Abstract
X chromosome inactivation (XCI) achieves dosage compensation between males and females for most X-linked genes in eutherian mammals. It is a whole-chromosome effect under the control of the XIST locus, although some genes escape inactivation. Marsupial XCI differs from the eutherian process, implying fundamental changes in the XCI mechanism during the evolution of the two lineages. There is no direct evidence for the existence of a marsupial XIST homologue. XCI has been studied for only a handful of genes in any marsupial, and none in the model kangaroo Macropus eugenii (the tammar wallaby). We have therefore studied the sequence, location and activity of a gene SLC16A2 (solute carrier, family 16, class A, member 2) that flanks XIST on the human and mouse X chromosomes. A BAC clone containing the marsupial SLC16A2 was mapped to the end of the long arm of the tammar X chromosome and used in RNA FISH experiments to determine whether one or both loci are transcribed in female cells. In male and female cells, only a single signal was found, indicating that the marsupial SLC16A2 gene is silenced on the inactivated X.
Keywords: marsupial, SLC16A2 gene, X chromosome evolution, X chromosome inactivation
Introduction
Comparative genomics is a valuable tool for exploring the organization, function and evolution of the mammalian genome. Comparison of gene location between distantly related species can provide information about genome rearrangement in evolution, identification of conserved sequences can reveal important functional sequence elements, and comparison of genetic control mechanisms can reveal how regulatory mechanisms evolved, and how they function. Studies of distantly related mammals such as human and kangaroo are particularly valuable in providing evolutionary depth for stringent analysis.
Comparative genomics has revealed the origin of the mammalian X chromosome. The gene content of the X is almost completely conserved across eutherian (‘placental’) mammals, but gene mapping and chromosome painting between the three groups of mammals (eutherians, marsupials and monotremes) show that part of the X is ancient, and part recently added (Graves 1995, Wilcox et al. 1996, Glas et al. 1999). Genes on the long arm of and the pericentric region of the human X are also X borne in marsupials, implying that this region (X conserved region XCR) has been part of the X chromosome since the divergence of marsupials and eutherians, at least 180 million years ago (Woodburne et al. 2003). However, most genes on the short arm of the human X are autosomal in marsupials and monotremes (X added region XAR), implying that they were added to the eutherian X chromosome between 105–180 million years ago (Graves 1995, Wilcox et al. 1996).
X chromosome inactivation (XCI) is a unique developmental regulatory mechanism that achieves dosage compensation of X-linked genes between males (XY) and females (XX) mammals (Lyon 1961). It occurs early in embryogenesis, is somatically heritable, and is effected by a multistep mechanism (Gartler et al. 1985, Plath et al. 2002). XCI is achieved by the transcriptional silencing of genes on one of the two X chromosomes in female cells (Graves & Gartler 1986), as predicted by Lyon (1961).
XCI in humans and mice requires a cis-acting locus for the initiation and propagation of the silencing signal. This X inactivation centre (XIC) is defined as the minimal region of overlap retained in a series of X-chromosome translocations and deletions that could undergo inactivation. In humans the XIC is a 680–1200-kb region within Xq13.2 (Brown et al. 1991, Leppig et al. 1993). The corresponding mouse XIC, based on sequence comparisons and functional analysis (located in syntenic region X42.0 cM) is smaller (Rastan 1983, Chureau et al. 2002), and is inverted relative to the centromere. However, the order and orientation of genes within the XIC is perfectly conserved except for SLC16A2, which is in the same location but in the inverse orientation in humans. The order of genes in humans is TEL, SLC16A2, XIST, CHIC1 (formerly, BRX), CDX4, NAPIL2 (formerly, BPX), and in mouse, CEN, Slc16a2, Xist, Chic1, Cdx4, NapIL2 (Chureau et al. 2002).
Surprisingly, 10–20% of genes on the human X chromosome, largely on the short arm, have been found to escape XCI (Carrel & Willard 2005), with differences in the number of escapee genes seen in mice (Disteche et al. 2002) The concentration of these escapee genes on the short arm of the human X chromosome is readily explained by the recent addition of this region to the X, and its progressive recruitment into the X inactivation system (Graves et al. 1998).
The XCI system of marsupials offers valuable variants to examine how and when the layers of this complex regulatory system evolved. Marsupial XCI differs from eutherian XCI in being paternal rather than random, incomplete and tissue-specific (Cooper et al. 1993), and there is little evidence of sex chromatin (McKay et al. 1987). Paternal X inactivation also occurs in early embryogenesis and in rodent extraembryonic tissue (Takagi et al. 1978, Huynh & Lee 2003, Okamoto et al. 2004), suggesting that the marsupial mechanism could be ancestral (discussed by Cooper 1971, Ohlsson et al. 2001). There are also differences in the molecular mechanism. Although late replication and histone deacetylation have been demonstrated in marsupials (Graves 1967, Wakefield et al. 1997), DNA methylation differences have not (Piper et al. 1993). This suggests that histone modification is a primary change shared by both groups, but DNA methylation evolved recently in eutherians as a late-acting locking mechanism.
There is no direct evidence for the existence of a marsupial XIST gene in marsupials, despite a ten-year search in this laboratory. As a starting point for a systematic search of the region syntenic to the human XIC in the tammar wallaby (Macropus eugenii), we describe here the isolation and characterization of the M. eugenii orthologue of the eutherian XIST-flanking gene SLC16A2, its physical location on the X chromosome and its X inactivation status.
