Abstract
Two empirical rules suggest that sex chromosomes play a special role in speciation. The first is Haldane's rule— the preferential sterility and inviability of species hybrids of the heterogametic (XY) sex. The second is the disproportionately large effect of the X chromosome in genetic analyses of hybrid sterility. Whereas the causes of Haldane's rule are well established, the causes of the ‘large X-effect’ have remained controversial. New genetic analyses in Drosophila confirm that the X is a hotspot for hybrid male sterility factors, providing a proximate explanation for the large X-effect. Several other new findings— on faster X evolution, X chromosome meiotic drive, and the regulation of the X chromosome in the male-germline— provide plausible evolutionary explanations for the large X-effect.
The two rules of speciation revisited
Speciation— the process by which new biological species arise— corresponds to the evolution of reproductive barriers that limit the potential for genetic exchange between populations [1, 2]. For geographically isolated populations, reproductive barriers evolve as incidental by-products of genetic divergence. Eventually, ‘good species’ come to be completely isolated by one or more reproductive barriers that take the form of, e.g., incompatible courtship signals that prevent mating (prezygotic isolation) or incompatible gene interactions that cause the sterility or lethality of species hybrids (postzygotic isolation; Box 1). The past decade has seen good progress in the molecular characterization of the ‘speciation genes’ involved in reproductive barriers. In particular, the recent identification of genes causing intrinsic postzygotic isolation has begun to provide important information about the functions of these genes within species and on the population genetic forces that shape their evolutionary history [3, 4]. I will not review the details of particular speciation genes here as they have been amply discussed elsewhere [3, 4]. Instead, this review will focus on new developments that concern an older but still controversial problem— the special role of sex chromosomes in the evolution of postzygotic reproductive isolation between animal species.
Box 1. The evolution and genetics of intrinsic postzygotic isolation.
The evolution of hybrid sterility and inviability corresponds to the gradual accumulation of incompatible genetic interactions between species. Dobzhansky [2] and Muller [76] first noted that geographically isolated— and hence independently evolving— populations inevitably fix new genetic variants that function well in one genetic background but not in the other. To understand why, consider two populations with the same two-locus genotype, aabb. As the populations diverge, mutation A might arise and become fixed in the first population (yielding AAbb), and B might arise and become fixed in the second (yielding aaBB). Importantly, as these substitutions occur, neither population passes through a sterile or inviable intermediate genotype. However, during their evolutionary history, the A and B alleles never occur in the same genetic background and, consequently, their interaction is never exposed to natural selection. If the A and B alleles are by chance functionally incompatible, disrupting hybrid gametogenesis or development, then hybrids might well be sterile or inviable. The Dobzhansky-Muller model thus shows how divergence between populations can incidentally give rise to epistatic hybrid incompatibilities that cause reproductive isolation.
The idea that sex chromosomes play a special role in speciation is based on two empirical rules that characterize speciation in animals: Haldane's rule and the so-called ‘large X-effect’ [5, 6]. Haldane's rule refers to the preferential sterility or inviability of species hybrids of the heterogametic (XY) sex: in crosses between many recently diverged species, XY hybrids are often sterile or inviable whereas their XX siblings are not [7]. In cases of unisexual hybrid sterility or inviability, Haldane's rule holds in 95% (n = 131) and 100% (n = 26) of species crosses in Drosophila and mammals, respectively, in which males are the XY sex, and in 97% (n = 87) and 96% (n = 114) of species crosses in birds and butterflies, respectively, in which females are the XY (or ZW) sex [1, 8, 9]. Thus, Haldane's rule depends not on sex per se but on sex chromosomes. The second, and related, rule is the large X-effect— the disproportionately large effect of the X chromosome versus autosomes in backcross genetic analyses of hybrid sterility and inviability (also known as “Coyne's rule” [10]; Figure 1). Evidence for large X-effects comes from a wide range of taxa including mouse, Drosophila, birds and Lepidoptera (i.e., moths and butterflies) [5, 6].
Figure 1. The large X-effect in Drosophila hybrids.
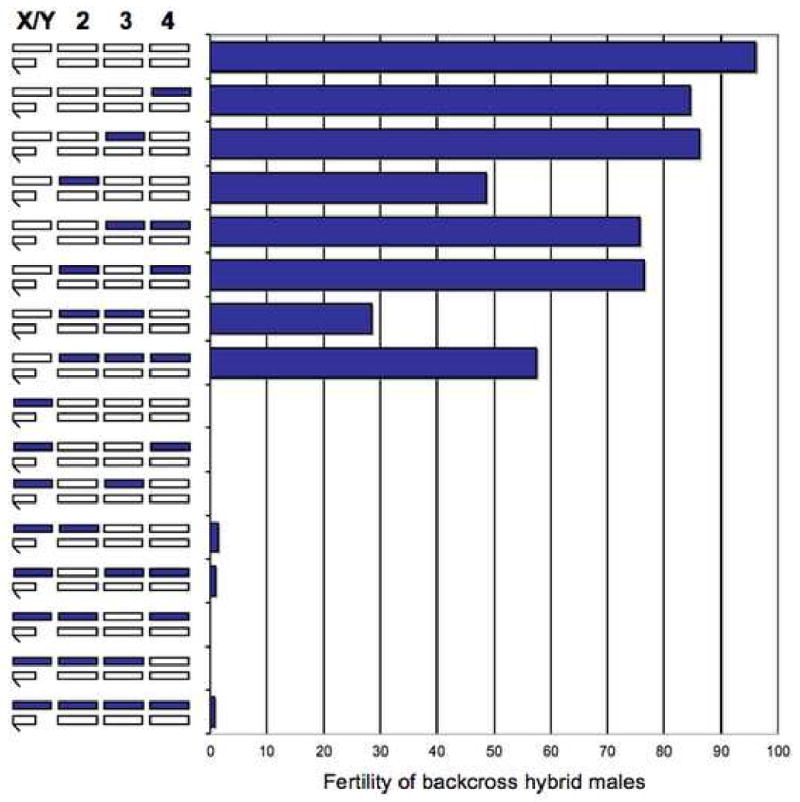
The X chromosome has a conspicuously large effect on the sterility of backcross hybrid males between D. pseudoobscura (white) and D. persimilis (purple; modified from [79]). The x axis shows the percentage of fertile male flies obtained in the different chromosomal backcrosses that are depicted on the y axis. Similar results have been obtained in backcross analyses of hybrid sterility between many other species pairs, including Drosophila, mammals, Lepidoptera and birds (reviewed in [6]).
