Abstract
Adipose triglyceride lipase (ATGL) is a recently described adipose-enriched protein with triglyceride-specific lipase activity. ATGL shares the greatest sequence homology with adiponutrin, a nutritionally regulated protein of unclear biological function. Here we present a functional analysis of ATGL and adiponutrin and describe their regulation by insulin. Retroviral-mediated overexpression of ATGL in 3T3-L1 adipocytes increased basal and isoproterenol-stimulated glycerol and nonesterified fatty acid (NEFA) release, whereas siRNA-mediated knockdown of ATGL had the opposite effect. In contrast, siRNA-mediated knockdown of adiponutrin in 3T3-L1 adipocytes had no effect on glycerol or NEFA release. In mice, both ATGL and adiponutrin are nutritionally regulated in adipose tissue, with ATGL being upregulated and adiponutrin being downregulated by fasting. In 3T3-L1 adipocytes, insulin decreased ATGL and increased adiponutrin expression in a dose- and time-dependent manner, suggesting that insulin directly mediates this nutritional regulation. In addition, adipose expression of ATGL was increased by insulin deficiency and decreased by insulin replacement in streptozotocin-induced diabetic mice and was increased in fat-specific insulin receptor knockout mice, whereas adiponutrin showed the opposite pattern. These data suggest that murine ATGL but not adiponutrin contributes to net adipocyte lipolysis and that ATGL and adiponutrin are oppositely regulated by insulin both in vitro and in vivo.
Adipose tissue triglycerides are the predominant form of energy storage in animals. The ability to store and release this energy in response to variable energy availability is advantageous to survival and requires a carefully regulated balance between triglyceride synthesis and hydrolysis. Dysregulation of these processes may result in metabolic disorders, such as obesity and lipodystrophy, which are associated with dyslipidemia, insulin resistance, and overt diabetes. Hence understanding the regulatory mechanisms underlying the storage and mobilization of triglycerides is essential to understanding the pathophysiology of obesity, diabetes, and related metabolic disorders.
Hormone-sensitive lipase (HSL) has traditionally been considered the key lipolytic enzyme in adipocytes (1–4). Lipolytic hormones such as catecholamines stimulate lipolysis primarily via cAMP-mediated activation of protein kinase (PK) A. PKA then phosphorylates HSL and perilipin A. Phosphorylation of perilipin A alleviates the barrier function of this protein and prompts its active participation in the lipolytic process. Phosphorylation of HSL results in translocation of HSL from the cytosol to the lipid droplet where it catalyzes the hydrolysis of tri-, di-, and monoglycerides; cholesteryl esters; and other substrates. Insulin acutely inhibits lipolysis, at least in part, via inhibition of the above cAMP-dependent pathway by PKB-dependent phosphorylation and activation of phosphodiesterase 3B, which in turn lowers cAMP levels (5). However, insulin also has long-term effects on lipolysis that are independent of HSL and cAMP and that remain poorly characterized (6).
Despite the central role of HSL in adipocyte lipolysis, the available evidence suggests that HSL is not the only hormone-responsive lipase in adipocytes. Mice with a targeted deletion of HSL (HSL-KO mice) have reduced catecholamine-sensitive adipocyte lipolysis but are not obese (7,8). Although HSL-KO mice do not accumulate triglycerides, they do accumulate diglycerides in various tissues, suggesting that HSL is rate limiting for diglyceride but not triglyceride hydrolysis (9). In addition, both adipose tissue and isolated adipocytes derived from mouse embryonic fibroblasts of HSL-KO mice have significant residual triglyceride lipase activity, suggesting the presence of one or more additional triglyceride-specific lipases (7,9,10). Furthermore, this triglyceride lipase activity is sensitive to regulation by various factors including catecholamines, tumor necrosis factor-α, and thiazolidinediones, suggesting that the triglyceride lipase activity is also hormone responsive (10).
A novel adipocyte triglyceride lipase has been recently reported, designated as adipose triglyceride lipase (ATGL) (11) or desnutrin (12) in mice and calcium-independent phospholipase A2 (iPLA2)ζ in humans (13). ATGL shares the greatest homology with adiponutrin, thus raising the possibility that these two molecules share function and/or regulation. Although the human homolog of adiponutrin (iPLA2ϵ) has acyl hydrolase activity in vitro (13), no lipolytic function has been reported for the murine homolog. Furthermore, despite insulin’s critical role in lipolysis, the ability of insulin to regulate ATGL and adiponutrin remains poorly characterized. Here we describe the functional analysis of ATGL and adiponutrin and the regulation of these two molecules by insulin.
RESEARCH DESIGN AND METHODS
Microarray analysis
Total RNA isolated from 3T3-L1 adipocytes after 0, 6, 12, 24, and 48 h and 8 days of differentiation was run as a single microarray at each time point using Affymetrix GeneChip technology on MOE 420A chips. RNA/cDNA preparation and hybridization were performed in accordance with the Affymetrix protocol. Computer data analysis was performed using DChip software with the PM/MM model for determining expression values (14,15).