The SLC16A2 gene (solute carrier, family 16, class A, member 2, also known as DSX128E, XPCT for X linked PEST box containing transporter and MCT8 for monocarboxylate cotransporter 8) is located on the long arm of the human X chromosome (Xq13.2) about 600 kb distal to the XIST gene (Lafreniere et al. 1993), and on the mouse X chromosome (X42.0 cM) about 300 kb proximal to the Xist gene (Debrand et al. 1998). Isolated by positional cloning in humans (Lafreniere et al. 1994), it covers 117 kb in humans and 147 kb in mouse (Chureau et al. 2002), with all six exon–intron boundaries perfectly conserved between the species (Debrand et al. 1998). Human SLC16A2 transcribes a 4.4-kb mRNA (Genbank accession number NM_006517) that encodes a protein of 613 amino acids. The 4.2-kb mouse cDNA (Genbank accession number NM_009197) encodes a predicted protein of 565 amino acids (Debrand et al. 1998). The SLC16A2 protein is one of 14 members of the monocarboxylate cotransporter family that mediates movement of lactate and pyruvate across membranes, and may be involved in thyroid hormone transport (Friesema et al. 2003). The protein contains 12 hydrophobic transmembrane domains and its most interesting feature is its N-terminal sequence containing a PEST motif (rich in proline/glutamic acid repeats), indicative of rapid SLC16A2 protein degeneration (Rechsteiner & Rogers 1996).
Expression studies have indicated that human SLC16A2 is subject to XCI, as it is expressed by the active X chromosome only (Lafreniere et al. 1994). X-inactivation of the mouse Slc16a2 gene has also been demonstrated (Debrand et al. 1998). We show here that a conserved tammar wallaby orthologue of SLC16A2 lies on the X chromosome and is also subject to X inactivation in a female cell line.
Materials and methods
Screening of a Macropus eugenii cDNA library
A M. eugenii pouch young cDNA library, constructed under contract by Clontech (USA) from RNA derived from one male and one female whole pouch young, was screened with a 2-kb human SLC16A2 cDNA clone. The probe was labelled with 32P-radiolabelled dATP using the Megaprime DNA labelling system (Amersham, Australia) according to the manufacturer's instructions. Hybridization was in Modified Church and Gilbert hybridization buffer (0.5 mmol/L phosphate buffer, pH 7.2, 7% (w/v) SDS, 10 mmol/L EDTA, 1% BSA), containing 100 mg/ml salmon sperm DNA, for 48 h at 55°C. Filters were first rinsed in 2× SSC and then washed three times with 0.1% SDS, 2× SSC for 10 min at 55°C and rinsed in 2× SSC at room temperature before exposing them to X-ray film for 16 h at −80°C. Positives clones were sequenced.
M. eugenii BAC library screen
The primers Me SLC16A2 F9: 5′-GCTCTGGGCT CCTCTGTTTA-3′ and Me SLC16A2 R1607: 5′-CTTCCCTGCCCTCATAACCT-3′ were designed from the isolated cDNA (above), to amplify the M. eugenii SLC16A2 3′-untranslated region (UTR, 1599 bp) from genomic DNA under the following cycling conditions: 94°C for 2 min × 1 cycle, followed by 35 cycles of 94°C for 30 s; 60°C for 30 s and 72°C for 1.5 min; with a final extension of 10 min at 72°C. PCR amplifications contained 1× PCR buffer containing MgCl2 (1.5 mmol/L, Promega, Sydney, Australia), 200 μmol/L dNTPs (Roche, Sydney, Australia), 1.0 μmol/L of each primer (Geneworks, Adelaide, Australia), 0.625 U Taq polymerase (Promega) in a 25-μl final reaction volume.
This 3′ UTR product was used to screen a M. eugenii bacterial artificial chromosome (BAC) library (Sankovic et al. 2005). The probe was labelled and BAC library filters hybridized and washed as described for the cDNA library screen; but hybridization was carried overnight at 65°C and washes performed at 65°C. Filters were exposed to X-ray film for up to a week at −80°C. DNA from positive BAC clones was prepared, and 50 ng were used for PCR reactions with primers Me SLC16A2 F9 and Me SLC16A2 R1607 to verify clones containing the SLC16A2 3′ UTR.
RNA isolation and reverse transcription PCR (RT-PCR)
Tissue was collected under The Australian National University Animal Experimentation Ethics Committee proposal number R.CG.08.03. RNA was isolated from 3-mm cubes of tissue using the RNAeasy Mini Kit (Qiagen, Melbourne, Australia) according to the manufacturer's instructions. Isolated RNA (1 μg) was reverse-transcribed using Expand Reverse Transcriptase (Roche, Sydney) with random decamers (Ambion, Adelaide, Australia) following the manufacturers' instructions (Ambion). ‘No reverse transcriptase’ control reactions were set up to make sure that amplified products did not originate from DNA contamination. The cDNA template (100 ng) was amplified with Me SLC16A2 F9 primer and Me SLC16A2 R9: 5′-CCTTCCCTGCCCTCATAACCTA-3′ primer, generating a 427-bp product. Relative quantitative RT-PCR was performed by multiplexing SLC16A2 amplification with Quantum RNA 18S Universal Internal Standards (Ambion). As advised by the manufacturer, we determined the linear range of amplification for SLC16A2 and the optimal ratio of 18S primer : competimer. PCR amplifications contained reagents as listed previously with the addition of 2 μl 3 : 7 18S primer : competimer mix in a 25-μl final reaction volume. Cycling parameters were also as described above, but only for 30 cycles. Images were analysed with the Image J software and relative quantitation was determined by measuring band brightness. Gene-specific values were divided by the corresponding 18S values. Experiments were performed in triplicate.
