, pGL-2 Basic) was incubated with TPZ (25–150 μM), NADPH (500 μM), cytochrome P450 reductase (0.03 mU/μL), catalase (100 μg/mL), superoxide dismutase (10 μg/mL), sodium phosphate buffer (50 mM, pH 7.0), and desferal (1 mM) under anaerobic conditions at 25 °C for 4 h, followed by agarose gel electrophoretic analysis. Lane 1, DNA alone (S = 0.24 ± 0.01); lane 2, NADPH (500 μM) + reductase (0.03 mU/μL) (S = 0.26 ± 0.03); lane 3, TPZ (150 μM) (S = 0.24 ± 0.02); lanes 4–9, NADPH (500 μM) + reductase (0.03 mU/μL) + TPZ (25 μM, lane 4) (S = 0.38 ± 0.01); (50 μM, lane 5) (S = 0.51 ± 0.03); (75 μM, lane 6) (S = 0.61 ± 0.03); (100 μM, lane 7) (S = 0.72 ± 0.02); (125 μM, lane 8) (S = 0.82 ± 0.05); (150 μM, lane 9) (S = 1.04 ± 0.01). The values, S, represent the mean number of strand breaks per plasmid molecule and were calculated using the equation S = −ln fI, where fI is the fraction of plasmid present in the supercoiled form I.
Acknowledgments
We are grateful to the NIH for support of this research (CA 100757), we acknowledge Dr. Nathan Leigh (University of Missouri Department of Chemistry) for assistance with mass spectrometric analysis, and we thank Earl Gates and Professor Marc Greenberg (Johns Hopkins University) for helpful comments on the manuscript.
References
- 1.Marco L, Olver I. Curr Clin Oncol. 2006;1:71–79. doi: 10.2174/157488406775268192. [DOI] [PubMed] [Google Scholar]
- 2.Horsman MR. Int J Radiat Oncol Biol Phys. 1998;42:701–704. doi: 10.1016/s0360-3016(98)00332-0. [DOI] [PubMed] [Google Scholar]
- 3.Siemann DW. Int J Radiat Oncol Biol Phys. 1998;42:697–699. doi: 10.1016/s0360-3016(98)00336-8. [DOI] [PubMed] [Google Scholar]
- 4.Vaupel P, Kallinowski F, Okunieff P. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 5.Wilson WR. Tumour hypoxia: challenges for cancer chemotherapy. In: Waring MJ, Ponder BAJ, editors. The Search For New Anticancer Drugs. Kluwer Academic; Lancaster: 1992. pp. 87–131. [Google Scholar]
- 6.Brown JM, Wilson WR. Nat Rev Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 7.Brown JM. Cancer Res. 1999;59:5863–5870. [PubMed] [Google Scholar]
- 8.Zeman EM, Brown JM, Lemmon MJ, Hirst VK, Lee WW. Int J Radiat Oncol Biol Phys. 1986;12:1239–1242. doi: 10.1016/0360-3016(86)90267-1. [DOI] [PubMed] [Google Scholar]
- 9.Laderoute KL, Wardman P, Rauth M. Biochem Pharmacol. 1988;37:1487–1495. doi: 10.1016/0006-2952(88)90010-x. [DOI] [PubMed] [Google Scholar]
- 10.Wardman P, Priyadarsini KI, Dennis MF, Everett SA, Naylor MA, Patel KB, Stratford IJ, Stratford MRL, Tracy M. Br J Cancer. 1996;74:S70–S74. [PMC free article] [PubMed] [Google Scholar]
- 11.Priyadarsini KI, Tracy M, Wardman P. Free Radic Res. 1996;25:393–399. doi: 10.3109/10715769609149061. [DOI] [PubMed] [Google Scholar]
- 12.Poole JS, Hadad CM, Platz MS, Fredin ZP, Pickard L, Guerrero EL, Kessler M, Chowdhury G, Kotandeniya D, Gates KS. Photochem Photobiol. 2002;75:339–345. doi: 10.1562/0031-8655(2002)075<0339:petrot>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Superoxide radical (O2•−) produced by this type of redox cycling can be cytotoxic, see Refs. 14–19. However, cellular enzymes including superoxide dismutase, catalase, glutathione peroxidase, and peroxiredoxins mitigate the deleterious effects of superoxide radical and its decomposition products (see refs. 18, 20, and 21) and, in the context of TPZ, back-oxidation of 2 clearly represents a detoxification process.