Retroviral constructs and stable cell lines
Full-length ATGL cDNA was amplified by PCR from pSPORT1 vector clone 30024535 (Open Biosystems) using forward primer 5′ GAAGATCTCCATGTTCCCGAGGGAG and reverse primer 5′ CCGCTCGAGTCAGCAAGGCGGGAG. The PCR product was digested with Bgl II and Xho I (New England Biolabs), gel purified (QIAquick gel extraction kit; Qiagen), and subcloned into Bgl II and Xho I sites of the pMSCVpuro retroviral vector (BD Biosciences/Clontech). The sequence was confirmed by sequence analysis followed by comparison with the reported sequence (GenBank BC064747). Packaging cells were transfected with 20 µg of retroviral construct containing full-length ATGL or control vector alone (16,17). Viral supernatants were used to infect 3T3-L1 cells, after which puromycin selection was conducted, as previously described (16,17). The resulting stably transfected 3T3-L1 cells were cultured and differentiated as described below, except that media were supplemented with 2.0 µg/ml puromycin.
Cell culture
3T3-L1 cells (American Type Culture Collection) were grown to 2 days postconfluence in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% calf serum (Invitrogen). Cells were induced to differentiate by changing the medium to DMEM containing 10% fetal bovine serum (FBS; Invitrogen), 0.5 mmol/l 3-isobutyl-1-methylxanthine (Sigma), 1 µmol/l dexamethasone (Sigma), and 10 µg/ml bovine insulin (Sigma) (16). After 48 h and every 2 days thereafter, the medium was replaced with maintenance medium containing DMEM with 10% FBS. For insulin regulation experiments, fully differentiated 3T3-L1 adipocytes were incubated in DMEM containing 10% FBS or serum-free DMEM containing 2% fatty acid–free BSA (Serologicals) with and without bovine insulin for the times and at the doses indicated.
RNA interference
RNA interference by small interfering RNA (siRNA) was performed as previously described (18). In brief, 3T3-L1 adipocytes were detached from culture dishes with 0.25% trypsin (Invitrogen) and 0.5 mg/ml collagenase D (Roche Diagnostics), washed twice, and resuspended in PBS. Control (siControl noninterfering control pool; Dharmacon) or gene-specific siRNAs for ATGL, adiponutrin, and/or HSL (siGenome SMARTselection duplexes; Dharmacon) were delivered into adipocytes (5 nmol siRNA/2.5 million cells) by electroporation using a Bio-Rad gene pulser II system set at 0.18 kV and 960 µF capacitance. Adipocytes were then mixed with DMEM containing 10% FBS and reseeded onto multiwell plates. Gene expression and glycerol and nonesterified fatty acid (NEFA) release were determined 48 h after electroporation. For each gene, four separate siRNAs were tested. Results were considered valid if data were reproducible with at least two of the four siRNAs tested. The siRNA sequences for data presented in relation to this study are as follows: ATGL: sense 3′ GCACAUUUAUCCCGGUGUAUU, anti-sense 5′P UACACCGGGAUAAAUGUGCUU; adiponutrin: sense 3′ GGAGAGCUGUGCUAUCAAGUU, antisense 5′P CUUGAUAGCACAGCUCUCCUU; and HSL: sense 3′ GAGCGUACUCUCUAAGUGUUU, antisense 5′P ACACUUAGAGAGUACGCUCUU.
RNA extraction, reverse transcription, and gene expression
Total RNA was extracted from homogenized tissues or cells using an RNeasy Lipid Tissue Mini kit with on-column DNase treatment (Qiagen). Reverse transcription of 1 µg total RNA was performed using random decamers (RETROscript Kit; Ambion). Gene expression was determined by quantitative real-time PCR (Q-PCR) (MX4000 Multiplex Quantitative PCR System; Stratagene). Reactions were performed in triplicate in 25 µl containing 2.5 µl of 1:100-diluted cDNA, 1× Taqman Universal PCR Master Mix (Applied Biosystems), and gene-specific primer probe sets (Taqman Gene Expression Assays; Applied Biosystems). Reactions were run at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Gene expression was determined by the standard curve method and normalized to the expression of 18S ribosomal RNA or hypoxanthine guanine phosphoribosyl transferase (HPRT) internal control genes. Appropriate analyses were performed to determine that the expression of control genes was unchanged under the experimental conditions described. The accuracy of RNA quantification was optimized by DNAse treatment of samples, the use of gene-specific primer probe sets that span intron-exon boundaries, and verification of the lack of amplification in no–reverse transcriptase and no-template controls.
Lipolysis and protein assays
For basal and stimulated lipolysis, adipocytes were washed twice with DMEM and then incubated for 2 h in serum-free DMEM containing 2% fatty acid–free BSA in the presence or absence of 2 µmol/l isoproterenol. Conditioned medium was then removed and stored at −80°C until analysis. Adipocytes were rinsed with PBS, harvested in lysis buffer containing 25 mmol/l Tris (pH 7.4), 50 mmol/l NaCl, 0.5% sodium deoxycholate, 2% NP-40, 0.2% SDS, and Complete Protease Inhibitor Cocktail (Roche) and stored at −80°C until analysis. The protein content of cell lysates was determined using the Bicinchoninic Acid Protein Assay kit (Pierce Chemical). Glycerol and NEFA content of conditioned medium and cell lysates was determined using the GPO-Trinder Reagent Set (Pointe Scientific) and the NEFA-C kit (Wako Chemicals), respectively.