Metaphase spread preparation
M. eugenii male fibroblast cells and female cornea cells were cultured at 35°C in an atmosphere of 5% CO2 in a 1 : 1 mix of Dulbecco's Modified Eagle's and AmnioMax C100 media (Gibco Invitrogen Corp., Carlsbad, CA, USA), supplemented with 10% fetal calf serum, l-glutamine (0.1 mg/ml), and gentamycin (10 μg/ml) (Gibco Invitrogen). Chromosome preparations were made by arresting cells at metaphase for 2 h with a 50 ng/ml final concentration of Colcemid (Roche). Cells were harvested and then swelled for 1 h in 0.075 mol/L warm (37°C) KCl and fixed in several changes of 3 : 1 methanol : acetic acid. The fixed cell suspensions were dropped onto water drops on cleaned microscope slides and air-dried. Slides were used for DNA FISH and for G-banding analysis.
DNA FISH
The Me SLC16A2 BAC (1 μg) was biotin-labelled by nick translation using the BioNick Labeling System (Invitrogen Life Technologies) following the manufacturer's instructions. The labelled probe was precipitated overnight (at −20°C) in the presence of 20 μg glycogen, 1 μg M. eugenii sheared genomic DNA and 3 volumes of 100% ethanol. The probe was resuspended by incubation in 15 μl hybridization buffer (50% v/v deionized formamide, 10% v/v dextran sulfate, 2× SSC, 40 mmol/L sodium phosphate, 1× Denhardt's solution) at 37°C for 30 min. M. eugenii male fibroblast metaphase spreads were placed in denaturing solution (2× SCC, 70% formamide) at 70°C for 1 min 40 s and, immediately after, in ice-cold 70% ethanol for 5 min, dehydrated to 100% ethanol at room temperature and air dried at 37°C until ready to use. The probe was denatured for 10 min at 70°C, quenched on ice for 2 min and allowed to preanneal for 20 min at 37°C. Probe (13 μl) was applied to a metaphase spread on a treated slide and sealed under a coverslip with rubber cement. Hybridization was for 48 h in a humid chamber at 37°C. After hybridization, slides were soaked in 2× SSC for 5 min, washed twice for 5 min in 0.5× SSC, 50% formamide at 42°C, twice for 5 min in 0.5× SSC at 42°C and once in 4× SST (4× SSC, 0.05% Tween 20) for 5 min at room temperature. They were blocked for 30 min in 3% BSA, 4× SST (100 μl) at 37°C under a parafilm coverslip. Probe was detected with three layers of antibodies (100 μl). Fluorescein isothiocyanate (FITC)-conjugated avidin was used in the first and last layer (1 : 200 dilution in 1% BSA, 4× SST) and biotin anti-avidin in the second layer (same dilution). Antibodies were incubated for 30 min each at 37°C and kept in the dark. Between layer applications slides were washed three times for 5 min in 4× SST at room temperature, except after the last antibody layer when four washes were performed. Slides were mounted using Vectashield (Vector Laboratories, California, USA) containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) to counterstain chromosomes and cell nuclei. Fluorescence was visualized with a Ziess Axioplan epifluorescence microscope and images recorded with a thermoelectronically-cooled charge-coupled device camera (RT Monochrome Spot, Diagnostic Instruments, Sterling Heights, MI, USA). IPLab imaging software (Scanalytics Inc., Fairfax, VA, USA) was used to capture grey-scale images and to convert them to coloured images that could be superimposed.
RNA FISH
RNA FISH was performed using the Me SLC16A2 BAC. The BAC (1 μg), labelled using a DIG nick-translation kit (Roche), was mixed with 45 μg total marsupial (M. eugenii) DNA, 3 μg salmon sperm DNA, 3 μg yeast tRNA, 10 μl 3-mol/L sodium acetate and 700 μl of 100% ethanol prior to overnight precipitation. Hybridization buffer (10 μl of 50% formamide, 2× SSC and 10% dextran sulfate) was added to the probe mix. Marsupial (M. eugenii) female fibroblasts derived from cornea and embryonic male fibroblasts were cultured on gelatin-coated Plastek coverslip kits (MatTek Corporations) in AmnioMAX media (Gibco Invitrogen). Coverslips were washed with PBS and cells were either permeabilized using Triton X in buffer (100 mmol/L NaCl, 300 mmol/L sucrose, 3 mmol/L MgCl2, 10 mmol/L Pipes, pH 6.8) prior to fixation in 4% paraformaldehyde (10 min) or fixed in 1 : 3 acetic acid : methanol fixative at room temperature twice for 2 min and once for 15 min and air-dried. There was no significant difference between results obtained with either fixation method. After denaturation of the labelled probe at 75°C for 5 min and preannealing at 37°C for 30–45 min, 6 μl of probe was added to the coverslips and hybridized overnight at 37°C in a humid chamber. Coverslips were washed in 2× SSC at 73°C for 5 min and in 2× SSC containing 0.03% Triton X-100 for 5 min to remove excess probe. Blocking buffer was added to coverslips (4× SSC with 0.4% nuclease-free BSA) and incubated for 20 min at 37°C. Coverslips were incubated with the primary antibody in detection buffer (4 μl mouse anti-DIG in 1 ml 4× SSC and 0.1% nuclease-free BSA) for 60 min. Coverslips were washed three times in 2× SSC with 0.03% Triton X-100 for 2 min each and then incubated with the secondary antibody in detection buffer (36 μl FITC anti-mouse in 964 μl 4× SSC and 0.1% nuclease-free BSA) for 30 min. Coverslips were again washed three times in 2× SSC with 0.03% Triton X-100 for 2 min each, and then 2 min with 2× SSC before being stained with 0.5 μg/ml Hoechst for 5 min. Coverslips were washed in PBS, then mounted using Vectashield (Vector Laboratories) and viewed under a fluorescence microscope. Two to five independent hybridizations were performed for each cell line, and two investigators examined the slides.
Results
Isolation of M. eugenii SLC16A2 cDNA and BAC clone
Screening of the M. eugenii pouch young cDNA library yielded 3797 bp of SLC16A2 mRNA sequence (Genbank accession AY735990). A total of five clones were isolated and restriction mapped. All contained shared bands, so the longest clone was sequenced.