- 14.Chen Y, Jungsuwadee P, Vore M, Butterfield DA, St Clair DK. Mol Interv. 2007;7:147–156. doi: 10.1124/mi.7.3.6. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell B, Gutteridge JMC. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 16.Bagley AC, Krall J, Lynch RE. Proc Natl Acad Sci USA. 1986;83:9189–9193. doi: 10.1073/pnas.83.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sánchez Sellero I, Lopez-Rivadulla Lamas M. Recent Res Dev Drug Metab Disp. 2002;1:275–313. [Google Scholar]
- 18.Finkel T, Holbrook NJ. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 19.Davis W, Jr, Ronai Z, Tew KD. J Pharm Exp Ther. 2001;296:1–6. [PubMed] [Google Scholar]
- 20.Sies H. Angew Chem Int Ed Eng. 1986;25:1058–1071. [Google Scholar]
- 21.Wood ZA, Poole LB, Karplus PA. Science. 300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 22.Kotandeniya D, Ganley B, Gates KS. Bioorg Med Chem Lett. 2002;12:2325–2329. doi: 10.1016/s0960-894x(02)00468-7. [DOI] [PubMed] [Google Scholar]
- 23.Birincioglu M, Jaruga P, Chowdhury G, Rodriguez H, Dizdaroglu M, Gates KS. J Am Chem Soc. 2003;125:11607–11615. doi: 10.1021/ja0352146. [DOI] [PubMed] [Google Scholar]
- 24.Chowdhury G, Junnutula V, Daniels JS, Greenberg MM, Gates KS. J Am Chem Soc. 2007;129:12870–12877. doi: 10.1021/ja074432m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzsimmons SA, Lewis AD, Riley RJ, Workman P. Carcinogenesis. 1994;15:1503–1510. doi: 10.1093/carcin/15.8.1503. [DOI] [PubMed] [Google Scholar]
- 26.Daniels JS, Gates KS. J Am Chem Soc. 1996;118:3380–3385. [Google Scholar]
- 27.Biedermann KA, Wang J, Graham RP. Br J Cancer. 1991;63:358–362. doi: 10.1038/bjc.1991.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siim BG, van Zijl PL, Brown JM. Br J Cancer. 1996;73:952–960. doi: 10.1038/bjc.1996.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zagorevski D, Yuan Y, Fuchs T, Gates KS, Song M, Breneman C, Greenlief CM. J Am Soc Mass Spec. 2003;14:881–892. doi: 10.1016/S1044-0305(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 30.Hydroxyl radical is a well known DNA-damaging agent, see refs 31–34.
- 31.Pogozelski WK, McNeese TJ, Tullius TD. J Am Chem Soc. 1995;117:6428–6433. [Google Scholar]
- 32.Pratviel G, Bernadou J, Meunier B. Angew Chem Int Ed Eng. 1995;34:746–769. [Google Scholar]
- 33.Breen AP, Murphy JA. Free Radic Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 34.Evans MD, Dizdaroglu M, Cooke MS. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Li LC, Zha D, Zhu YQ, Xu MH, Wong NB. Chem Phys Lett. 2005;408:329–334. [Google Scholar]
- 36.The possibilty that activated TPZ releases hydroxyl radical was first considered by Laderoute and coworkers (ref. 9) and, later, the analogous mechanism was favored by Hecht and coworkers in the context of reductively-activated phenazine di-N-oxides: Nagai K, Carter BJ, Xu J, Hecht SM. J Am Chem Soc. 1991;113:5099–5100.