Animals
Mice were housed individually under standard conditions at 25°C with a 14:10-h light:dark cycle (lights from 6:00 a.m. to 8:00 p.m.) with free access to standard diet (Harlan Teklad RD 8664) and water. Animals were handled in accordance with the guidelines established by the National Institutes of Health. Experimental procedures were approved by the Institutional Animal Care and Use Committee. Fat-specific insulin receptor knockout (FIRKO) mice were obtained from a colony maintained at Joslin Diabetes Center (19). C57BL/6J mice were obtained from The Jackson Laboratories (Bar Harbor, ME). FVB mice were obtained from Taconic (Germantown, NY). To determine gene expression in various tissues, ad libitum–fed male C57BL/6J mice (n = 6) were killed at age 10 weeks. To determine the effect of nutritional status, FVB mice age 8 weeks were either fed ad libitum or fasted for 6, 12, 18, or 24 h with the onset of the light cycle (6:00 a.m.) as time 0 (n = 10 per group). Two additional groups of mice were re-fed for 12 or 24 h after the 24-h fast (n = 10 per group). To determine the effects of insulin deficiency and replacement on gene expression, male C57BL/6J mice age 10 weeks were intraperitoneally given 50 mmol/l sodium citrate (control group, n = 8) or 250 mg/kg body wt streptozotocin (STZ) dissolved in 50 mmol/l sodium citrate (STZ group, n = 20). Blood glucose was determined twice daily using a OneTouch Glucometer until blood glucose was >450 on two consecutive measurements, at which time STZ-administered animals were randomly allocated into two groups (n = 10 per group): 1) the STZ group, which was followed for an additional 24 h with no insulin therapy, or 2) the STZ + INS group, which received Ultralente insulin at 2–4 units, s.c. per mouse every 6–8 h for 24 h. To control for the effects of stress, mice of the control and STZ groups also had their blood glucose measured and received an equal volume of PBS subcutaneously every 6–8 h for 24 h. All mice were killed by CO2 inhalation, after which their tissues were collected and stored at −80°C until analysis.
Statistical analysis
Data are given as means ± SE. Comparisons were made by unpaired two-tailed Student’s t tests or one- or two-way ANOVA, as appropriate. For ANOVAs, post hoc analysis was performed by Bonferroni’s multiple comparison test.
RESULTS
ATGL and adiponutrin are induced during adipogenesis
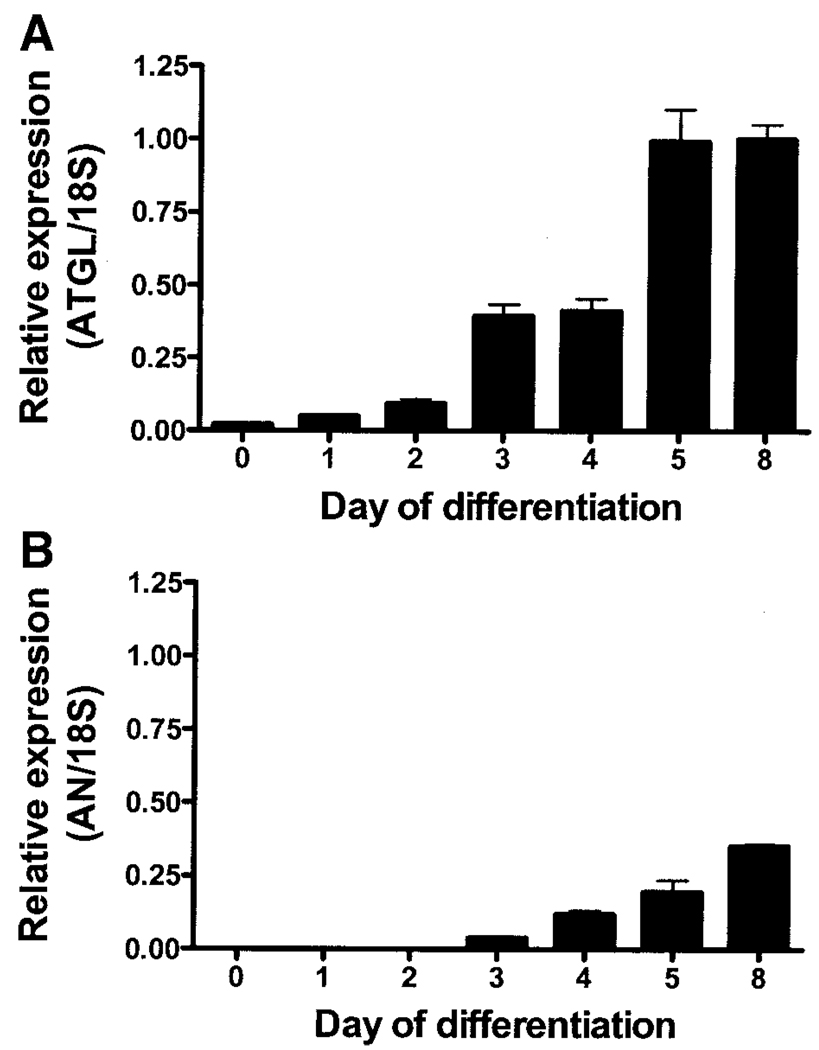
Affymetrix Microarray analysis revealed a 30-fold increase in the expression of ATGL (Riken 0610039C21, GenBank NM_025802) in 3T3-L1 adipocytes from day 0 through day 8 of differentiation. Q-PCR analysis in a separate set of differentiating 3T3-L1 adipocytes confirmed that ATGL was expressed at low but detectable levels early in differentiation (days 0–2), increased significantly by days 3–4, and reached maximum levels by day 5 (P < 0.0001) (Fig. 1A). The temporal expression pattern of ATGL was similar to that of HSL (data not shown). Adiponutrin expression was not detectable until day 3 of differentiation and reached maximum levels by day 8 (P < 0.0001) (Fig. 1B).