The CLUSTAL W program (Thompson et al. 1994), available through Biomanager (ANGIS – http://www.angis.com.au), was used to compare the M. eugenii and human (Genbank accession U05315) SLC16A2 sequences. This comparison revealed strong conservation in the coding region of the gene (77% sequence identity), dramatically decreasing over the 3′ UTR (47%). The M. eugenii predicted SLC16A2 protein is 495 amino acids and shares high homology with the human protein (Figure 1).
Figure 1.
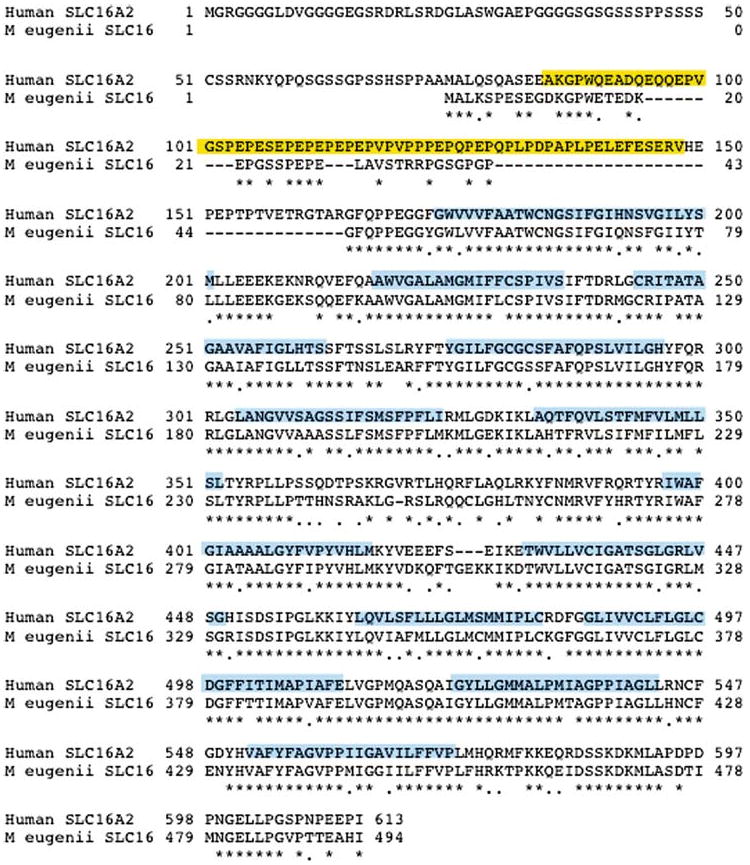
Alignment of M. eugenii and human SLC16A2 proteins. An asterisk below the two sequences indicates homology; dots indicate conservative substitutions. Human PEST domain is boxed yellow and hydrophobic transmembrane domains are boxed blue.
A maximum likelihood protein tree (Whelan & Goldman 2001) using an empirical model for globular proteins was constructed using the PyEvolve molecular evolution toolkit (Butterfield et al. 2004). The SLC16A2 protein phylogenetic tree (Figure 2) shows the M. eugenii SLC16A2 protein clustering with all the SLC16A2 proteins available for different species, further supporting the identification of a SLC16A2 orthologue rather than a pseudogene or homologue of other family members. The topology of the tree is in good agreement with the accepted phylogenetic relationship of the species used.
Figure 2.
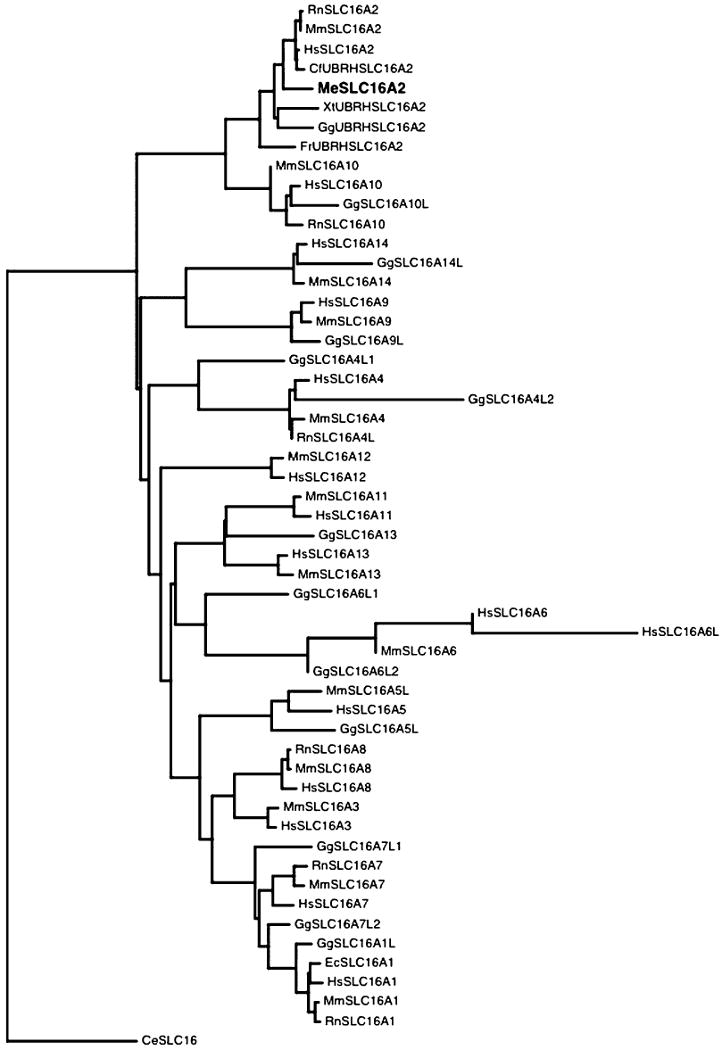
Phylogenetic tree based on SLC16A family protein sequences. Species abbreviations: Hs = Homo sapiens, Rn = Rattus norvegicus, Mm = Mus musculus, Cf = Canis familiaris, Xl = Xenopus laevis, Gg = Gallus gallus, Fr = Fugu rubripes, Ec = Equus caballus. Genbank accession numbers used in analysis: Me SLC16A2 AY735990, CfUBRHSLC16A2 : ENSCAFP00000025324, GgUBRHSLC16A2:ENSGALP00000012547, XtUBRHSLC16A2:ENSXETP00000009960, FrUBRHSLC16A2:SINFRUP00000142054, RnSLC16A1:NP_036848.1, RnSLC16A2:NP_671749.1, RnSLC16A4L:XP_215673.2, RnSLC16A7:NP_058998.1, RnSLC16A8:NP_113932.1, RnSLC16A10:NP_620186.1, HsSLC16A1:NP_003042.2, HsSLC16A2:NP_006508.1, HsSLC16A3:NP_004198.1, HsSLC16A4:NP_004687.1, HsSLC16A5:NP_004686.1, HsSLC16A6:NP_004685.2, HsSLC16A6L:XP_496241.1, HsSLC16A7:NP_004722.2, HsSLC16A8:NP_037488.1, HsSLC16A9:NP_919274.1, HsSLC16A10:NP_061063.2, HsSLC16A11:NP_699188.1, HsSLC16A12:ENSG00000152779, HsSLC16A13:NP_963860.1, HsSLC16A14:NP_689740.2, MmSLC16A1:NP_033222.1, MmSLC16A2:NP_033223.1, MmSLC16A3:NP_109621.1, MmSLC16A4:NP_666248.1, MmSLC16A5L:XP_126601.4, MmSLC16A