Abstract
The yeast, Saccharomyces cerevisiae, is a model system for the study of eukaryotic mRNA degradation. In this organism, a variety of methods have been developed to measure mRNA decay rates, trap intermediates in the mRNA degradation process, and establish precursor–product relationships. In addition, the use of mutant strains lacking specific enzymes involved in mRNA destruction, or key regulatory proteins, allows one to determine the mechanisms by which individual mRNAs are degraded. In this chapter, we discuss methods for analyzing mRNA degradation in S. cerevisiae.
1. INTRODUCTION
An important step in the control of eukaryotic gene expression is the process of mRNA turnover in the cytoplasm. The study of mRNA turnover often requires knowledge of the rates of mRNA degradation and the pathway by which a particular mRNA is being degraded. Therefore, in this chapter we describe experimental procedures that can be used to determine the rates of mRNA turnover in yeast, as well as the specific pathway of degradation for any given transcript. Although the methods described here are routinely used to examine cytoplasmic mRNA decay events, they can also be used in similar manners to monitor nuclear mRNA turnover.
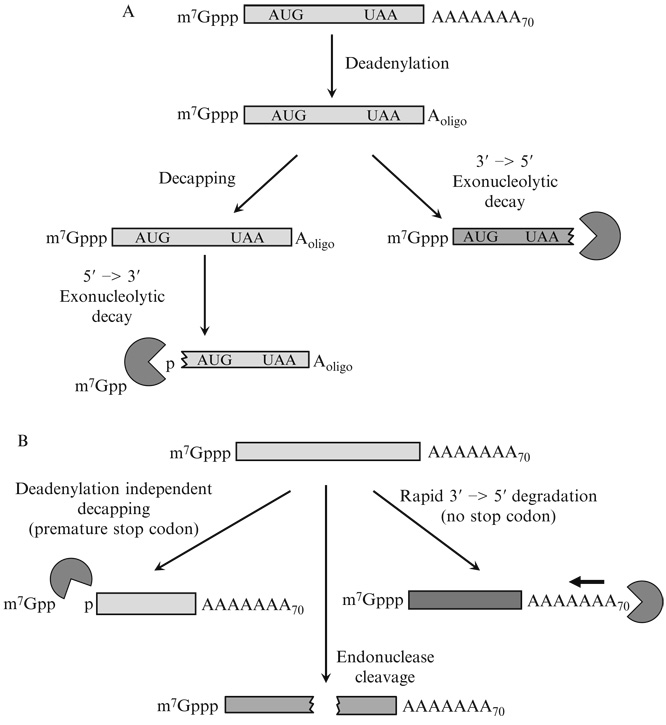
An understanding of the pathways of mRNA degradation in yeast, and other eukaryotes, has facilitated these approaches (Fig. 20.1) (reviewed in Parker and Song, 2004). In yeast, mRNA decay is generally initiated by shortening of the 3′-poly(A) tail in a process referred to as deadenylation. Deadenylation is primarily followed by decapping and rapid 5′ to 3′-exonucleolytic decay. Alternately, cytoplasmic transcripts can be subject to 3′ to 5′-exonucleolytic decay after deadenylation.
Figure 20.1.
Pathways of mRNA decay in yeast. (A) Model showing the two major routes for general mRNA decay in yeast. Normally, mRNAs are initially subjected to deadenylation followed either by decapping and 5′ to 3′-decay by Xrn1p or by 3′ to 5′-degradation by the exosome. (B) Three pathways known in yeast for bypassing the deadenylation requirement for mRNA turnover include nonsense-mediated mRNA decay (left), which causes deadenylation independent decapping, no-go decay (middle), which leads to endonucleolytic cleavage, and nonstop decay (right), which leads to rapid and continuous 3′ to 5′-degradation.
Additional specialized pathways of decay exist that target subsets of mRNAs, including those with translational abnormalities (Fig. 20.1B). First, mRNAs with premature termination codons or extended 3′ UTRs are degraded by decapping in a deadenylation-independent manner by rapid 5′ to 3′-decay (Muhlrad and Parker, 1994). In addition, some normal mRNAs can also be degraded independently of deadenylation (Badis et al., 2004). Second, mRNAs lacking translation termination codons recruit the exosome complex for very rapid 3′ to 5′-degradation without a distinct and slow deadenylation process (Van Hoof et al., 2002). Finally, mRNAs with strong stalls to translation elongation can be subject to endonucleolytic cleavage (Doma and Parker, 2006). An unresolved issue is how many normal yeast mRNAs are subject to these deadenylation-independent pathways of mRNA degradation. Thus, investigating the mechanism of mRNA degradation can include determining whether a given transcript is subject to any of these specialized mechanisms of mRNA degradation.
2. MEASURING mRNA HALF-LIFE
An mRNA half-life is routinely measured for an mRNA of interest such that the overall stability of that mRNA can be analyzed and quantitated. Several different experimental procedures can be used to measure the decay rates of individual mRNAs in the yeast Saccharomyces cerevisiae. In the following sections, we discuss the main procedures developed to determine the mRNA half-lives in yeast and their advantages and disadvantages (summarized in Table 20.1).