FIG. 1.
ATGL and adiponutrin (AN) expression during adipogenesis. ATGL (A) and adiponutrin (B) expression were determined by Q-PCR in 3T3-L1 adipocytes on days 0, 2, 3, 4, 5, and 8 of differentiation (n = 3 per time point). Gene expression was normalized to 18S ribosomal RNA and expressed relative to ATGL expression in day 8 adipocytes.
ATGL and adiponutrin are enriched in adipose tissue
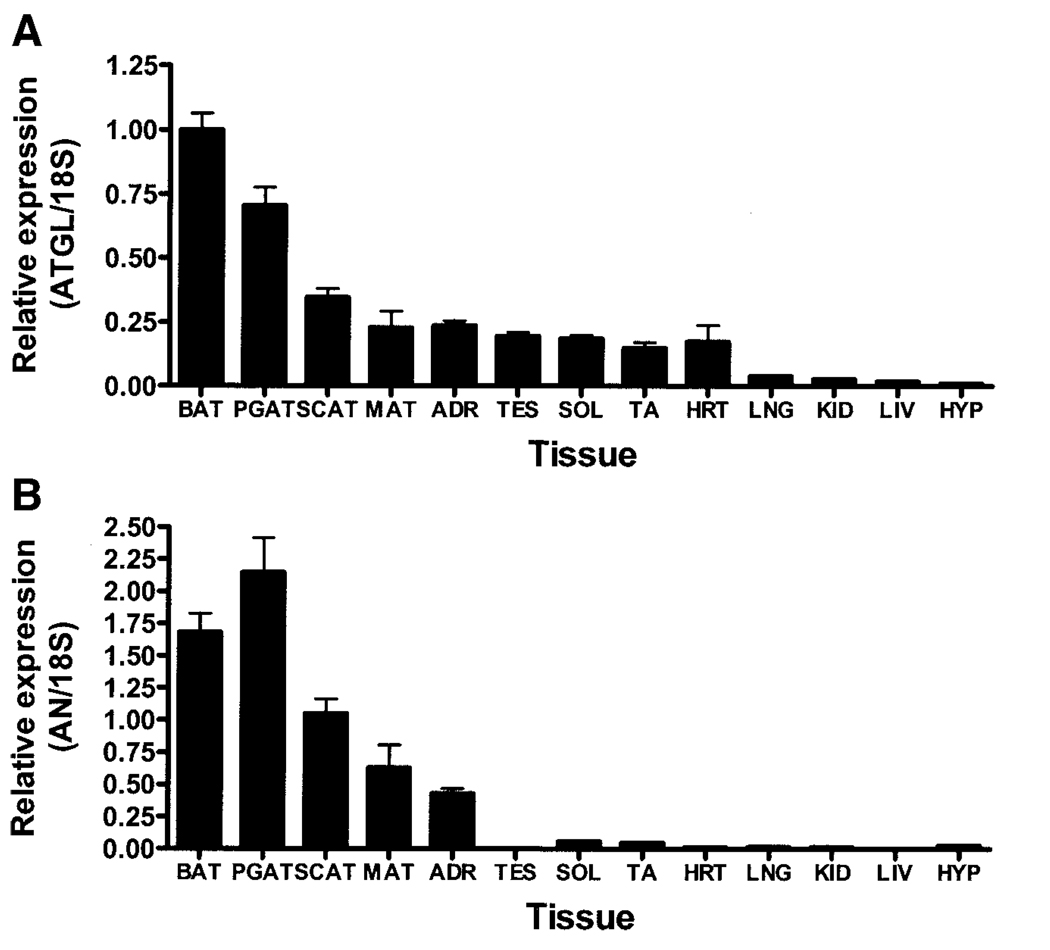
Q-PCR analysis of tissue-specific gene expression in mal C57BL/6J mice at age 10 weeks revealed that ATGL and adiponutrin were highly enriched in adipose tissue (Fig. 2A and B). Although adiponutrin expression was predominantly confined to adipose tissue and the adrenal gland, ATGL had a broader expression pattern, with moderate expression in the adrenal gland, testes, cardiac muscle, and skeletal muscle and low but detectable levels in the lung, kidney, liver, and hypothalamus. Adipose tissue expression of ATGL and adiponutrin was depot specific, with higher expression in brown adipose tissue (BAT) and perigonadal adipose tissue (PGAT) depots relative to other depots.