6:NP_598799.1, MmSLC16A7:NP_035521.1, MmSLC16A8:NP_065262.1, MmSLC16A9:NP_080083.2, MmSLC16A10:NP_082523.1, MmSLC16A11:NP_694721.1, MmSLC16A12:NP_766426.1, MmSLC16A13:NP_758959.1, MmSLC16A14:NP_082197.1, GgSLC16A13:XP_423563.1, GgSLC16A1L:XP_418002.1, GgSLC16A4L1:XP_417946.1, GgSLC16A4L2:XP_417945.1, GgSLC16A5L:XP_420120.1, GgSLC16A6L1:XP_429154.1, GgSLC16A6L2:XP_415687.1, GgSLC16A7L1:XP_414421.1, GgSLC16A7L2:XP_416057.1, GgSLC16A9L:XP_421548.1, GgSLC16A10L:XP_419783.1, GgSLC16A14L:XP_422602.1, EcSLC16A1:AAR21622.1, CeSLC16:NP_510225.2.
The isolated M. eugenii SLC16A2 3′ UTR sequence was used as a probe to screen the M. eugenii BAC library. One positive BAC clone (121C8) was confirmed by amplification of SLC16A2 3′ UTR and sequencing of the product. The full BAC clone sequence (37 kb) is available under Genbank accession number CR385048. The BAC clone contains exons 4–6 of the marsupial SLC16A2, and the two terminal exons of the RNF12 (putative ring zinc finger protein Ny-REN-43 antigen) gene. Comparisons of the genomic sequence (BAC clone 128C1) with human and mouse sequence showed no conserved intronic or extragenic regions.
Expression analysis of the marsupial SLC16A2
RT-PCR was performed to determine whether the marsupial SLC16A2 gene was expressed in adult female tissues (brain, lung, heart, muscle, liver, kidney), adult testis and female pouch young brain (Figure 3). Internal 18S rDNA universal primers were used to relatively quantitate expression. The marsupial SLC16A2 gene was highly expressed in pouch young brain and adult kidney, moderately expressed in adult brain, lung, heart and muscle, and not, or barely expressed in liver and testis.
Figure 3.
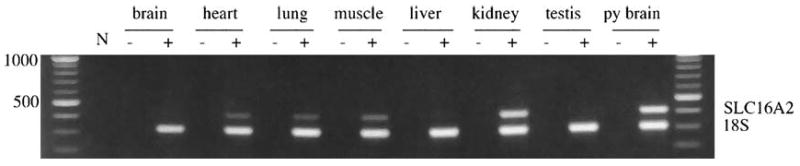
Expression of SLC16A2. RT-PCR experiments showing M. eugenii SLC16A2 gene expression across different adult female tissues, testis and female pouch young brain. N = PCR negative control, − = no RT control.
Mapping marsupial SLC16A2
Fluorescence in-situ hybridization (DNA FISH) using the Me SLC16A2 BAC clone 121C8 mapped the M. eugenii SLC16A2 gene to the end of the long arm of the X chromosome (Figure 4). Twenty male cells were examined, and all showed signal in this position on both chromatids.
Figure 4.
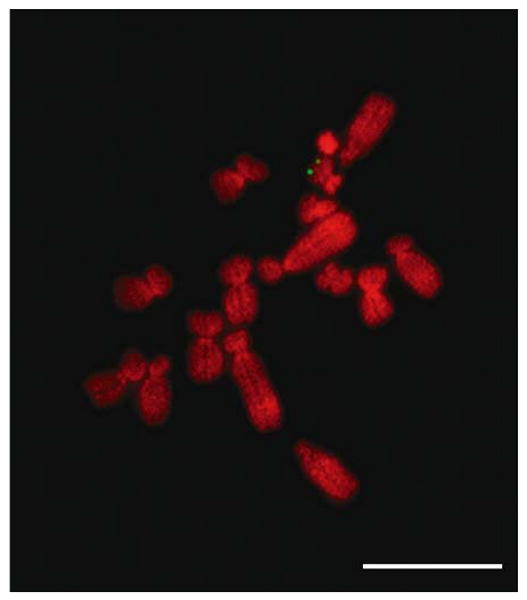
Mapping of SLC16A2 by DNA FISH. M. eugenii male metaphase spread showing signals on the long arm of the X chromosome after DNA FISH using the marsupial SLC16A2 BAC clone. Bar represents 10 μm.