Table 20.1.
Methods for measuring mRNA half-lives in yeast
| Method | Advantage | Disadvantage | |
|---|---|---|---|
| In vivo labeling | Approach to steady-state pulse-chase |
Minimal cell perturbation No need for special strain or construct Can monitor the half-life of many mRNAs simultaneously |
Requires large amounts of radioactive material Poor signal-to-noise ratio for mRNA of low mRNA abundance |
| Transcriptional inhibition |
With drugs (thiolutin, 1,10- phenanthroline) |
No need for special strain or construct Can monitor the half-life of many mRNAs simultaneously |
May cause a loss of labile factors May alter the decay of specific mRNAs May alter other cellular pathways (i.e., transcription or translation) |
| Transcriptional inhibition |
Temperature-sensitive RNA polymerase II mutant (rpb1-1 mutation) |
No need for special construct Can monitor the half-life of many mRNAs simultaneously |
May cause a loss of labile factors Not useful with other conditional mutants Possible secondary complications caused by heat shock Requires a special strain |
| Transcriptional control |
Regulatable GAL promoter |
Minimal cell perturbation Applicable to transcriptional pulse–chase experiments |
Requires a special construct Allows only mRNAs under control of GAL promoter to be analyzed Changing the carbon source may alter mRNA stability |
| Transcriptional control |
Regulatable ‘‘Tet-off’’ |
Minimal cell perturbation Applicable to transcriptional pulse–chase experiments Allows an accurate control of inhibition or induction |
Requires a special construct Allows only mRNAs under control of Tet-promoter to be analyzed The antibiotics used may alter cellular metabolism, influencing the gene under control |
2.1. In vivo labeling
Transcripts can be radiolabeled in vivo and the rate of mRNA decay determined from either the disappearance of specific mRNAs during a chase (pulse chase) or from the kinetics of the initial labeling (approach to steady state). The main advantage of the in vivo labeling methods is the limited cell perturbation. Nevertheless, several disadvantages include a requirement for large quantities of radioactively labeled nucleic acid precursors, poor signal-to-noise ratios for low abundance mRNAs, and a failure to provide information on mRNA integrity during the course of an experiment (Herrick et al., 1990). Given such disadvantages, we will emphasize protocols that measure mRNA decay rates in yeast with methods to control transcription.
2.2. General inhibition of transcription with drugs or mutations
A simple way to measure mRNA decay rates is to use methods to inhibit transcription and then follow the loss of the mRNA of interest over time. In the following section, we discuss procedures by which mRNA synthesis can be inhibited, either in general or for specific genes, and, at various times after such inhibition, the abundance of particular mRNAs is monitored by simple techniques (e.g., Northern blotting or RNase protection). Inhibition of total mRNA synthesis is accomplished by the use of either drugs that globally inhibit transcription in yeast (thiolutin; 1,10-phenanthroline) or with a strain with a temperature-sensitive allele of RNA polymerase II and shifting the culture to the restrictive temperature (Herrick et al., 1990; see also chapter 14 by Coller). These approaches are generally applicable, straightforward, and also provide information on mRNA integrity. The potential disadvantages of these protocols include the nonspecific side effects of drugs used to inhibit transcription and the potential loss of labile turnover factors in the absence of ongoing transcription. It should be noted that actinomycin D, a drug commonly used to inhibit transcription in mammalian cells, does not effectively inhibit transcription in yeast at reasonable doses, presumably because it does not efficiently cross the cell wall. The ability to globally repress transcription in Saccharomyces cerevisiae can also been used in conjunction with microarrays to analyze the rates of decay of the entire mRNA transcriptome (Duttagupta et al., 2005; Wang et al., 2002).
2.3. Regulatable promoter systems
2.3.1. Use of the galactose promoter
An alternate approach to the use of global transcriptional repression is to control the transcription of the mRNA of interest, which can be done by placing the corresponding gene under the control of a regulatable promoter. The GAL promoter is often used because it is one of the strongest inducible promoters in S. cerevisiae. In the presence of galactose, the GAL promoter is induced, and the mRNA under its control is highly expressed. Conversely, in the presence of glucose, transcription is promptly inhibited, allowing a quick switch between induction and inhibition of the mRNA to be studied. In addition, genes regulated by the GAL promoter can be used in the transcriptional pulse–chase protocol as described later. One disadvantage of the use of the GAL promoter to control transcription is that this method subjects the cells to a change in carbon source, which might alter cell physiology and thereby affect mRNA decay processes.
A procedure for measuring the half-life of an mRNA with the GAL promoter, which can also be used for the use of the temperature-sensitive allele of RNA polymerase II (rpb1-1), is described as follows:
Grow 200 ml of cells in the appropriate medium (for the GAL promoter, the medium should contain 2% galactose) until the cells reach midlog phase (OD600 0.3 to 0.4).