It can safely be argued that modern speciation genetics was launched by attempts to explain these two rules (e.g., [11]) and doing so remains an important task as the existence of these rules suggests that generalities underlie the evolutionary and genetic basis of speciation in all animals [5, 6]. Although most speciation geneticists agree on the causes of Haldane's rule (Box 2), the causes and significance of the large X-effect have been questioned— so much so that the large X-effect is now rarely discussed, even by its original proponents [1]. However, several recent developments suggest that the time is right to reconsider the large X-effect. Here, I review new findings bearing on its existence, its significance, and its possible causes in Drosophila, the source of most of the new data. In particular, I review new genetic analyses that show that hybrid male sterility genes are conspicuously concentrated on the X chromosome, and I consider three models that might explain why.
Box 2. The causes of Haldane's rule.
Haldane's rule— the preferential sterility or inviability of hybrids of the XY sex— has multiple causes [14, 20, 77]. First, the dominance theory posits that the alleles causing hybrid sterility and inviability typically behave as recessives. Thus, XY hybrids suffer disproportionately because they experience the full effects of any X-linked recessive hybrid sterility or inviability factors whereas their XX siblings are mostly protected. Several genetic analyses provide evidence that recessive incompatibility alleles accumulate faster between species than dominant alleles [16, 19]. Second, the faster-male theory posits that incompatibility factors causing hybrid male sterility accumulate much faster than those causing other hybrid fitness problems [14]. Faster-male evolution will therefore contribute to Haldane's rule in species with XY males. Genetic analyses confirm that hybrid male sterility genes accumulate much faster than other kinds of incompatibilities [13, 16, 18, 19], but the reasons are not entirely clear. One possibility is that sexual selection drives the rapid divergence of male-specific fertility factors, giving rise to male-specific incompatibilities. Consistent with this idea, Drosophila genes with male-biased expression patterns show faster rates of DNA sequence evolution and gene expression divergence [78]. It is too soon, however, to exclude the second possibility that spermatogenesis is particularly sensitive to genetic perturbations and is thus easily disrupted in hybrids [14, 60]. Third, growing evidence makes it increasingly plausible that repeated bouts of sex chromosome meiotic drive and suppression could contribute to Haldane's rule [44-48].
Significance of the large X-effect
Coyne and colleagues first called attention to the importance of the X chromosome in speciation [5, 6, 11, 12]. Although the evidence for large X-effects in backcross analyses has never been disputed, its meaning has. On the one hand, the large X-effect could signify something special about sex chromosomes during speciation, e.g., that the X chromosome is a hotspot for speciation genes. On the other, the large X-effect could be a trivial consequence of the general recessivity of speciation genes, signifying nothing about the X per se [13, 14]. As Wu and Davis [14] correctly note, backcross analyses provide an unfair contrast between hemizygous X effects and heterozygous autosomal efects (Figure 1): recessive speciation genes on the X are fully expressed in backcross hybrid males whereas those on autosomes are largely masked. Thus, because any X vs. autosome difference in the density of speciation genes is confounded by dominance, the large X-effect might tell us more about the limits of backcross analyses than about the X chromosome.
To separate the effects of density versus dominance, high-resolution chromosomal mapping data are necessary in which the effects of dominance are controlled. Over the last 15 years, high-resolution mapping data have accumulated steadily as backcross analyses have given way to fine-scale introgression studies (Figure 2). In these analyses, small chromosomal segments are moved from one species into the genetic background of another by repeated backcrossing, made homozygous and then tested for hybrid fertility and viability effects (Figure 2). Introgression analyses therefore contrast the fitness effects of hemizygous X-linked segments with homozygous autosomal segments [14]. In some studies, the sizes of chromosomal segments have been determined using either cytological [15] or molecular markers. The results from early analyses between Drosophila species were mixed. Whereas some studies found evidence consistent with a higher density of hybrid male sterility genes on the X chromosome [15-19], others did not [13]. Part of the difficulty in settling this seemingly straightforward question is that confounding technical artifacts could not always be excluded (e.g., previous studies could not exclude possible X vs. autosome differences in introgression size or publication bias; reviewed in [16, 20]). To date, only one study has phenotyped and estimated the sizes of a large collection of X-linked and autosomal introgressions in a single experiment. In a genetic analysis between two island endemic species that began diverging ∼400,000 years ago [21], Masly and Presgraves [16] assayed the hybrid fitness effects of 142 introgressions from D. mauritiana in an otherwise D. sechellia genome. The data show that the X chromosome harbors roughly four times as many hybrid male sterility factors as an average, comparably sized, autosomal arm (Figure 3). These results confirm previous analyses [15, 18, 19] involving other Drosophila species pairs and provide strong evidence that the X chromosome harbors a higher density of hybrid male sterility factors than the autosomes.