Inactivation status of the marsupial SLC16A2
To determine the X inactivation status of SLC16A2, we performed RNA FISH using the Me SLC16A2-BAC clone 121C8 on both female and male fibroblasts cells (Figure 5). As shown in Table 1, we found a single signal in most nuclei from both sexes (84% of 296 cells scored in male from two experiments, and 76% of 561 cells in female from five experiments). There was no significant difference between results obtained from nuclei fixed in paraformaldehyde (75% of 435 female cells with one signal) and nuclei fixed in methanol : acetic acid (82% of 126 female cells with one signal). Cells with no signals probably represent inefficient hybridization. Of the female cells, 17% had two signals, and could potentially represent cells in which SLC16A2 escaped X inactivation, but the additional signal might also represent background fluorescence, and/or tetraploid cells. By cytogenetic analysis we determined that 6.4% and 3.6% of metaphase cells were tetraploid in the female cell line and in the male cell line, respectively. Our results indicate that the marsupial SLC16A2 gene is subject to X inactivation, at least in a large majority of cells.
Figure 5.
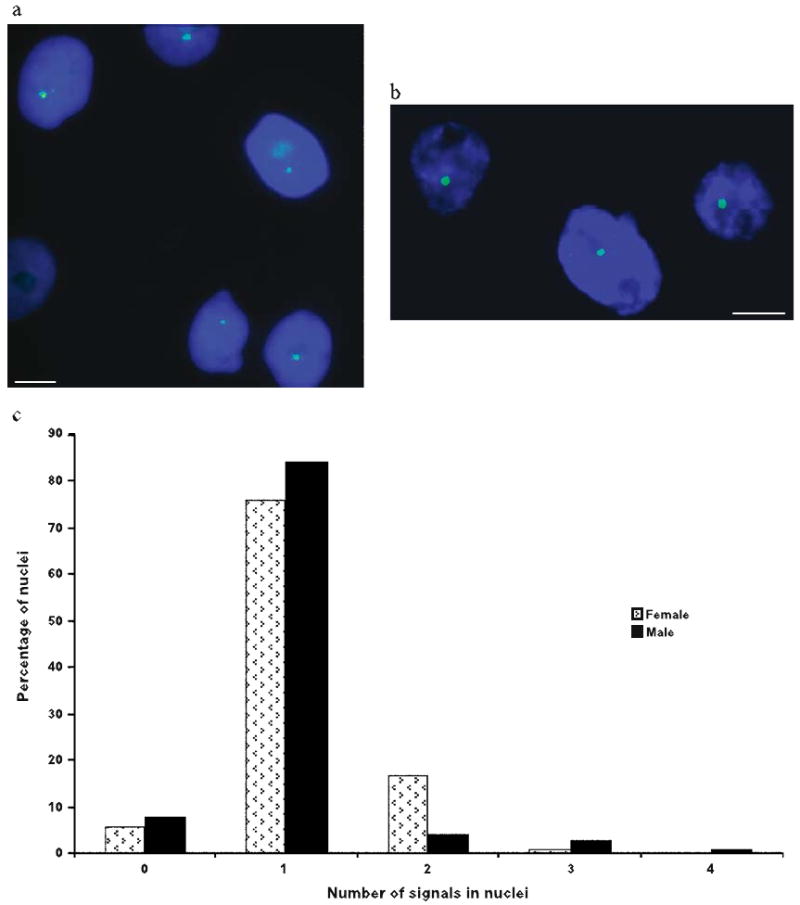
X inactivation of SLC16A2 determined by RNA FISH. (a) Hybridization signals (green) in nuclei from a female cell line. Only one signal is visible per nucleus. (b) Hybridization signals (green) in nuclei from a male fibroblast cell line. Only one signal is visible per nucleus. Nuclei are counterstained with Hoechst stain (blue). Bars represent 10 μm. (c) Histogram summarizing results shown in Table 1.
Table 1.
RNA FISH analysis of M. eugenii SLC16A2.
| Number of signals in interphase nuclei | |||||
|---|---|---|---|---|---|
| Cell line | 0 | 1 | 2 | 3 | 4 |
| Female | 31 (6%)a | 429 (76%) | 93 (17%) | 8 (1%) | 0 (0%) |
| Male | 23 (8%) | 248 (84%) | 13 (4%) | 10 (3%) | 2 (1%) |
Number of cells and % of total with a given number of signals.
Discussion
The informative variation between eutherian and marsupial XCI, and the prediction that marsupial XCI represents the ancestral mechanism, make it vital to compare the elements of X inactivation. Comparison of the physical organization and evolution of the region in and surrounding the putative marsupial XIC will provide us with a starting point to search for the elusive marsupial XIST homologue. We have therefore isolated a full-length cDNA clone, and a partial genomic clone, of the marsupial SLC16A2 gene, which flanks XIST in human and mouse, and can therefore compare its sequence, expression, location and activity between marsupials and eutherians.