Harvest the cells by centrifugation and resuspend in appropriate medium. For rpb1-1 shutoffs, resuspend in 20 ml of medium preheated to 37 ° C. For shutoffs of the GAL promoter, use medium containing 2% glucose. Immediately remove a 2-ml aliquot of cells as a zero time point. This and all subsequent aliquots are centrifuged for 10 sec in a microfuge, the medium supernatant is removed by suction, and the resulting cell pellet is quickly frozen in dry ice.
Continue growing the cells taking subsequent time points as described by the individual protocol. Initially, time points at 5, 10, 20, 30, 40, 50, and 60 min are commonly used.
RNA is isolated from these cells (Caponigro et al., 1993). Northern blotting, dot blotting, or RNase protection procedures are useful for quantitating the amount of a particular mRNA at each time point. A semilog plot of the percent of mRNA remaining versus time allows for the assessment of mRNA half-life (Kim and Warner, 1983; Losson and Lacroute, 1979; Parker et al., 1991).
2.3.2. Use of the tetracycline-regulatable promoter system
An alternate method for repressing transcription of a specific yeast mRNA and thereby measure of its decay rate is to use the "Tet-Off" system, which allows the addition of the antibiotic tetracycline (Tc) or its derivative doxycycline (Dox) to selectively repress transcription of an mRNA placed under the control of the "Tet-off" promoter (Hilleren and Parker, 2003). The principle of this technique is based on the regulatory operon system present in Escherichia coli, which has been adapted to the yeast system (Gari et al., 1997). The main advantage of the "Tet-off" system over the use of the GAL promoter is that addition of Dox to cells does not elicit a nonspecific cellular response, and, therefore, the transcriptional repression is specific to only the genes expressed from the "Tet-off" promoter. However, it should be noted that a few yeast strains are sensitive to Tc, and this method should not be used in those genetic backgrounds (Blackburn and Avery, 2003). An experimental procedure with the Tet-off system to analyze mRNA decay rates is described as follows:
Prepare a construct with the gene of interest under the control of the Tet-off promoter. Note that if is the 5′ UTR portion of the mRNA is to be analyzed, care must be taken to ensure that the normal site of transcription is used.
Grow strains expressing the mRNA under the control of the Tet-off promoter in 200 ml of medium containing 2% glucose to midlog (OD600 of 0.3 to 0.4).
Pellet the cells and resuspend in 20 ml of the same medium used to grow the cells. This allows the cells to be more concentrated and facilitates the rapid harvesting of aliquots.
Add Dox to a final concentration of 2 µg/ml to block transcription.
Immediately remove a 2-ml aliquot of cells as a zero time point. This and all subsequent aliquots should be centrifuged for 10 sec in a microfuge, the medium supernatant removed by suction, and the resulting cell pellet quickly frozen in dry ice.
Continue growing the cells taking subsequent time points as described by the individual protocol. Initially, time points at 5, 10, 20, 30, 40, 50, and 60 min are commonly used. If the mRNA of interest is highly unstable, then time points of shorter duration should be used.
RNA is isolated from these cells (Caponigro et al., 1993). Northern blotting, dot blotting, or RNase protection procedures are useful for quantitating the amount of a particular mRNA at each time point. A semilog plot of the percent of mRNA remaining versus time allows for the assessment of mRNA half-life (Kim and Warner, 1983; Losson and Lacroute, 1979; Parker et al., 1991).
3. DETERMINATION OF mRNA DECAY PATHWAYS
The experimental procedures that were used to define the pathways of mRNA turnover in yeast can now be applied to any transcript to determine the specific pathway(s) of degradation an mRNA undergoes. Three main experimental approaches allow for determining the mRNA decay mechanism. First, inserting a strong secondary structure within the mRNA allows for trapping mRNA decay intermediates, which are useful for determining the directionality of decay. Second, putting the gene of interest under a regulatable promoter allows for pulse–chase experiments, thereby allowing the determination of precursor-product relationships during the mRNA decay process. Third, examining mRNA decay in various strains defective in individual steps in the mRNA turnover pathways can help define the specific turnover pathway(s) that target the mRNA of interest. Although each experimental approach has its limitations, when used in combination, clear information about the mechanism through which a transcript is degraded can often be obtained.
3.1. Trapping mRNA decay intermediates
Understanding the mechanism responsible for degrading a specific mRNA can be facilitated through trapping and structurally analyzing mRNA decay intermediates. Normally, mRNA transcripts are rapidly degraded after an initiating event (e.g., decapping), and decay intermediates cannot be detected. However, introduction of a strong secondary structure into a transcript that inhibits exonucleases traps intermediates in mRNA turnover (Fig. 20.2; Beelman and Parker, 1994; Decker and Parker, 1993; Muhlrad et al., 1994; 1995 Vreken and Raue, 1992).
Figure 20.2.