- 37.Barton DHR, Crich D, Motherwell WB. Tetrahedron. 1985;41:3901–3924. [Google Scholar]
- 38.Boivin J, Crepon E, Zard SZ. Tetrahedron Lett. 1990;31:6869–6872. [Google Scholar]
- 39.Adam W, Ballmaier D, Epe B, Grimm GN, Saha-Moller CR. Angew Chem Int Ed Eng. 1995;34:2156–2158. [Google Scholar]
- 40.Barton DHR, Jasberenyi JC, Morrell AI. Tetrahedron Lett. 1991;32:311–314. [Google Scholar]
- 41.Aveline BM, Kochevar IE, Redmond RW. J Am Chem Soc. 1996;118:289–290. [Google Scholar]
- 42.For generation of hydroxyl radical via photoreduction of a pyridine N-oxide, see: Nakanishi I, Nishizawa C, Ohkubo K, Takeshita K, Suzuki KT, Ozawa T, Hecht SM, Tanno M, Sueyoshi S, Miyata N, Okuda H, Fukuzumi S, Ikota N, Fukuhara K. Org Biomol Chem. 2005;3:3263–3265. doi: 10.1039/b509447j.
- 43.Wölfle I, Lodays J, Sauerwein B, Schuster GB. J Am Chem Soc. 1992;114:9304–9309. [Google Scholar]
- 44.Lorance ED, Kramer WH, Gould IR. J Am Chem Soc. 2002;124:15225–15238. doi: 10.1021/ja020768e. [DOI] [PubMed] [Google Scholar]
- 45.Bockman TM, Lee KY, Kochi JK. J Chem Soc Perkin. 2;1992:1581–1594. [Google Scholar]
- 46.Karki SB, Dinnocenzo JP, Jones JP, Korzekwa KR. J Am Chem Soc. 1995;117:3657–3664. [Google Scholar]
- 47.Anderson RF, Shinde SS, Hay MP, Gamage SA, Denny WA. J Am Chem Soc. 2003;125:748–756. doi: 10.1021/ja0209363. [DOI] [PubMed] [Google Scholar]
- 48.Shinde SS, Anderson RF, Hay MP, Gamage SA, Denny WA. J Am Chem Soc. 2004;126:7865–7874. doi: 10.1021/ja048740l. [DOI] [PubMed] [Google Scholar]
- 49.Anderson RF, Shinde SS, Hay MP, Gamage SA, Denny WA. Org Biomol Chem. 2005;3:2167–2174. doi: 10.1039/b502586a. [DOI] [PubMed] [Google Scholar]
- 50.Anderson RF, Shinde SS, Hay MP, Denny WA. J Am Chem Soc. 2006;128:245–249. doi: 10.1021/ja0559101. [DOI] [PubMed] [Google Scholar]
- 51.Mvula E, Schuchmann MN, von Sonntag C. J Chem Soc Perkin. 2;2001:264–268. [Google Scholar]
- 52.Christensen HC, Sehested K, Hart EJ. J Phys Chem. 1973;77:983–987. [Google Scholar]
- 53.Fuchs T, Chowdhury G, Barnes CL, Gates KS. J Org Chem. 2001;66:107–114. doi: 10.1021/jo001232j. [DOI] [PubMed] [Google Scholar]
- 54.Kelson AB, McNamara JP, Pandey A, Ryan KJ, Dorie MJ, McAfee PA, Menke DR, Brown JM, Tracy M. Anti-Cancer Drug Des. 1998;13:575–592. [PubMed] [Google Scholar]
- 55.Attallah RH, Nazer MZ. Tetrahedron. 1982;38:1793–1796. [Google Scholar]
- 56.Khodja M, Moulay S, Boutoumi H, Wilde H. Heteroatom Chem. 2006;17:166–172. [Google Scholar]
- 57.Delahoussaye YM, Evans JM, Brown JM. Biochem Pharmacol. 2001;62:1201–1209. doi: 10.1016/s0006-2952(01)00784-5. [DOI] [PubMed] [Google Scholar]
- 58.Patterson AV, Saunders MP, Chinje EC, Patterson LH, Stratford IJ. Anti-Cancer Drug Des. 1998;13:541–573. [PubMed] [Google Scholar]
- 59.Walton MI, Wolf CR, Workman P. Biochem Pharmacol. 1992;44:251–259. doi: 10.1016/0006-2952(92)90007-6. [DOI] [PubMed] [Google Scholar]