Figure 2. Introgression analyses of hybrid incompatibilities.
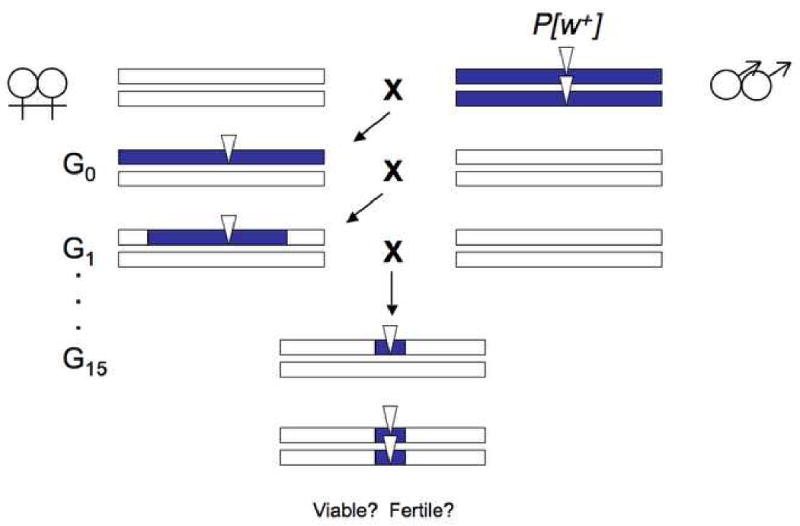
Introgression studies move small chromosomal regions from one species (purple bars) into the genomic background of a closely related sister species (white bars) by repeated backcrossing through fertile hybrid females. Introgressed chromosomal segments typically bear a selectable dominant genetic marker (e.g., a P-element bearing the w+ eye color marker) and become progressively smaller with each generation of backcrossing. After several generations of backcrossing, the foreign introgression is made homozygous and tested for its effects on hybrid fitness.
Figure 3. The X chromosome is a hotspot for hybrid male sterility factors.
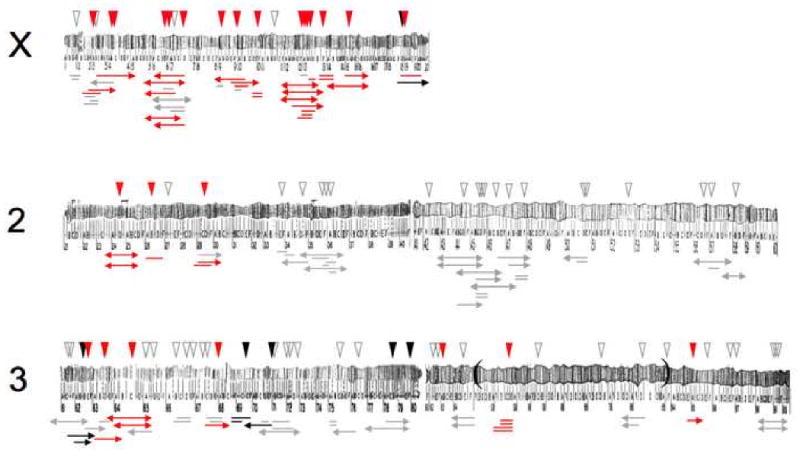
The density of introgressions from D. mauritiana that cause hybrid male sterility (red triangles) in an otherwise D. sechellia genomic background is four times higher on the X chromosome than on chromosomes 2 and 3. A chromosomal inversion is bracketed by parentheses on chromosome arm 3R. Gray and black triangles mark fertile and hybrid lethal introgressions, respectively. Introgression sizes are depicted beneath the chromosomes (modified from [16]).
It is important to be clear about what these findings say about the large X-effect. First, while the new findings show that the large X-effect is not a trivial methodological consequence of dominance, they do not exclude a role for dominance. Both dominance and the higher density of hybrid male sterility factors might contribute to the large X-effect. Importantly, however, theory shows that as the rate at which hybrid male steriles accumulate on the X increases, dominance quickly becomes unimportant [22, 23]. Second, the large X-effect was originally believed to apply to all forms of intrinsic postzygotic isolation— hybrid sterility in the XY sex, hybrid sterility in the XX sex, and hybrid inviability [5, 6]. Despite earlier claims, evidence for large X-effects for hybrid inviability or for hybrid sterility in the XX sex is meager. Until more evidence accrues for other forms of isolation, the large X-effect— as a “rule of speciation”— is best limited to the sterility of the XY sex [22, 23]. The newer analyses relate only to hybrid male sterility— and to the XY sex in Drosophila, in particular. Fine-scale genetic analyses in non-Drosophila species are needed to determine if the large X-effects seen in other taxa (e.g., mammals, birds, Lepidoptera) also reflect a higher density of X-linked hybrid sterility factors.
Evolutionary causes of the large X-effect for hybrid male sterility
The genetics of speciation now faces the challenge of determining why the X chromosome accumulates hybrid male sterility factors so rapidly. The simplest explanation— that the large X-effect reflects a greater concentration of fertility genes on the X— can be ruled out immediately. Classical mutagenesis studies show that loci mutable to male sterility are randomly distributed throughout the genome [24], and whole-genome expression analyses show that genes with male-biased expression are, if anything, under-represented on the Drosophila X chromosome [25, 26]. At least three other kinds of evolutionary explanations for the large X-effect remain, and important developments bearing on the plausibility of each have appeared recently.