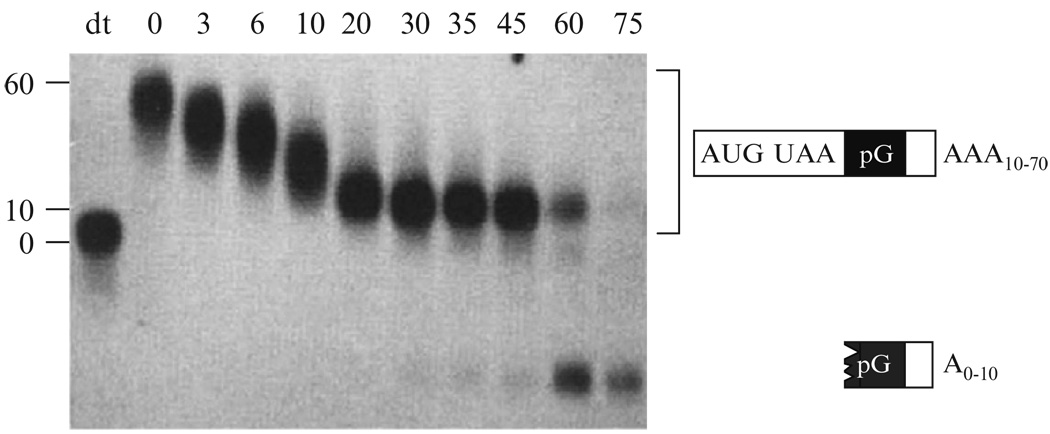
Example of a transcriptional pulse–chase experiment. The figure shows Northern blot analysis of a transcriptional pulse–chase experiment with PGK1 mRNA containing a poly(G) tract inserted into its 3′ UTR. After transcriptional repression, samples were taken at different times shown on the top. The first lane (dT) specifies sample treated with oligo d(T) and RNase H to remove the poly(A) tail. At later time points, an intermediate of mRNA decay trapped by the presence of the poly(G) tract in the 3′ UTR can be seen to accumulate.
Introducing a poly(G) tract of at least 18 G nucleotides into a yeast mRNA is commonly used as a means for introducing secondary structure (Beelman and Parker, 1994; Decker and Parker, 1993; Muhlrad et al., 1994; 1995). The strong secondary structure conferred by the addition of a series of G nucleotides is likely a result of the formation of a G-quartet structure arising from hydrogen-bonding of four G nucleotides in a planar array (Williamson et al., 1989). The poly(G) tract serves as a partial block to both 5′ to 3′ and 3′ to 5′-exonucleases, thus causing the accumulation of distinct mRNA decay intermediates (see Fig. 20.2, last two lanes, 60 and 75 min).
3.1.1. Determining the directionality of a decay pathway
The exact nature of the accumulated mRNA decay intermediate depends on the directionality of mRNA degradation. Any appropriate combination of Northern blot analyses, RNase protection assays, and primer extension assays can be used to determine the structure of a trapped intermediate and infer the nucleolytic steps that produced the intermediate. For example, an mRNA undergoing 5′ to 3′-decay will accumulate an mRNA fragment beginning with the poly(G) tract and extending to the 3′-end of that RNA (Fig. 20.2) (Decker and Parker, 1993; Muhlrad et al., 1994; 1995). Alternately, an RNA undergoing 3′ to 5′-decay will accumulate an mRNA fragment from the 5′-end to the 3′-side of the poly(G) tract (Anderson and Parker, 1998).
The placement of a poly(G) tract within an mRNA requires careful consideration. Placing the poly(G) tract in the 5′ UTR blocks ribosome scanning and alters translation of the mRNA. Because translation and mRNA degradation are interrelated (Coller and Parker, 2004), an inhibition of translation is likely to alter the decay of that mRNA and is, therefore, inappropriate for the analysis of mRNA turnover. A poly(G) tract placed within the open reading frame of an mRNA may actually stall translation elongation and trigger no-go decay, on which the mRNA is then subject to endonucleolytic cleavage (Doma and Parker, 2006). Given these complications with the 5′ UTR and coding regions, the poly(G) tract is often placed in 3′ UTR, because this positioning does not generally interfere with mRNA translation or turnover. However, additional analyses are necessary to ensure that insertion of the poly(G) tract does not alter the rate or mechanism of mRNA decay by possibly disrupting an important regulatory element found in the 3′ UTR.
A limitation of inferring decay mechanisms from the structures of trapped intermediates is that the initial nucleolytic event cannot be identified. For example, a poly(G)→3′-end fragment can be generated by decapping and then 5′ to 3′-decay (Muhlrad et al., 1994) or by an initial endonucleolytic cleavage followed by 5′ to 3′-degradation (Doma and Parker, 2006). To distinguish between these two possibilities, one needs to capture early decay intermediates to examine the initial nucleolytic events. One way to capture early decay intermediates is to insert multiple poly(G) tracts (Muhlrad and Parker, 1994). When a transcript is degraded first by decapping and followed by 5′ to 3′-degradation, the transcript will first be degraded to the 5′-poly(G) tract. In contrast, an initial endonucleolytic cleavage between the poly(G) tracts will produce two different fragments. Therefore, these two events can be distinguished by examining the structures of the intermediates derived from a transcript with two poly(G) insertions. Importantly, it should be noted that different mechanisms leading to the same decay intermediates could also be distinguished with specific trans-acting mutations (see section 3.3).