FIG. 2.
ATGL and adiponutrin (AN) expression in various murine tissues. ATGL (A) and adiponutrin (B) expression were determined by Q-PCR in BAT, PGAT, subcutaneous adipose tissue (SCAT), mesenteric adipose tissue (MAT), adrenal gland (ADR), testes (TES), soleus (SOL), tibialis anterior (TA), heart (HRT), lung (LNG), kidney (KID), liver (LIV), and hypothalamus (HYP) of 10-week-old male C57BL/6J mice (n = 6 per group). Gene expression was normalized to 18S ribosomal RNA and expressed relative to ATGL expression in BAT.
Retroviral-mediated overexpression of ATGL increases lipolysis
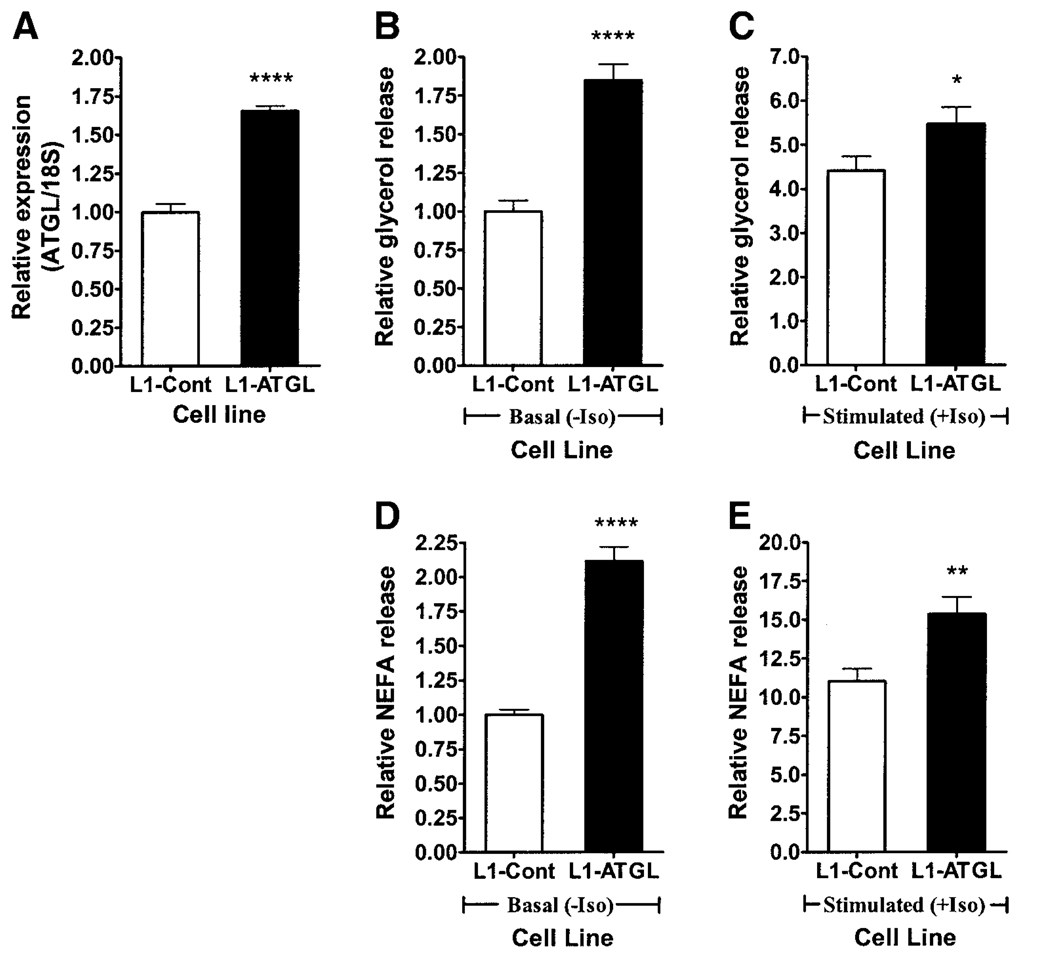
3T3-L1 adipocytes stably transfected with a retrovirus containing full-length ATGL cDNA demonstrated higher ATGL expression than control cells throughout adipogenesis, with the greatest difference at day 4 (1.65-fold higher) (Fig. 3A), without affecting HSL or adiponutrin expression (data not shown). This retroviral-mediated overexpression of ATGL was associated with a 1.85-fold increase in basal (Fig. 3B) and a 1.24-fold increase in stimulated (Fig. 3C) glycerol release. Basal (Fig. 3D) and stimulated (Fig. 3E) NEFA release were similarly increased (by 2.12- and 1.39-fold, respectively).