Faster X evolution
There are several reasons why X-linked loci might evolve faster than autosomal loci [27]. X-linked and autosomal loci can differ in effective population size, mutation rate, and the efficacy of natural selection. In one of the original attempts to explain the large X-effect, Charlesworth et al. [12] showed that X-linked loci will evolve faster than autosomal loci so long as newly arising beneficial mutations are, on average, partially recessive (see also [6]). (Note here that the dominance of favorable mutations within species says nothing about the dominance of their incidentally deleterious side effects in hybrids.) Although the expected signature of ‘faster X evolution’ is clear, empirical tests in Drosophila have proven to be frustratingly inconclusive. As Table 1 shows, different analyses have arrived at different conclusions [28-38; J. Parsch, pers. comm.].
Table 1. Tests for faster X evolution in Drosophila.
| Species | Number of Genes | Analysis | Result | References |
|---|---|---|---|---|
| D. melanogaster - D. simulansa | 254 genes | dNb and dN/dSc | X ≈ A | [30] |
| D. melanogaster | 1,841 duplicate genes | dN/dS | X > A | [38] |
| D. melanogaster | 13 duplicate genes | MKd | X > A | [39] |
| D. pseudoobscura - D. melanogaster | 9,184 genes | dN | X ≈ A | [35] |
| D. pseudoobscura - D. melanogaster | 8,453 genes | dN | X > A | [33] |
| D. pseudoobscura - D. melanogaster | 110 genes | dN/dS | XR > 3Le | [32] |
| D. pseudoobscura - D. melanogaster | 2,646 genes | dN/dS | XR ≈ 3L | [37] |
| D. melanogaster lineage | 98 genes | MK | X ≈ A | [34] |
| D. melanogaster lineage | 337 genes | MK | X ≈ A | [31] |
| D. melanogaster - D. simulans | 50 genes with male-biased expression | MK | X > A | [28] |
| D. melanogaster - D. simulans | 41 genes with female-biased expression | MK | X ≈ A | [28] |
| D. melanogaster - D. simulans | 45 genes with no sex-biased expression | MK | X ≈ A | [28] |
| D. melanogaster - D. simulans and D. melanogaster-D. yakuba | 597 genes with male-biased expression | dN/dS | X > A | [28] |
| D. melanogaster - D. simulans and D. melanogaster-D. yakuba | 645 genes with female-biased expression | dN/dS | X ≈ A | [28] |
| D. melanogaster - D. simulans and D. melanogaster-D. yakuba | 3,254 genes with no sex-biased expression | dN/dS | X > A | [28] |
| D. simulans, D. melanogaster and D. yakuba lineages | genome-wide, 50 kb windows | divergence at all sitesf | X > A | [29] |
| D. melanogaster, D. sechellia, D. erecta, D. ananassae, D. pseudoobscura, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi lineages | 6,698 genes | amino acid divergence | X ≈ A | [36] |
| D. persimilis lineage | 6,698 genes | amino acid divergence | X > A | [36] |
| D. yakuba and D. simulans lineages | 6,698 genes | amino acid divergence | A > X | [36] |
| D. melanogaster, D. sechellia, D. simulans, D. yakuba and D. erecta | 8,510 genes | dN/dS | X ≈ A | [36] |
| D. sechellia and D. simulans lineages | 8,510 genes | dN/dS | X > A | [36] |
| D. melanogaster-D. simulans (3L) vs. D. pseudoobscura-D. persimilis (XR) | 2,392 | dN/dS | XR > 3L | [36] |
Molecular divergence can be estimated between two species (e.g., D. melanogaster - D. simulans) or along a single lineage (e.g., D. melanogaster).
dN = number of nonsynonymous differences per nonsynonymous site.
dN/dS = nonsynonymous differences per nonsynonymous site standardized by synonymous (neutral) differences per synonymous sites.
The McDonald-Kreitman (MK) test contrasts the numbers of replacement and synonymous fixed differences with the numbers of replacement and synonymous polymorphisms.
Orthologous genes that are X-linked (XR) in D. pseudoobscura and close relatives but autosomal (3L) in D. melanogaster.
For dS, X ≈ A in the D. simulans lineage.
Genome-wide analyses of lineage-specific evolution— DNA sequence evolution in seven Drosophila lineages [28, 32, 34-36] and protein evolution across 12 Drosophila lineages [35]— together suggest weakly elevated substitution rates on the X chromosome in some lineages but not in others. It is not clear to what extent this lineage-specific faster X evolution reflects X vs. autosome differences in mutation rate or in the efficacy of natural selection [28, 35]. Statistical tests that distinguish mutation and selection by combining data on DNA polymorphism and divergence do not detect an excess of genes with histories of recurrent adaptive evolution on the X chromosome relative to the autosomes in D. melanogaster or in D. simulans [28, 30]. However, a new analysis that accounts for sex-specific gene expression finds that genes with male-biased expression patterns experience moderately higher rates of adaptive protein evolution when X-linked (J. Parsch, pers. comm.). This is a potentially important finding as nearly every speciation gene identified to date shows individually significant evidence of recurrent adaptive evolution [39-42; but see 43]. However, given the paucity of genes with male-biased expression on the X [25, 26], it seems unlikely that their moderately higher rate of adaptive evolution can, by itself, explain the 2.5- to 4-fold excess of hybrid male sterility genes on the X [16, 18].