Sequence comparisons of SLC16A2
The phylogenetic relationship that we established between the marsupial SLC16A2 protein and all other SLC16A2 protein sequences available to date is in accordance with the evolutionary divergence of the species. The full-length cDNA sequence of the M. eugenii SLC6A2 gene isolated from a male + female pouch young cDNA library shows high homology to its human orthologue over the coding sequence. At the protein level, the human and marsupial SLC6A2 amino acid sequences are particularly conserved over the 12 hydrophobic transmembrane domains (Lafreniere et al. 1994), reflecting important evolutionary constraints on the structure of the SLC16A2 protein. However, the N-terminal PEST domain, which is thought to be important for the turnover of the protein, is less conserved than it is between the human and murine proteins. This is consistent with the proposal (Debrand et al. 1998) that overall structural conservation of the PEST domain is sufficient, without a requirement for exact homology at the amino acid level, although it remains possible that sequence divergence of this domain reflects a real functional divergence between the species.
Expression profile of marsupial SLC16A2
The expression profile of SLC16A2 was somewhat different between M. eugenii and human/mouse. In tammar, expression was highest in the pouch young brain and adult kidney, whereas in humans it is highest in liver; expression in mouse liver and kidney was also high, with very low expression in adult brain tissues (Lafreniere et al. 1994, Debrand et al. 1998). The high expression in brain of the developing tammar is interesting because it may relate to the function of SLC16A2 in thyroid hormone transport. SLC16A2 has been described as the most active and very specific thyroid hormone transporter known to date (Friesema et al. 2003). Thyroid hormones are important for tissue development and metabolic function, and have been shown to be essential for normal neonatal brain development in both humans and rodents (Porterfield & Hendrich 2005). Mutations in the human gene have been described in two unrelated families. Affected males showed abnormal thyroid phenotypes with severe neurological abnormalities. Related affected females showed a milder thyroid phenotype and no neurological defects (Dumitrescu et al. 2004), presumably because random X inactivation results in the presence of some normal protein in heterozygotes.
Location of marsupial SLC16A2
Human SLC16A2 (human Xq 13.2) maps within the X conserved region on the long arm of the human X at Xq13.2, close to the XIST gene. We determined that the M. eugenii SLC16A2 maps to the distal end of the long arm of the X chromosome. The X location of the tammar SLC16A2 is consistent with the theory that the marsupial X chromosome represents the ancient mammalian X, represented by human Xq, which was enlarged by the addition of an autosomal region (Graves 1995). However, the M. eugenii G6PD gene (located at human Xq28) maps just below the centromere on the long arm of the tammar X chromosome (Koina & Graves 2005), suggesting at least one large rearrangement between the X chromosomes of M. eugenii and humans.
Inactivation of marsupial SLC16A2
The human SLC16A2 gene was shown to be subject to XCI by recording expression (RT-PCR) from the active and inactive human X chromosomes isolated in somatic cell hybrids. Products could be amplified only from hybrids containing an active X (Lafreniere et al. 1994). In mouse, Debrand et al. (1998) used SSCP analysis to distinguish allelic transcripts that differ in the 3′ UTR in F1 hybrid females that undergo non-random XCI such that genes subject to XCI are expressed only from one allele (Rougeulle & Avner 1996). There was only monoallelic SLC16A2 expression in the F1 females, indicating the mouse SLC16A2 gene is also subject to XCI.
The analysis of whether or not genes are subject to X inactivation is more difficult in species such as the tammar for which somatic cell hybrids or extensive expressed polymorphisms and clonal cell lines are not available, or are more difficult to obtain. The marsupial X inactivation status of classical X-linked genes like G6PD and PGK1 were determined by analysing isozyme expression in hybrids (Samollow et al. 1986). The isolation of a marsupial BAC clone containing the SLC16A2 gene allowed us to performed RNA FISH experiments to determine the X inactivation status of the gene. Our RNA FISH results are consistent with the SLC16A2 gene being subject to X inactivation in marsupials.
The marsupial X inactivation system is thought to be ancestral to all mammals, so it is important to identify the components of the complex regulatory system that are shared with eutherians, and therefore ancient. Identification of marsupial XIST, or the unambiguous demonstration that there is no marsupial XIST, is therefore important for our understanding of eutherian X inactivation. The impending marsupial genomic sequencing (Wakefield & Graves 2003) will allow us to perform a more complete comparative analysis, including the identification of additional transcripts and their X inactivation status, providing a framework for generating and testing different hypotheses to explain how X chromosome inactivation evolved, and how it functions.
Acknowledgments
This work was supported by funding from the Australian Research Council to the Centre for Kangaroo Genomics (to JAMG), and a grant from the National Institutes of Health GM 61948 (to CMD).