FIG. 3.
Overexpression of ATGL in 3T3-L1 adipocytes. Retroviral-mediated overexpression of ATGL in 3T3-L1 adipocytes was achieved via stable transfection with vector alone (L1-Cont) or vector containing full-length ATGL cDNA (L1-ATGL) (three experiments, n = 3 each). ATGL expression (A), basal (B) and stimulated (C) glycerol release, and basal (D) and stimulated (E) NEFA release were then evaluated at day 4 of differentiation. Gene expression was normalized to 18S ribosomal RNA and expressed relative to ATGL expression in the control group. Glycerol and NEFA release were normalized to total cellular protein and expressed relative to the nonstimulated control group. Iso, isoproterenol. *P < 0.05; **P < 0.01; ****P < 0.0001.
siRNA-mediated knockdown of ATGL decreases lipolysis
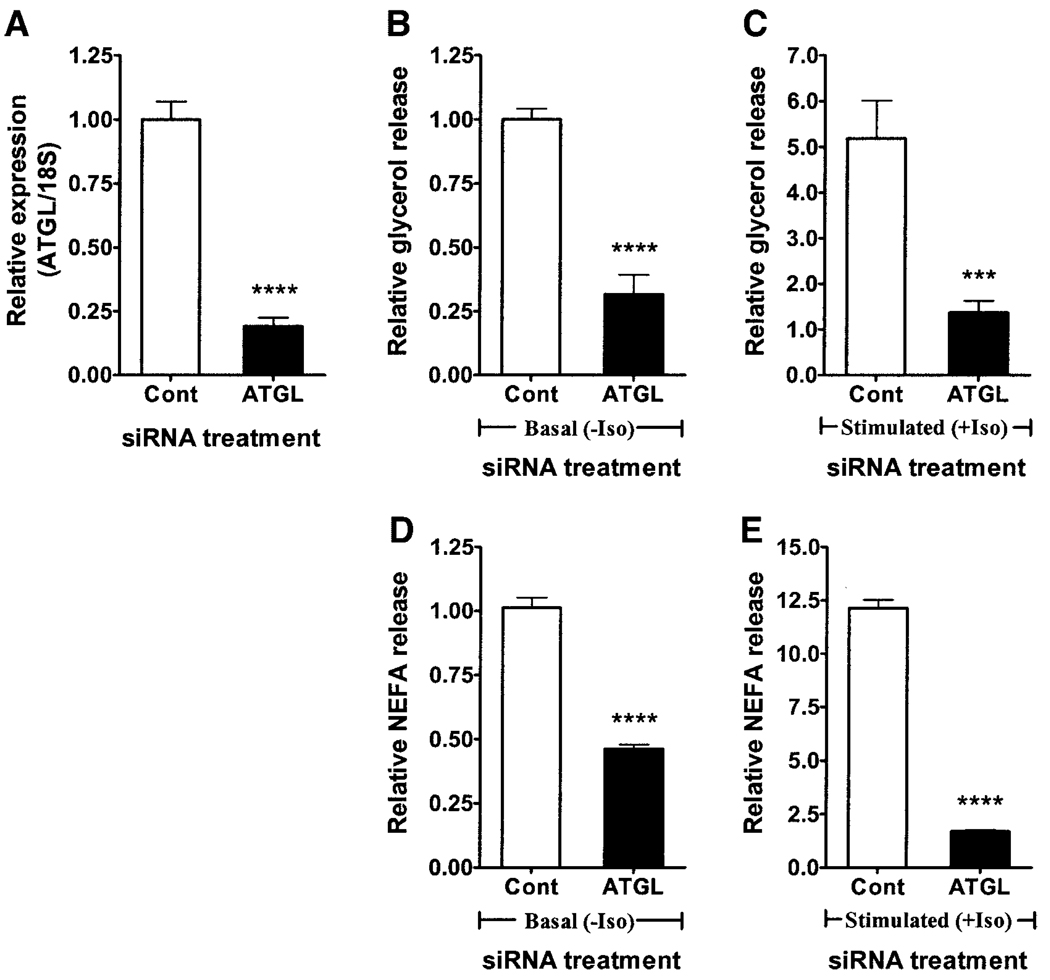
Treatment of 3T3-L1 adipocytes with ATGL-specific siRNA at day 4 of differentiation resulted in 81% knockdown of ATGL mRNA at day 6 (Fig. 4A) but had no effect on HSL or adiponutrin expression (data not shown). This siRNA-mediated knockdown of ATGL was associated with a 68% decrease in basal (Fig. 4B) and 74% decrease in stimulated (Fig. 4C) glycerol release. Basal (Fig. 4D) and stimulated (Fig. 4E) NEFA release were similarly decreased by 55 and 86%, respectively.
FIG. 4.