Sex ratio meiotic drive
A second class of explanations for the large X-effect involves recurrent bouts of evolutionary conflict [44, 45]. Mendelian transmission through the male germline is often subverted by selfish meiotic drive (or segregation distorter) elements that kill or incapacitate sperm bearing allelic rivals so that drive element-bearing sperm are preferentially transmitted. Because drive elements impose direct fitness costs on their targets and indirect fertility costs on their bearers, they often elicit the evolution of unlinked suppressors. Coevolutionary arms races between drivers and their suppressors can, in principle, cause the evolution of hybrid male sterility in several ways. For instance, different species can become fixed for different driver-suppressor systems that, in hybrids, could be unleashed (if, e.g., suppressors are incompletely dominant) causing mutual sperm destruction. In addition, drive elements could show aberrant expression in hybrids, triggering meiotic or spermatogenic checkpoint arrests that result in sterility. Last, evolutionary conflict between drivers and suppressors could cause the rapid, lineage-specific evolution of sperm-related genes, giving rise to hybrid incompatibilities that disrupt spermatogenesis.
Two recent studies provide evidence that genes involved in drive or suppression within species can contribute to hybrid male sterility. Tao and colleagues [46] showed that the D. mauritiana allele of the autosomal gene too much yin (tmy), when introgressed into an otherwise D. simulans genome, unleashes a cryptic driver on the D. simulans X chromosome; in combination with other genes, tmy also contributes to hybrid male sterility. Similarly, Orr and Irving [47] discovered X-chromosome drive in the very weakly fertile F1 hybrid males of two young subspecies, D. pseudoobscura bogatana and D. p. pseudoobscura. Subsequent genetic analyses show that the factors causing hybrid male sterility and hybrid drive co-localize and have the same patterns of complex epistasis, strongly suggesting that the same genes are involved [47, 48]. It seems clear, then, that the genes involved in cryptic meiotic drive and suppression can contribute to hybrid male sterility.
For recurrent drive and suppression to explain the large X-effect, two things must be true. First, species must harbor multiple fixed, but otherwise cryptic, drive elements. The failure to unmask cryptic drive in species hybrids is one reason why the drive hypothesis was initially discarded [49-51]. However, it now appears that the choice of genotypes and species in these early surveys was simply unlucky as multiple Drosophila species are now known to carry cryptic drive systems, and some carry more than one. D. simulans, for instance, harbors at least three cryptic X-chromosome drive systems, each involving different suppressor loci [46, 52-54]. Whether this situation is typical remains unclear, however, as some species subject to similarly intense genetic scrutiny have been shown to harbor cryptic drive (one in D. pseudoobscura bogatana; [47]) whereas others have not (e.g., D. melanogaster and D. sechellia; reviewed in [55]).
Second, to explain the large X-effect, drive elements must preferentially accumulate on the X chromosome. Population genetic theory shows that, under most conditions, X- and Y-linked drive elements can invade populations more readily than autosomal elements [45]. Consistent with the theory, X-linked drive has been identified in at least a dozen Drosophila species [56], whereas autosomal drive has been observed just once. Unfortunately, this discrepancy could reflect a clear observational bias: sex chromosome drive distorts sex ratios whereas autosomal drive, unless associated by chance with a genetic marker, is usually undetectable [56, 57]. Other complications exist. For drive elements to contribute to speciation they must not only invade; they also must become fixed. Although X-linked drivers enjoy better probabilities of invasion than autosomal drivers, there is good reason to believe that the reverse is true for probabilities of fixation [55-57]. In addition, unlinked suppressors of X-linked drive can accumulate on the Y or on the autosomes. If X-linked drivers are typically silenced by 3 or 4 autosomal suppressors then, assuming a Drosophila-like karyotype, there would be little or no preferential accumulation of divergence on the X chromosome: for every X-linked driver fixed, each autosomal arm, on average, would fix a suppressor.
The idea that meiotic drive contributes to Drosophila speciation seems increasingly plausible. However it remains to be seen if the fixation of cryptic drive elements on the X chromosome— as observed in D. simulans and in D. pseudoobscura bogatana— is sufficiently common to explain Haldane's rule and the large X-effect.
Regulation of the X chromosome in the male germline
The third class of explanation for the large X-effect involves the regulation of the X chromosome in the male germline. During the early stages of spermatogenesis in male heterogametic taxa, the X chromosome undergoes transcriptional inactivation and chromosomal condensation before the autosomes (reviewed in [58]). (The normally inactive W chromosome of female heterogametic species [i.e., ZZ males and ZW females] becomes active during oogenesis [59].) In a classic paper, Lifschytz and Lindsley [60] argued that X chromosome inactivation is a critical stage of spermatogenesis that, if disrupted, causes male sterility. As evidence, they cited cytological observations and showed that 75% (85/110) of reciprocal X;autosome translocations in D. melanogaster cause dominant male sterility whereas autosome;autosome and Y;autosome translocations do not (females remain fertile regardless of translocation type). Although compelling, their genetic data provide only indirect evidence, leading some to question the existence of X inactivation in Drosophila [26, 61-63]. (Spermatogenic X inactivation is well-established in marsupials, eutherian mammals, grasshoppers and nematode worms [64-67].) However, new evidence from D. melanogaster should settle the matter. Hense et al. [68] generated 47 transgenic lines, each harboring a construct comprising the lacZ reporter driven by the testis-specific promoter of an autosomal gene, ocnus. The 27 autosomal insertions behaved as expected, producing robust testis-specific expression; the 20 X-linked insertions, however, showed dramatically reduced expression. The uniformly low expression of X-linked insertions— regardless of their position on the X chromosome— supports the idea of chromosome-wide transcriptional inactivation of the X during spermatogenesis.