References
- Brown CJ, Lafreniere RG, Powers VE, et al. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- Butterfield A, Vedagiri V, Lang E, Lawrence C, Wakefield MJ, Isaev A. PyEvolve: a toolkit for statistical modelling of molecular evolution. BMC Bioinformatics. 2004;5:1. doi: 10.1186/1471-2105-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, et al. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DW. Directed genetic change model for X chromosome inactivation in eutherian mammals. Nature. 1971;230:292–294. doi: 10.1038/230292a0. [DOI] [PubMed] [Google Scholar]
- Cooper DW, Johnston PG, Graves JAM. X-inactivation in marsupials and monotremes. Sem Dev Biol. 1993;4:117–128. [Google Scholar]
- Debrand E, Heard E, Avner P. Cloning and localization of the murine Xpct gene: evidence for complex rearrangements during the evolution of the region around the Xist gene. Genomics. 1998;48:296–303. doi: 10.1006/geno.1997.5173. [DOI] [PubMed] [Google Scholar]
- Disteche CM, Filippova GM, Tsuchiya KD. Escape from X inactivation. Cytogenet Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao X, Best TB, Brockman K, Refetoff S. A novel syndrome combining thyroid and neurological abnormalities is associated with mutations in a monocarboxylate transporter gene. Am J Hum Genet. 2004;74:168–175. doi: 10.1086/380999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesema EC, Ganguly S, Abdalla A, Manning Fox JE, Halestrap AP, Visser TJ. Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter. J Biol Chem. 2003;278:40128–40135. doi: 10.1074/jbc.M300909200. [DOI] [PubMed] [Google Scholar]
- Gartler SM, Dyer KA, Graves JAM, Rocchi M. In: Biochemistry and Biology of DNA Methylation. Cantoni SL, Razin A, editors. AR Liss; 1985. pp. 223–235. [Google Scholar]
- Glas R, Graves JAM, Toder R, Ferguson-Smith M, O'Brien PC. Cross-species chromosome painting between human and marsupial directly demonstrates the ancient region of the mammalian X. Mamm Genome. 1999;10:1115–1116. doi: 10.1007/s003359901174. [DOI] [PubMed] [Google Scholar]
- Graves JAM. DNA synthesis in chromosomes of cultured leucocytes from two marsupial species. Exp Cell Res. 1967;46:37–47. doi: 10.1016/0014-4827(67)90407-7. [DOI] [PubMed] [Google Scholar]
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes – an evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Gartler SM. Mammalian X chromosome inactivation: testing the hyphothesis of transcriptional control. Somat Cell Molec Genet. 1986;12:275–280. doi: 10.1007/BF01570786. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Disteche CM, Toder R. Gene dosage in the evolution and function of mammalian sex chromosomes. Cytogenet Cell Genet. 1998;80:94–103. doi: 10.1159/000014963. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- Koina E, Graves JAM. Assignment of the glucose-6-phosphate dehydrogenase (G6PD) gene to tammar wallaby chromosome Xq by fluorescence in situ hybridization with a BAC clone. Cytogenet Genome Res. 2005;108:362. doi: 10.1159/000081535. [DOI] [PubMed] [Google Scholar]
- Lafreniere RG, Brown CJ, Rider S, et al. 2.6 Mb YAC contig of the human X inactivation center region in Xq13: Physical linkage of the RPS4X, PHKA1, XIST and DXS128E genes. Hum Mol Genet. 1993;2:1105–1115. doi: 10.1093/hmg/2.8.1105. [DOI] [PubMed] [Google Scholar]
- Lafreniere RG, Carrel L, Willard HF. A novel transmembrane transporter encoded by the XPCT gene in Xq13.2. Hum Mol Genet. 1994;3:1133–1139. doi: 10.1093/hmg/3.7.1133. [DOI] [PubMed] [Google Scholar]
- Leppig KA, Brown CJ, Bressler SL, et al. Mapping of the distal boundary of the X inactivation center in a rearranged X chromosome from a female expressing XIST. Hum Mol Genet. 1993;2:883–887. doi: 10.1093/hmg/2.7.883. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- McKay LM, Wrigley JM, Graves JAM. Evolution of mammalian X-chromosome inactivation: Sex chromatin in monotremes and marsupials. Aust J Biol Sci. 1987;40:397–404. doi: 10.1071/bi9870397. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Paldi A, Graves JAM. Did genomic imprinting and X chromosome inactivation arise from stochasti expression. Trends Genet. 2001;17:136–141. doi: 10.1016/s0168-9525(00)02211-3. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Piper AA, Bennett AM, Noyce L, Swanton MK, Cooper DW. Isolation of a clone partially encoding hill kangaroo X-linked hypoxanthine phosphoribosyltransferase: sex differences in methylation in the body of the gene. Somat Cell Mol Genet. 1993;19:141–159. doi: 10.1007/BF01233530. [DOI] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- Porterfield SP, Hendrich CE. The role of thyroid hormones in prenatal and neonatal neurological development – current perspectives. Endocr Rev. 2005;14:94–106. doi: 10.1210/edrv-14-1-94. [DOI] [PubMed] [Google Scholar]
- Rastan S. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos – location of the inactivation centre. J Embryol Exp Morphol. 1983;78:1–22. [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Rougeulle C, Avner P. Cloning and chracaterization of a murine brain specific gene Bpx and its human homolog lying within the Xic candidate region. Hum Mol Genet. 1996;5:41–49. doi: 10.1093/hmg/5.1.41. [DOI] [PubMed] [Google Scholar]
- Samollow PB, Ford AL, VandeBerg JL. X-chromosome dosage compensation in the Virginia opossum. Differential expression of the paternally derived Gpd and Pgk-A loci. Genetics. 1986;115:185–195. doi: 10.1093/genetics/115.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankovic N, Bawden W, Martyn J, Graves JAM, Zuelke K. Construction of a marsupial BAC library from the model Australian marsupial Tammar Wallaby (Macropus eugenii) Aust J Zool. 2005 in press. [Google Scholar]
- Takagi N, Wake N, Sasaki M. Cytologic evidence for preferential inactivation of the paternally derived X chromosome in XX mouse blastocysts. Cytogenet Cell Genet. 1978;20:240–248. doi: 10.1159/000130856. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield MJ, Graves JAM. The kangaroo genome. Leaps and bounds in comparative genomics. EMBO Rep. 2003;4:143–147. doi: 10.1038/sj.embor.embor739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield MJ, Keohane AM, Turner BM, Graves JAM. Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc Natl Acad Sci USA. 1997;94:9665–9668. doi: 10.1073/pnas.94.18.9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wilcox SA, Watson JM, Spencer JA, Graves JAM. Comparative mapping identifies the fusion point of an ancient mammalian X-autosomal rearrangement. Genomics. 1996;35:66–70. doi: 10.1006/geno.1996.0323. [DOI] [PubMed] [Google Scholar]
- Woodburne MO, Rich TH, Springer MS. The evolution of tribospheny and the antiquity of mammalian clades. Mol Phylogenet Evol. 2003;28:360–385. doi: 10.1016/s1055-7903(03)00113-1. [DOI] [PubMed] [Google Scholar]