Knockdown of ATGL in 3T3-L1 adipocytes. siRNA-mediated knockdown of ATGL in 3T3-L1 adipocytes was achieved by electroporating 3T3-L1 adipocytes with either control siRNA (Cont) or ATGL-specific siRNA (ATGL) at day 4 of differentiation (three experiments, n = 3 each). ATGL expression (A), basal (B) and stimulated (C) glycerol release, and basal (D) and stimulated (E) NEFA release were then evaluated at day 6 of differentiation. Gene expression was normalized to 18S ribosomal RNA and expressed relative to ATGL expression in the control group. Glycerol and NEFA release were normalized to total cellular protein and expressed relative to the nonstimulated control group. Iso, isoproterenol. ***P < 0.001; ****P < 0.0001.
Comparison of siRNA-mediated knockdown of ATGL, adiponutrin, and HSL in fully differentiated 3T3-L1 adipocytes
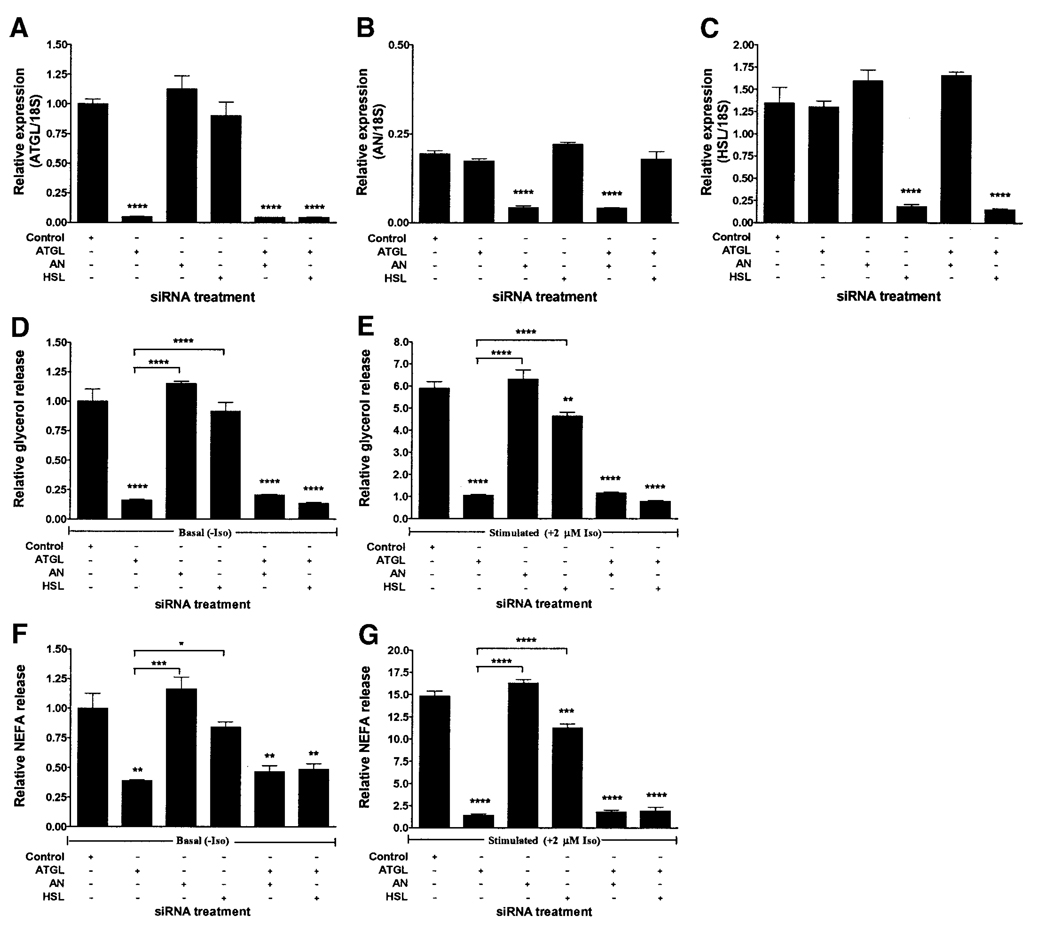
The treatment of 3T3-L1 adipocytes with siRNA specific for ATGL, adiponutrin, or HSL at day 7 of differentiation resulted in 96, 78, and 83% average knockdown of ATGL (Fig. 5A), adiponutrin (Fig. 5B), and HSL (Fig. 5C) mRNA, respectively, at day 9 of differentiation. Gene-specific mRNA expression was altered in those groups treated with the corresponding gene-specific siRNA but not in other groups. Basal glycerol and NEFA release were decreased only by ATGL knockdown and not by knockdown of adiponutrin or HSL alone, although there was a small reproducible decrease after HSL knockdown that did not reach statistical significance (Fig. 5D and F). Stimulated glycerol and NEFA release were decreased by knockdown of ATGL as well as HSL but not by knockdown of adiponutrin alone (Fig. 5E and G). Neither adiponutrin nor HSL knockdown had an additive effect on basal or stimulated glycerol or NEFA release when performed in combination with knockdown of ATGL. ATGL knockdown had a significantly greater effect on both basal and stimulated glycerol and NEFA release than did either HSL or adiponutrin knockdown.
FIG. 5.