Given the ubiquity of X inactivation in male heterogametic species, Lifschytz and Lindsley [60] suggested that its possible disruption in species hybrids could contribute to Haldane's rule [69, 70]. Their original argument can be extended to the high density of hybrid male sterility factors on the X chromosome: if ‘foreign’ X-linked introgressions are recognized as ‘non-X’ by the X inactivation machinery, they might mimic X;autosome translocations thereby causing sterility [16]. Moreover, if X inactivation is disrupted in hybrids, then X-linked genes that are normally silenced during spermatogenesis should show aberrant over-expression. Interestingly, this is the pattern that has emerged from recent surveys of gene misexpression in the testis, but not the whole bodies, of Drosophila species hybrids [71, 72]. Only a small fraction, ∼9%, of autosomal genes that are misexpressed in hybrid testis are overexpressed (the remaining 91% are underexpressed). Consistent with the idea of disrupted X chromosome inactivation, the X chromosome is significantly enriched for genes that are overexpressed in hybrid testis (34-55%) [72].
The other difference in the regulation of the X and the autosomes in the Drosophila male germline involves dosage compensation. In somatic cells, the X chromosome is hypertranscribed so that the ratio of transcripts from X-linked and autosomal genes in males roughly equals that in females [73]. But, like X inactivation, the existence of dosage compensation in the Drosophila male germline has been uncertain [63]. Using whole-genome microarrays, Gupta et al. [61] have now shown that dosage compensation occurs in the male germline of D. melanogaster. However, somatic and germline dosage compensation appear to occur via distinct mechanisms in Drosophila as only one of the five dosage compensation complex (DCC) proteins, maleless, is present in male germline cells where, unlike in somatic cells, it shows no association with the X chromosome [63].
Spermatogenic X inactivation and dosage compensation provide two processes that distinguish the X chromosome from autosomes and male from female fertility. Both processes require that the X chromosome is recognized— probably via cis-acting X chromosome-specific sequence motifs— and appropriately regulated by RNA and/or protein complexes. If the molecular basis for recognition and regulation diverges between species, then incompatibilities disrupting either process could cause hybrid male sterility that maps disproportionately to the X chromosome. Interestingly, recent findings show that four of the five protein-coding genes of the somatic DCC have histories of recurrent adaptive evolution in the D. melanogaster lineage [74, 75]. It will be important to determine if the genes involved in germline dosage compensation have a similar history. For the moment, the overexpression of X-linked genes in hybrid testis suggests that, if germline dosage compensation is disrupted, it does not cause the loss of X chromosome hypertranscription [72]. Clearly, further experimental work is needed to determine the molecular bases of spermatogenic X inactivation and male germline dosage compensation, to test if either is disrupted in hybrids and, if so, to determine why.
Concluding remarks
The findings reviewed here leave little doubt that the X chromosome plays a special role in speciation. In Drosophila, the generally recessive behavior of hybrid male sterility factors and their exceptionally high density on the X chromosome provide proximate genetic explanations for the large X-effect and, in turn, for Haldane's rule. Speciation genetics must now determine why hybrid male sterility evolves so rapidly on the X chromosome. The answer is important not just for explaining the large X-effect but also for what it says about evolution within species. For example, is the recurrent fixation of selfish sex ratio meiotic drive systems a regular feature of the evolutionary history of species with sex chromosomes? If X inactivation or germline dosage compensation is disrupted, then we must ask why the molecular bases of these processes should diverge between species so quickly? The answers could come from straightforward experiments that assay X inactivation and germline dosage compensation in species hybrids. Or the answers might have to wait for the considerably harder task of identifying a large sample of the DNA sequences that cause hybrid male sterility. Like Haldane's rule (Box 2), it is entirely possible that the large X-effect will be a composite phenomenon with multiple genetic and evolutionary causes.
Acknowledgments
I thank Victoria Cattani, John Baines, Pierre Gerard, J.P. Masly, Mohamed Noor, Allen Orr, John Parsch, Shanwu Tang and three anonymous reviewers for discussions and/or comments on earlier drafts of the manuscript. Work in my laboratory is supported by funds from the University of Rochester and the National Institutes of Health (GM79543).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coyne JA, Orr HA. Speciation. Sunderland, Massachusetts: Sinauer; 2004. [Google Scholar]
- 2.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- 3.Orr HA, Masly JP, Presgraves DC. Speciation genes. Current Opinion in Genetics & Development. 2004;14:675–679. doi: 10.1016/j.gde.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Wu CI, Ting CT. Genes and speciation. Nature Reviews Genetics. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- 5.Coyne J. Genetics and speciation. Nature. 1992;355:511–515. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- 6.Coyne JA, Orr HA. Two rules of speciation. In: Otte D, Endler J, editors. Speciation and Its Consequences. Sunderland, MA: Sinauer Associates; 1989. pp. 180–207. [Google Scholar]
- 7.Haldane JBS. Sex ratio and unisexual sterility in animal hybrids. Journal of Genetics. 1922;12:101–109. [Google Scholar]
- 8.Presgraves DC. Patterns of postzygotic isolation in Lepidoptera. Evolution. 2002;56:1168–1183. doi: 10.1111/j.0014-3820.2002.tb01430.x. [DOI] [PubMed] [Google Scholar]
- 9.Price TD, Bouvier MM. The evolution of F1 postzygotic incompatibilities in birds. Evolution. 2002;56:2083–2089. [PubMed] [Google Scholar]