3.1.2. The use of decay intermediates to determine the role of deadenylation in decay
Trapped decay intermediates can also be examined directly to determine the role of deadenylation in mRNA decay. The predominant decay pathway accumulates poly(G)→3′-end fragment with an oligo (short) A tail. This is because turnover proceeds through first deadenylation, then decapping, and finally rapid 5′ to 3′-decay. However, decapping can occur without poly (A)-tail shortening, as has been seen with aberrant mRNAs containing premature translation termination codons (Cao and Parker, 2003; Muhlrad and Parker, 1994). In this case, because decapping occurs before deadenylation, the trapped intermediate initially contains a longer poly(A)-tail, as determined by measuring the poly(A)-tail length. Poly(A)-tail length is measured by hybridizing a portion of isolated RNA with oligo d(T), followed by RNase H cleavage; this removes the poly(A)-tail that is not hybridized to oligo d(T) (Fig. 20.2, lane dT). Separation, through a 6% polyacrylamide/8 M urea gel, of isolated RNAs both treated and untreated with oligo d(T)/RNase H allows for direct determination of the poly(A)-tail state of the RNA (Decker and Parker, 1993).
It should be noted that mRNA fragments with long poly(A) tails can be hard to detect in a steady-state population for the following reason. After their production, the poly(G)→3′-end fragments with a long poly(A) tail undergo both deadenylation and 3′ to 5′-decay (Muhlrad and Parker, 1994). Because the 3′ to 5′-decay rate is typically slower than deadenylation rate, most fragments originally with a long poly(A) tail will deadenylate rapidly and accumulate at steady state with short or no poly(A) tails. Given this limitation, the best experiment to examine whether an mRNA decay fragment is initially produced with a long poly(A) tail is to use a transcriptional induction or pulse-chase (see later) to generate a pool of newly synthesized poly(G)→3′-end fragments, whose poly(A) tails lengths can then be examined.
3.1.3. Trapped decay intermediates as a simple assay for 3′ to 5′-mRNA decay
Examining 3′ to 5′-mRNA decay can be specifically examined with the poly(G)→3′-end fragment. Investigating 3′ to 5′-decay on the full-length mRNA can be difficult, because 3′ to 5′-decay occurs slower than 5′ to 3′-decay, and very little 5′-end→poly(G) fragment is formed (Anderson and Parker, 1998; Beelman and Parker, 1994; Decker and Parker, 1993; Muhlrad et al., 1994; 1995; Williamson et al., 1989). Thus, the poly(G)→3′-end fragment can be used as a starting mRNA species for monitoring 3′ to 5′ -mRNA decay; decay of this RNA will result in a fragment containing only the poly(G) tract (Anderson and Parker, 1998). However, determining the rate of 3′ to 5′-decay on the poly(G)→3′-end fragment requires that the production of this RNA species be inhibited (as discussed in section I for determining mRNA half-life). Production of the poly(G)→3′-end fragment can be stopped by inhibiting decapping through the addition of 0.1 mg/ml cycloheximide to cultures. Cycloheximide added to cultures at a concentration of 0.1 mg/ml also inhibits translation elongation; the inhibition of mRNA decapping is likely caused by an indirect effect of cycloheximide addition (Beelman and Parker, 1994). Cycloheximide can be added at the time mRNA transcription is inhibited, and cells can be collected as indicated previously (Beelman and Parker, 1994). Alternately, the dcp1-2 temperature-sensitive allele of the decapping enzyme can be used to rapidly block decapping at the nonpermissive temperature (Tharun and Parker, 1999).
3.1.4. Limitations in the interpretations of trapped decay intermediates
Trapping and analyzing mRNA decay intermediates can ultimately lead to an understanding of the mechanism responsible for mRNA degradation, but this method does have limitations. The trapped intermediates may not reflect the major mRNA decay pathway for a particular mRNA. Specifically, mRNAs that undergo decay through more than one pathway may accumulate intermediates from a minor pathway. Two main aspects should be considered in such a case. First, the relative amounts of an accumulated intermediate should be proportional to the initial amount of full-length mRNA, although this can be misleading if the intermediate itself is very unstable. Second, analyzing the decay rate of an mRNA in cells where one decay mechanism is defective (see section 3.3 ) will aid in understanding the predominant decay mechanism. A mutation within the main pathway responsible for degrading an mRNA results in a more stable mRNA than mutations that alter a minor or secondary decay pathway. Another limitation of the methods described previously is that the intermediates trapped in the steady state provide limited information about how an individual transcript is degraded. The experiments described in the following overcome this limitation by revealing precursor-product–relationships during mRNA decay.
3.2. Determining precursor–product relationships in a transcriptional pulse–chase
A powerful method to analyze the pathway of mRNA degradation is a transcriptional pulse–chase experiment. In this experiment a regulatable promoter is used to produce a homogeneous population of mRNA transcripts, which are then followed during the subsequent steps in mRNA degradation. This reveals the order in which decay events occur and precursor-product relationships.
