Summary
To increase compatibility between different generations of Affymetrix GeneChip arrays, we propose a method of filtering probes based on their sequences. Our method is implemented as a web-based service for downloading necessary materials for converting the raw data files (*.CEL) for comparative analysis. The user can specify the appropriate level of filtering by setting the criteria for the minimum overlap length between probe sequences and the minimum number of usable probe pairs per probe set. Our website supports a within-species comparison for human and mouse GeneChip arrays.
INTRODUCTION
Microarray analysis involving different array types is a challenging task. While the importance of a comparative analysis involving related data in various repositories is recognized, many difficulties currently hinder such analysis. The several array platforms available are very different in probe design, hybridization protocols, and data processing. As a result, the variability due to platform is often greater than the biological variability and the data generated from different platforms cannot be combined efficiently. Moreover, even the data from different generations of the same platform suffer from the same problem (Hwang et al. (2004)). Due to the still-evolving nature of genomic sequence information and technological advances in probe design, the probe sequences for the same transcripts change, and this can result in significant discrepancies in expression measurements from previous ones. These difficulties have resulted in various levels of discordance in array comparisons so far, e.g., Kuo et al. (2002); Nimgaonkar et al. (2003); Hwang et al. (2004).
As a preliminary step to the resolution of this issue, we have implemented a method for enhancing the comparability between different generations of Affymetrix GeneChip arrays. It has been shown that the similarity of probe sets is significantly related to their reproducibility across different generations of arrays (Mecham et al. (2004)) and that simple matching of the most similar probe sets alone is inadequate for comparative analysis (Hwang et al. (2004)). Our solution is to increase the similarity between probe sets by filtering probes based on their sequences. For this purpose, the minimum overlap length between probe sequences is used as the basic criterion for probe filtering. Another criterion is the minimum number of usable probe pairs per probe set, as each probe set contains multiple probe pairs. There is a trade-off between compatibility and gene coverage here: more stringent values will result in more comparable and stable expression values across arrays but for fewer probe sets.
IMPLEMENTATION
The website generates a mask file for the platforms and parameters specified by the user and provides a Java program to modify the raw data files (*.CEL) accordingly. The motivation and methodological justification for this work are described in our previous investigation with HG-U95Av2 and HG-U133A data (Hwang et al. (2004))
Probe set matching
Array Comparison Spreadsheets from Affymetrix web site (http://www.affymetrix.com/support/technical/comparison_spreadsheets.affx) were used for probe set matching. The “Best Match” table was adopted when available. In order to apply the criteria on probe filtering, we focused on one-to-one matches from the match tables.
Probe alignment
All probes were aligned to human genome sequence Build 34 (July 2003 freeze) or mouse genome sequence Build 32 (Oct. 2003 freeze), available at UCSC Genome Bioinformatics (http://genome.ucsc.edu/). The alignment was efficiently performed using the BLAT search tool (build version 26). Probes aligned to multiple regions on genomic sequences were excluded from further analysis because of the possibility of cross hybridization.
Probe filtering
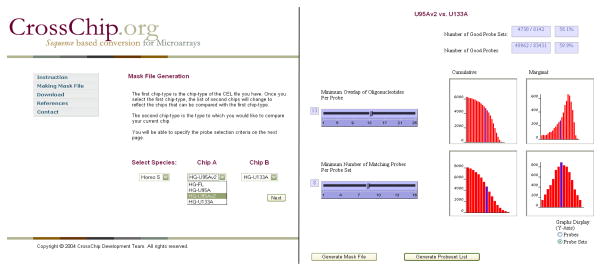
First, the user specifies the species and the platforms to be compared, as well as the minimum sequence overlap length and the minimum probes per probe set, as shown in Figure 1. The sequence overlap can range from 1 to 25 since the probes are 25-mers and the minimum probes can range from 1 to 11, 16, or 20, depending on the chip type. In order to guide the user in choosing the appropriate parameters, four graphs dynamically display the number of probes and probe sets satisfying the criteria.
Fig. 1.
The mask file generation page of http://www.crosschip.org. The user generates mask files for the two chip types to be compared, according to the criteria on the minimum sequence overlap length and the minimum number of probe pairs per probe set.
Our method of probe filtering is carried out by masking out the filtered probes from the raw data files. The website generates the mask file for the platforms of interest according to the user-specified criteria and provides a Java application for converting CEL files. After these two files are downloaded, the Java program on the user’s computer augments the CEL files with the mask information. (Due to their large sizes, we have avoided having to upload the CEL files to our website.) After the modification, the user can reprocess the CEL files using Microarray Analysis Suite from Affymetrix or any other program that computes probe-set level expression levels from probe level data.
CONCLUSION
The CrossChip website (http://www.crosschip.org) supports comparative analysis between different generations of Affymetrix GeneChip arrays by sequence-based filtering of probes. The mask files generated by this website allow the user to obtain a new set of expression values that are amenable to cross-platform analysis.
Acknowledgments
SWK was supported by 5U01HL066582-04 from NIH; PJP was supported by K25-GM67825 from NIH. KBH and BTZ were supported by Korean Ministry of Science and Technology under the NRL project.
Footnotes
Availability: http://www.crosschip.org
References
- Hwang KB, Kong SW, Greenberg SA, Park PJ. Combining gene expression data from different generations of oligonucleotide arrays. BMC Bioinformatics. 2004;5:159. doi: 10.1186/1471-2105-5-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18(3):405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- Mecham BH, Klus GT, Strovel J, Augustus M, Byrne D, Bozso P, Wetmore DZ, Mariani TJ, Kohane IS, Szallasi Z. Sequence-matched probes produce increased cross-platform consistency and more reproducible biological results in microarray-based gene expression measurements. Nucleic Acids Res. 2004;32(9):e74. doi: 10.1093/nar/gnh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimgaonkar A, Sanoudou D, Butte AJ, Haslett JN, Kunkel LM, Beggs AH, Kohane IS. Reproducibility of gene expression across generations of affymetrix microarrays. BMC Bioinformatics. 2003;4(1):27. doi: 10.1186/1471-2105-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]