- 10.Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne JA. The genetic basis of Haldane's rule. Nature. 1985;314:736–738. doi: 10.1038/314736a0. [DOI] [PubMed] [Google Scholar]
- 12.Charlesworth B, Coyne JA, Barton NH. The Relative Rates of Evolution of Sex-Chromosomes and Autosomes. American Naturalist. 1987;130:113–146. [Google Scholar]
- 13.Hollocher H, Wu CI. The genetics of reproductive isolation in the Drosophila simulans clade: X vs. autosomal effects and male vs. female effects. Genetics. 1996;143:1243–1255. doi: 10.1093/genetics/143.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CI, Davis AW. Evolution of postmating reproductive isolation: the composite nature of Haldane's rule and its genetic bases. American Naturalist. 1993;142:187–212. doi: 10.1086/285534. [DOI] [PubMed] [Google Scholar]
- 15.Naveira H, Fontdevila A. The evolutionary history of Drosophila buzzatii. XII. The genetic basis of sterility in hybrids between D. buzzatii and its sibling D. serido from Argentina. Genetics. 1986;114:841–857. doi: 10.1093/genetics/114.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. Public Library of Science Biology. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naveira HF, Maside XR. The genetics of hybrid male sterility in Drosophila. In: Howard DJ, Berlocher SH, editors. Endless Forms. Oxford: Oxford University Press; 1998. pp. 330–338. [Google Scholar]
- 18.Tao Y, Chen S, Hartl DL, Laurie CC. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics. 2003;164:1383–1397. doi: 10.1093/genetics/164.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.True JR, Weir BS, Laurie CC. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics. 1996;142:819–837. doi: 10.1093/genetics/142.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurie CC. The weaker sex is heterogametic: 75 years of Haldane's rule. Genetics. 1997;147:937–951. doi: 10.1093/genetics/147.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kliman RM, Andolfatto P, Coyne JA, Depaulis F, Kreitman M, Berry AJ, McCarter J, Wakeley J, Hey J. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics. 2000;156:1913–1931. doi: 10.1093/genetics/156.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turelli M, Orr HA. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naveira HF. On the relative roles of faster-X evolution and dominance in the establishment of intrinsic postzygotic isolating barriers. Genetica. 2003;118:41–50. doi: 10.1023/a:1022978222021. [DOI] [PubMed] [Google Scholar]
- 24.Lindsley DL, Lifschytz E. The genetic control of spermatogenesis in Drosophila. In: Beatty RA, Gluecksohn-Waelsh S, editors. Proceedings of the International Symposium on the Genetic of the Spermatozoon. Copenhagen: Bogtrykkeriet Forum; 1972. [Google Scholar]
- 25.Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nature Reviews Genetics. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 28.Begun DJ, Holloway AK, S K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, Pachter L, Myers E, Langley CH. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans Public Library of Science Biology. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betancourt AJ, Presgraves DC, Swanson WJ. A test for faster X evolution in Drosophila. Molecular Biology and Evolution. 2002;19:1816–1819. doi: 10.1093/oxfordjournals.molbev.a004006. [DOI] [PubMed] [Google Scholar]
- 30.Connallon T. Adaptive protein evolution of X-linked and autosomal genes in Drosophila: Implications for faster-X hypotheses. Molecular Biology and Evolution. 2007;24:2566–2572. doi: 10.1093/molbev/msm199. [DOI] [PubMed] [Google Scholar]
- 31.Counterman BA, Ortiz-Barrientos D, Noor MAF. Using comparative genomic data to test for faster-X evolution. Evolution. 2004;58:656–660. [PubMed] [Google Scholar]
- 32.Musters H, Huntley MA, Singh RS. A genomic comparison of faster-sex, faster-X, and faster-male evolution between Drosophila melanogaster and Drosophila pseudoobscura. Journal of Molecular Evolution. 2006;62:693–700. doi: 10.1007/s00239-005-0165-5. [DOI] [PubMed] [Google Scholar]
- 33.Presgraves DC. Recombination enhances protein adaptation in Drosophila melanogaster. Current Biology. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. [DOI] [PubMed] [Google Scholar]
- 34.Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Sujun H, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MAF, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Research. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh ND, Larracuente AM, Clark AG. Contrasting the efficacy of selection on the X and autosomes in Drosophila. Molecular Biology and Evolution. 2007;25:454–467. doi: 10.1093/molbev/msm275. [DOI] [PubMed] [Google Scholar]
- 36.Thornton K, Bachtrog D, Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: No evidence for faster-X protein evolution. Genome Research. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornton K, Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Molecular Biology and Evolution. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- 38.Thornton K, Long M. Excess amino acid substitutions relative to polymorphism between X-linked duplications in Drosophila melanogaster. Molecular Biology and Evolution. 2005;22:273–284. doi: 10.1093/molbev/msi015. [DOI] [PubMed] [Google Scholar]
- 39.Barbash DA, Awadalla P, Tarone AM. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. Public Library of Science Biology. 2004;2:839–848. doi: 10.1371/journal.pbio.0020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. Two Dobzhansky-Muller Genes Interact to Cause Hybrid Lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 41.Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- 42.Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 43.Masly JP, Jones CD, Noor MAF, Locke J, Orr HA. Gene transposition as a novel cause of hybrid male sterility. Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 44.Frank SH. Divergence of meiotic drive-suppressors as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 45.Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proceedings of the National Acadamy of Sciences. 2001;98:13183–13188. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orr HA, Irving S. Segregation Distortion in Hybrids Between the Bogota and USA Subspecies of Drosophila pseudoobscura. Genetics. 2005;169:671–682. doi: 10.1534/genetics.104.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orr HA, Masly JP, Phadnis N. Speciation in Drosophila: From phenotypes to molecules. Journal of Heredity. 2007;98:103–110. doi: 10.1093/jhered/esl060. [DOI] [PubMed] [Google Scholar]