In S. cerevisiae, the GAL promoter has been used to perform transcriptional pulse–chase experiments (Decker and Parker, 1993; Muhlrad and Parker, 1994; Muhlrad et al., 1994; 1995). A gene of interest is put under GAL control so that a short period of transcription can be accomplished by changing carbon source. This is performed by first growing cells in medium containing a neutral carbon source, usually raffinose, which does not suppress or induce the GAL promoter. Subsequently, the promoter is induced by addition of galactose followed by suppression by glucose after a short time. Raffinose can breakdown to yield galactose at low pH, so when pregrowing cultures in raffinose, care must be taken to keep the pH above 6.0. This is mostly an issue when yeast cells are grown in synthetic media, which can be addressed by adjusting the pH of the medium (see later). For galactose induction, yeast cells can also be pregrown in medium with sucrose as a carbon source, which generally allows better growth and limits preinduction because of raffinose breakdown. However, sucrose partially represses the GAL promoter so the level of induction after galactose addition is not optimal.
After a brief period of transcriptional induction, the GAL promoter is inhibited by the addition of glucose. This creates a "pulse" of relatively homogeneous transcripts whose pathway of degradation can then be analyzed as a sequence of events. For instance, poly(A) shortening and disappearance of the full-length mRNA can be monitored to calculate the deadenylation rate and whether mRNA degradation begins before or after deadenylation. When transcripts with a poly(G) tract are used as mRNA decay precursors, the fragments trapped by the poly(G) tract will provide even more information. The level of full length mRNA and poly(G)→3′-end fragments can be monitored as can the appearance and disappearance of poly(G)→3′-end fragments and the poly(A) tail length of mRNA whose transcribed region is intact. These data provide information about the precursor-product relationships and thus the sequence of events. A detailed experimental procedure is outlined in the following.
Pregrow cells in 5 ml of medium containing 2% raffinose or 2% sucrose. Use this culture to inoculate 200 ml of medium containing 2% raffinose or 2% sucrose until cells reach early-log phase (OD600: 0.3 to 0.4). Because low pH results in acid hydrolysis of raffinose into galactose and sucrose, the pH of the medium should be adjusted to pH 6.5 with NH4OH. In some cases, overgrowth of the culture grown in raffinose can result in a lowering of the pH and, therefore, premature induction of transcription.
Pellet cells in four 50-ml Falcon tubes by spinning for 2 min at top speed in a tabletop centrifuge, resuspend cells in 10 ml of medium containing 2% raffinose (or 2% raffinose + 2% sucrose), return to incubator and shake for 10 min.
Transcriptional induction is accomplished by adding 0.5 ml of 40% galactose (a final concentration of 2%).
Immediately after adding galactose, transfer an aliquot of cells (usually 1 to 2 ml) to a 2-ml Eppendorf tube, briefly centrifuge 10 sec at top speed in a microcentrifuge, and remove the medium/supernatant by aspiration. Rapidly freeze cell pellets in crushed dry ice. This is the preinduction sample.
After a short time of induction (typically 8 to 10 min), an equal amount of medium containing 4% glucose is added. Immediately remove and quickly harvest an aliquot of cells as described above. This is the t0 sample. (Transcriptional repression can also be carried out by temperature shift from 24 °C to 36 °C with a strain harboring a Rpb1-1 mutation [a temperature-sensitive allele of RNA polymerase II, as described in section 1].) With Rpb1-1 mutants, addition of glucose to repress the GAL promoter and temperature shift to repress global transcription can be applied simultaneously to achieve tighter transcriptional repression (Decker and Parker, 1993).
Additional cell aliquots are harvested at different time points (as described previously).
mRNA is isolated from the cells. Typically 10 to 40 µg of total-cell RNA is separated through a 6% polyacrylamide/8 M urea gel (this gel is 20-cm long and 1-mm thick), which is typically subject to 300 volts for 7.5 h at room temperature. These gels allow direct determination of the poly(A) tail length by comparing the size of the RNA before and after hybridizing to oligo d(T) and treating with RNase H. RNAs too long for polyacrylamide gel analysis can be cleaved with oligonucleotides that specifically anneal to the 3′-end and subsequently treated with RNase H before loading.
Note: RNase H digestion of RNAs requires 10 µg of RNA plus 300 ng of oligo dried in a speed vac. Resuspend the pellet in 10 µl Hyb mix (25 mM Tris [pH 7.5], 1 mM EDTA, 50 mM NaCl), heat for 10 min at 68 °C, cool slowly to 30 °C and spin down. Next, add 10 µl of 2× RNase H buffer (40 mM Tris [pH 7.5], 20 mM MgCl2, 100 mM NaCl, 2 mM DTT, 60 µg/ml BSA, and 1 unit of RNase H), and incubate 30 °C for 1 h. Stop the reaction by adding 130 µl of stop mix (0.04 mg/ml tRNA, 20 mM EDTA and 300 mM NaOAC). Prepare the RNA for electrophoresis through a polyacrylamide gel by extracting with phenol/chloroform and, subsequently, chloroform, precipitating with ethanol, washing the pellet with 70% ethanol and resuspending in 10 µl of formamide gel loading dye (samples are heated to 100 °C for 3 min before loading).