- 60.Jones GDD, Weinfeld M. Cancer Res. 1996;56:1584–1590. [PubMed] [Google Scholar]
- 61.Chowdhury G, Kotandeniya D, Barnes CL, Gates KS. Chem Res Toxicol. 2004;17:1399–1405. doi: 10.1021/tx049836w. [DOI] [PubMed] [Google Scholar]
- 62.Ganley B, Chowdhury G, Bhansali J, Daniels JS, Gates KS. Bioorg Med Chem. 2001;9:2395–2401. doi: 10.1016/s0968-0896(01)00163-8. [DOI] [PubMed] [Google Scholar]
- 63.Hintermann G, Fischer HM, Crameri R, Hutter R. Plasmid. 1981;5:371–373. doi: 10.1016/0147-619x(81)90012-3. [DOI] [PubMed] [Google Scholar]
- 64.Jonson PH, Grossman LI. Biochemistry. 1977;16:4217–4224. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- 65.Mirabelli CK, Huang CH, Fenwick RG, Crooke ST. Antimicrob Agents Chemother. 1985;27:460–467. doi: 10.1128/aac.27.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greenberg MM. Org Biomol Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 67.Greenberg MM. Chem Res Toxicol. 1998;11:1235–1248. doi: 10.1021/tx980174i. [DOI] [PubMed] [Google Scholar]
- 68.Pogozelski WK, Tullius TD. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 69.Gates KS. Chemical reactions of DNA damage and degradation. In: Platz MSMRA, Jones M Jr, editors. Reviews of Reactive Intermediates. John Wiley and Sons, Inc; Hoboken: 2007. pp. 333–378. [Google Scholar]
- 70.(a) Burrows CJ, Muller JG. Chem Rev. 1998;98:1109–1151. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]; (b) Pratviel G, Meunier B. Chem Eur J. 2006;12:6018–6030. doi: 10.1002/chem.200600539. [DOI] [PubMed] [Google Scholar]
- 71.Burrows CJ, Muller JG, Kornyushyna O, Luo W, Duarte V, Leipold MD, David SS. Environ Health Perspect Suppl. 2002;110:713–717. doi: 10.1289/ehp.02110s5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laderoute K, Rauth AM. Biochem Pharmacol. 1986;35:3417–3420. doi: 10.1016/0006-2952(86)90448-x. [DOI] [PubMed] [Google Scholar]
- 73.Baker MA, Zeman EM, Hirst VK, Brown JM. Cancer Res. 1988;48:5947–5952. [PubMed] [Google Scholar]
- 74.Junnotula V, Sarkar U, Barnes CL, Thallapally PK, Gates KS. J Chem Cryst. 2006;36:557–561. [Google Scholar]
- 75.Steiner MG, Babbs CF. Arch Biochem Biophys. 1990;278:478–481. doi: 10.1016/0003-9861(90)90288-a. [DOI] [PubMed] [Google Scholar]
- 76.Klein SM, Cohen G, Cederbaum AI. Biochemistry. 1981;20:6006–6012. doi: 10.1021/bi00524a013. [DOI] [PubMed] [Google Scholar]
- 77.Fukui S, Hanasaki Y, Ogawa S. J Chromatogr. 1993;630:187–193. doi: 10.1016/0021-9673(93)80455-h. [DOI] [PubMed] [Google Scholar]
- 78.Babbs C, Steiner MG. Methods Enzymol. 1990;186:137–147. doi: 10.1016/0076-6879(90)86103-3. [DOI] [PubMed] [Google Scholar]
- 79.Halliwell B, Kaur H. Free Rad Res Commun. 1997;27:239–244. doi: 10.3109/10715769709065762. [DOI] [PubMed] [Google Scholar]
- 80.Halliwell B, Whiteman M. Br J Pharmacol. 2004;142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaur H, Halliwell B. Methods Enzymol. 1994;233:67–82. doi: 10.1016/s0076-6879(94)33009-3. [DOI] [PubMed] [Google Scholar]