- 49.Coyne JA. Meiotic segregation and male recombination in interspecific hybrids of Drosophila. Genetics. 1986;114:485–494. doi: 10.1093/genetics/114.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson NA, Wu CI. An empirical test of the meiotic drive models of hybrid sterility: sex ratio data from hybrids between Drosophila simulans and Drosophila sechellia. Genetics. 1992;130:507–511. doi: 10.1093/genetics/130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coyne JA, Orr HA. Further evidence against meiotic-drive models of hybrid sterility. Evolution. 1993;47:685–687. doi: 10.1111/j.1558-5646.1993.tb02123.x. [DOI] [PubMed] [Google Scholar]
- 52.Mercot H, Atlan A, Jacques M, Montchamp-Moreau C. Sex-ratio distortion in Drosophila simulans: co-occurrence of a meiotic drive and a suppressor of drive. Journal of Evolutionary Biology. 1995;8:283–300. [Google Scholar]
- 53.Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, Hartl DL. A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. Public Library of Science Biology. 2007;5:e293. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL. A sex-ratio meiotic drive system in Drosophila simulans. I: An autosomal suppressor. Public Library of Science Biology. 2007;5:e292. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Presgraves DC. Drive and sperm: evolution and genetics of male meiotic drive. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Elsevier Press; 2008. Volume In press. [Google Scholar]
- 56.Jaenike J. Sex chromosome meiotic drive. Annual Review of Ecology and Systematics. 2001;32:25–49. [Google Scholar]
- 57.Lyttle TW. Segregation Distorters. Annual Review of Genetics. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- 58.Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 59.Jablonka E, Lamb MJ. Meiotic pairing constraints and the activity of sex chromosomes. Journal of Theoretical Biology. 1988;133:23–36. doi: 10.1016/s0022-5193(88)80022-5. [DOI] [PubMed] [Google Scholar]
- 60.Lifschytz E, Lindsley DL. The role of the X-chromosome inactivation during spermatogenesis. Proceedings of the National Academy of Sciences. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. Journal of Biology. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKee BD, Handel MA. Sex chromosomes, recombination, and chromatin conformation. Chromosoma. 1993;102:71–80. doi: 10.1007/BF00356023. [DOI] [PubMed] [Google Scholar]
- 63.Rastelli L, Kuroda MI. An analysis of maleless and histone H4 acetylation in Drosophila melanogaster spermatogenesis. Mechanisms of Development. 1998;71:107–117. doi: 10.1016/s0925-4773(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 64.Kelly WG, Schaner CE, Dernburg AF, Lee MH, Kim SK, Villeneuve AM, Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarrey JR, Watson C, Atencio J, Ostermeier GC, Marahrens Y, Jaenisch R, Krawetz SA. X-chromosome inactivation during spermatogenesis is regulated by an Xist/Tsix-independent mechanism in the mouse. Genesis. 2002;34:257–266. doi: 10.1002/gene.10163. [DOI] [PubMed] [Google Scholar]
- 66.Namekawa SH, VandeBerg JL, McCarrey JR, Lee JT. Sex chromosome silencing in the marsupial male germ line. Proceedings of the National Academy of Sciences. 2007;104:9730–9735. doi: 10.1073/pnas.0700323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cabrero J, Teruel M, Carmona FD, Jimenez R, Camacho JPM. Histone H3 lysine acetylation pattern suggests that X and B chromosomes are silenced during entire male meiosis in a grasshopper. Cytogenetic and Genome Research. 2007;119:135–142. doi: 10.1159/000109630. [DOI] [PubMed] [Google Scholar]
- 68.Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. Public Library of Science Biology. 2007;5:2288–2295. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Forejt J. Hybrid sterility in the mouse. Trends in Genetics. 1996;12:412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- 70.Jablonka E, Lamb MJ. Sex chromosomes and speciation. Proc R Soc Lond B. 1991;243:203–208. doi: 10.1098/rspb.1991.0032. [DOI] [PubMed] [Google Scholar]
- 71.Michalak P, Noor MAF. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Molecular Biology and Evolution. 2003;20:1070–1076. doi: 10.1093/molbev/msg119. [DOI] [PubMed] [Google Scholar]
- 72.Moehring AJ, Teeter KC, Noor MAF. Genome-wide patterns of expression in Drosophila pure species and hybrid males. II. Examination of multiple-species hybridizations, platforms, and life cycle stages. Molecular Biology and Evolution. 2007;24:137–145. doi: 10.1093/molbev/msl142. [DOI] [PubMed] [Google Scholar]
- 73.Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nature Reviews Genetics. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- 74.Levine MT, Holloway AK, Arshad U, Begun DJ. Pervasive and largely lineage-specific adaptive protein evolution in the dosage compensation complex of Drosophila melanogaster. Genetics. 2007;177:1959–1962. doi: 10.1534/genetics.107.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodriguez MA, Vermaak D, Bayes JJ, Malik HS. Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proceedings of the National Academy of Sciences. 2007;104:15412–15417. doi: 10.1073/pnas.0707445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muller HJ. Isolating mechanisms, evolution, and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- 77.Orr HA. Haldane's rule. Annual Review of Ecology and Systematics. 1997;28:195–218. doi: 10.1146/annurev.ecolsys.28.1.85. [DOI] [PubMed] [Google Scholar]
- 78.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nature Reviews Genetics. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 79.Orr HA. Genetics of sterility in hybrids between two subspecies of Drosophila. Evolution. 1989;43:180–189. doi: 10.1111/j.1558-5646.1989.tb04216.x. [DOI] [PubMed] [Google Scholar]