Northern blot analysis with a [32P]-labeled oligonucleotide probe requires transfer of the RNA from the gel to a nitrocellulose membrane. After transfer, the membrane is washed in 0.1× SSC/0.1% SDS for 1 h at 65 °C. The blot is incubated for at least 1 h with prehybridization buffer (10× Denhardt’s, 6× SSC, 0.1% SDS) at a temperature 15 °C below the Tm of the oligonucleotide probe. Then the blot is hybridized to the labeled probe at the same temperature for at least 6 h. The blot is washed three times with 6× SSC, 0.1% SDS for 5 min at room temperature, and once for 20 min at 10 °C below the Tm of the oligonucleotide probe, dried, and exposed to X-ray film or a PhosphorImager.
3.3. Analysis of decay pathways through mutations in trans-acting factors
With S. cerevisiae as a model organism, the stability of a particular mRNA can be examined in a strain deficient in a specific mechanism of decay. An alteration of an mRNAs half-life caused by a specific mRNA decay defect directly implicates the defective mRNA turnover pathway in the decay of the mRNA of interest. Analysis of mRNA stability in a strain deficient for a particular mRNA decay pathway should be coupled with direct analysis of the decaying mRNA. The combined data will limit any possible indirect defects on mRNA decay.
3.3.1. Strains defective in deadenylation
Specific yeast mutants can be used to determine whether an mRNA requires deadenylation for degradation. This is based on the demonstration that the major cytoplasmic mRNA deadenylase requires the products of the CCR4 and CAF1 genes. In either ccr4Δ or caf1Δ strains, mRNAs that require deadenylation show slower rates of mRNA turnover, although it is most pronounced in ccr4Δ strains (Tucker et al., 2001). In contrast, mRNAs that undergo deadenylation-independent decapping show no alteration in mRNA turnover (Badis et al., 2004). In addition, it is possible that some mRNAs will be primarily deadenylated by a second deadenylase known as Pan2 (Brown et al., 1996). Thus, examining whether deadenylation is required for the degradation of a specific mRNA should involve examining the decay rate of the transcript in both ccr4Δ and pan2Δ mutant strains.
3.3.2. Strains defective in 5′ to 3′-decay
Mutations in several genes has been identified that affect mRNA decapping or 5′ to 3′-exonucleolytic decay (Table 20.2). These genes encode Dcp2p, which is the catalytic subunit of the decapping enzyme (Dunckley and Parker, 1999; Steiger et al., 2003), and Dcp1p, which directly activates Dcp2p (She et al., 2006). Furthermore, several proteins, including Dhh1p, Pat1p, Edc3p, and the Lsm1-7 complex, have been identified to enhance the rate of decapping of some, if not most, mRNAs (Coller and Parker, 2004). In addition, Xrn1p is the nuclease responsible for 5′ to 3′-degradation of mRNAs after decapping. Mutations in any of the corresponding genes cause a stabilization of transcripts that are degraded by decapping followed by 5′ to 3′-exonuclease digestion. Because Dcp2p and Xrn1p have clear biochemical roles, an ideal first experiment to see if an mRNA is subject to 5′ to 3′-degradation is to determine its rate of degradation in dcp2Δ and xrn1Δ strains compared with a wild-type strain. Such an experiment can also be followed up by determining whether degradation of the mRNA is affected in dhh1Δ, pat1Δ, lsm1Δ, and edc3Δ strains, which will provide information as to the decapping activators that are most effective on that particular mRNA.
Table 20.2.
Genes that affect mRNA decay
| Pathway | Gene | Function of protein | Accumulating intermediate in loss of function (mutant) |
|---|---|---|---|
| Deadenylation | CCR4, CAF1, Pan2 | Major deadenylases Minor deadenylase |
Adenylated capped mRNA |
| 5′ to 3′-Decay | Dcp2 | Decapping enzyme | |
| Dcp1 Dhh1 Lsm1-7 Pat1 | Activator of decapping enzyme |
Deadenylated capped mRNA | |
| Xrn1 | 5′ to 3′-exonuclease | Deadenylated decapped mRNA | |
| 3′ to 5′-Decay | Ski6 (RRP41), Ski4 (CSL4) RRP4 |
Components of exosome | Poly(G)-3′-end fragment and a series of 3′ to 5′- decay intermediates |
| Ski2, Ski3, Ski7, Ski8 | Modulate 3′ to 5′- exonuclease activity |
||
| Specialized mRNA decay |
|||
| Upf1 Upf2 Upf3 | Required for nonsense-mediated decay |
||
| Hbs1 Dom34 | Required for no-go decay | ||
| Ski7 | Specifically blocks nonstop decay |
In principle, xrn1Δ and dcp2Δ mutants could be used to distinguish endonucleolytic cleavage followed by 5′ to 3′-decay from decapping followed by 5′ to 3′-decay. In a case of endonucleolytic cleavage, a dcp2Δ mutation would be predicted to have no effect, and an xrn1Δ mutant would accumulate the products of the initial cleavage. In contrast, in a case of decapping, a dcp2Δ mutation would stabilize the transcript of interest, and an xrn1Δ mutant would accumulate full-length decapped RNA.