- 82.Jiang F, Yang B, Fan L, He Q, Hu Y. Bioorg Med Chem Lett. 2006;16:4209–4213. doi: 10.1016/j.bmcl.2006.05.095. [DOI] [PubMed] [Google Scholar]
- 83.Zeman EM, Baker MA, Lemmon MJ, Pearson BA, Adams JA, Brown JM, Lee WW, Tracy M. Int J Radiat Oncol Biol Phys. 1989;16:977–981. doi: 10.1016/0360-3016(89)90899-7. [DOI] [PubMed] [Google Scholar]
- 84.(a) Hay MP, Gamage SA, Kovacs MS, Pruijn FB, Anderson RF, Patterson AV, Wilson WR, Brown JM, Denny WA. J Med Chem. 2003;46:169–182. doi: 10.1021/jm020367+. [DOI] [PubMed] [Google Scholar]; (b) Kovacs MS, Patterson AV, Wilson WR, Brown JM, Denny WA. J Med Chem. 2004;47:475–488. doi: 10.1021/jm030399c. [DOI] [PubMed] [Google Scholar]
- 85.Minchinton AI, Lemmon MJ, Tracy M, Pollart DJ, Martinez AP, Tosto LM, Brown JM. Int J Radiat Oncol Biol Phys. 1992;22:701–705. doi: 10.1016/0360-3016(92)90507-e. [DOI] [PubMed] [Google Scholar]
- 86.Cerecetto H, Gonzalez M, Onetto S, Risso M, Saenz P, Seoane G, Bruno AM, Alarcon J, Olea-Azar C, Lopez De Cerain A, Ezpeleta O, Monge A. Med Chem Res. 2001;10:328–337. [Google Scholar]
- 87.Parrick J, Mehta LK. J Heterocylic Chem. 1993;30:323–327. [Google Scholar]
- 88.Daniels JS, Chatterji T, MacGillivray LR, Gates KS. J Org Chem. 1998;63:10027–10030. [Google Scholar]
- 89.Fuchs T, Gates KS, Hwang JT, Greenberg MM. Chem Res Toxicol. 1999;12:1190–1194. doi: 10.1021/tx990149s. [DOI] [PubMed] [Google Scholar]
- 90.Solano B, Junnotula V, Marin A, Villar R, Burguete A, Vicente E, Perez-Silanes S, Monge A, Dutta S, Sarkar U, Gates KS. J Med Chem. 2007;50:5485–5492. doi: 10.1021/jm0703993. [DOI] [PubMed] [Google Scholar]
- 91.Hay MP, Pchalek K, Pruijn FB, Hicks KO, Siim BG, Anderson MM, Shinde SS, Denny WA, Wilson WR. J Med Chem. 2007;50:6654–6664. doi: 10.1021/jm701037w. [DOI] [PubMed] [Google Scholar]
- 92.Hay MP, Hicks KO, Pruijn FB, Pchalek K, Siim BG, Wilson WR, Denny WA. J Med Chem. 2007;50:6392–6404. doi: 10.1021/jm070670g. [DOI] [PubMed] [Google Scholar]
- 93.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Lab Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 94.Bauer W, Vinograd J. J Mol Biol. 1968;33:141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- 95.Maxam AM, Gilbert W. Methods Enzymol. 1980;65:499. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 96.Ahern MF, Leopold A, Beadle JR, Gokel GW. J Am Chem Soc. 1982;104:548–554. [Google Scholar]
- 97.Macgregor RB., Jr Anal Biochem. 1992;204:324–327. doi: 10.1016/0003-2697(92)90246-4. [DOI] [PubMed] [Google Scholar]
- 98.Daniels JS, Gates KS, Tronche C, Greenberg MM. Chem Res Toxicol. 1998;11:1254–1257. doi: 10.1021/tx980184j. [DOI] [PubMed] [Google Scholar]
- 99.Hwang JT, Greenberg MM, Fuchs T, Gates KS. Biochemistry. 1999;38:14248–14255. doi: 10.1021/bi991488n. [DOI] [PubMed] [Google Scholar]