3.3.3. Strains defective in 3′ to 5′-decay
It is also possible to use trans-acting mutations to determine whether a transcript is primarily degraded in a 3′ to 5′-direction. Several proteins have been identified that are required for efficient cytoplasmic 3′ to 5′-decay of mRNA, including the Ski2p, Ski3p, Ski4p, Ski6p/Rrp41p, Ski7p, Ski8p, and Rrp4p (reviewed in Parker and Song, 2004). Rrp4p, Skip6/Rrp41p, and Ski4p/Csl4p are components of a multiprotein complex termed the exosome (Mitchell et al., 1997) and are likely to be part of the actual nucleolytic complex that can degrade the mRNA body 3′ to 5′- (Anderson and Parker, 1998). Ski2p, Ski3p, Ski7p, and Ski8p do not seem to be nucleases and are likely to modulate the exosome activity on mRNA substrates. Although all transcripts examined to date are not stabilized significantly in mutants solely defective in 3′ to 5′-decay, it is possible that there will be specific mRNAs, or specific conditions, wherein the 3′ to 5′-decay pathway is predominant. Thus, to determine whether an mRNA is primarily degraded 3′ to 5′, its decay rate should be examined in some combination of the ski2, ski3, ski4, ski7, ski8, rrp4, and ski6 mutants.
3.3.4. Strains specifically affecting specialized mRNA decay pathways
Yeast strains exist that allow the determination of whether a specific mRNA is targeted to one of the specialized pathways of mRNA decay (Fig. 20.1B). For example, in yeast, the Upf1p, Upf2p, and Upf3p have been shown to be specifically required for the nonsense-mediated mRNA decay (NMD) pathway (He and Jacobson, 1995; Lee and Culbertson, 1995; Leeds et al., 1991; 1992). Thus, to determine whether an mRNA is subject to NMD, its rate of degradation can be examined in Upf mutant strains compared with wild-type strains. Whether an mRNA is subject to nonstop decay (NSD) can also be determined on the basis of the finding that NSD requires a GTPase domain in the Ski7p that is not required for 3′ to 5′-degradation of normal mRNAs (Van Hoof et al., 2002). Thus, mRNAs that are stabilized in a ski7Δc mutant are, in theory, candidate NSD substrates. Finally, mRNAs that are subject to no-go decay (NGD) require Hbs1p and Dom34p for efficient endonucleolytic cleavage (Doma and Parker, 2006). Given this, mRNAs that are stabilized in these mutant strains are NGD candidates. Taken together, these mutant strains provide a mechanism for determining whether an mRNA is subject to any of these specialized pathways of destruction.
4. COMBINATION OF THE PRECEDING APPROACHES
A combination of the preceding approaches provides a powerful analysis of the mechanism responsible for the decay of a specific mRNA. A useful first step in combining the experimental approaches described is to place the gene of interest under a regulatable promoter. This allows for both easy measurements of the mRNA decay rate and the ability to perform transcriptional pulse–chase experiments if the GAL promoter is used. Next, a poly(G) tract can be inserted into the 3′ UTR. Insertion of the poly(G) tract allows for the detection of mRNA decay intermediates, which enables analysis of the role of deadenylation and the directionality of decay. Finally, examining the rate and mechanism of mRNA decay in yeast strains defective for specific mechanisms of mRNA decay can link the turnover to a specialized decay pathway. However, it should be noted that all the approaches described here are specific for defining the decay of an mRNA of interest within the context of the known mRNA decay pathways. The discovery of new mRNA decay pathways will rely on different experimental approaches and thereby will increase the number of variables to consider.
REFERENCES
- Anderson JSJ, Parker R. The 3′ to 5′-degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′-exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Blackburn AS, Avery SV. Genome-wide screening of Saccharomyces cerevisiae to identify genes required for antibiotic insusceptibility of eukaryotes. Antimicrob. Agents Chemother. 2003;47:676. doi: 10.1128/AAC.47.2.676-681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994;269:9687. [PubMed] [Google Scholar]
- Brown CE, Tarun SZ, Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;133:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: A stimulatory role for rare codons. Mol. Cell. Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2004;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta R, Tian B, Wilutsz CJ, Khounh DT, Soteropoulos P, Ouyang M, Dougherty JP, Peltz SW. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 2005;25:5499–5513. doi: 10.1128/MCB.25.13.5499-5513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldeia M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- Kim CH, Warner JR. Messenger RNA for ribosomal proteins in yeast. J. Mol. Biol. 1983;165:79–89. doi: 10.1016/s0022-2836(83)80243-5. [DOI] [PubMed] [Google Scholar]
- Lee BS, Culbertson MR. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc. Natl. Acad. Sci. USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Mol. Cell. Biol. 1991;12:2165–2177. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ to 5′-exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ to 3′-digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Parker R, Herrick D, Peltz SW. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:415–423. doi: 10.1016/0076-6879(91)94032-8. [DOI] [PubMed] [Google Scholar]
- She M, Decker CJ, Chen N, Tumati S, Parker R, Song H. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2006;13:163–170. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Vreken P, Raue HA. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′-terminal part of the coding region. Mol. Cell. Biol. 1992;12:2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schimid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]