Comparison of siRNA-mediated knockdown of ATGL, adiponutrin (AN), and HSL in fully differentiated 3T3-L1 adipocytes. 3T3-L1 adipocytes were electroporated in the presence of control siRNA or siRNAs specific for ATGL, adiponutrin, or HSL, alone or in combination at day 7 of differentiation (n = 3, representative one of two experiments). ATGL (A), adiponutrin (B), and HSL (C) expression were determined by Q-PCR 48 h later. Gene expression was normalized to 18S ribosomal RNA and expressed relative to ATGL expression in the control group. Basal (D) and stimulated (E) glycerol release and basal (F) and stimulated (G) NEFA release were determined at day 9 of differentiation. Glycerol and NEFA release was normalized to total cell protein and expressed relative to the nonstimulated control group. Iso, isoproterenol. *P < 0.05; **P < 0.01; ***P < 0.001; **** P < 0.0001.
ATGL and adiponutrin are oppositely regulated by nutritional status
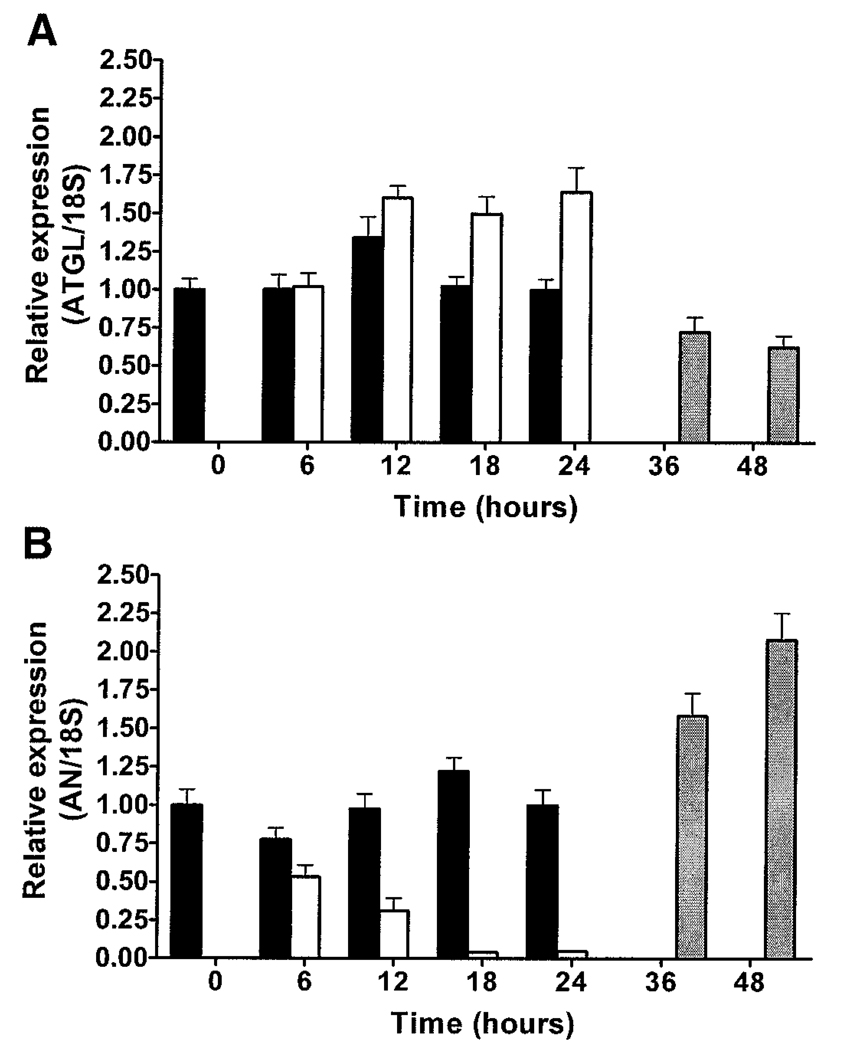
ATGL expression was increased by fasting (P < 0.0001) and decreased by re-feeding after a 24-h fast (P < 0.0001) (Fig. 6A). In contrast, adiponutrin expression was decreased by fasting (P < 0.0001) and increased by re-feeding (P < 0.0001) (Fig. 6B). A significant effect of time of day on gene expression was observed in ad libitum–fed animals for both ATGL (P < 0.05) and adiponutrin (P < 0.05), characterized by an increase in ATGL and a decrease in adiponutrin during the light cycle when animals consume less food and the opposite pattern during the dark cycle when animals consume most of their food. Similar expression patterns for ATGL and adiponutrin were observed in subcutaneous adipose tissue and BAT (data not shown). HSL expression was not significantly affected by fasting or re-feeding (data not shown).
FIG. 6.
Regulation of ATGL and adiponutrin (AN) expression by nutritional status. ATGL (A) and adiponutrin (B) mRNA expression were determined by Q-PCR in PGAT of 8-week-old male FVB mice that were fed ad libitum (■), fasted (6, 12, 18, or 24 h) (□), or re-fed (12 or 24 h after a 24-h fast) ( ) (n = 10 per group) with the onset of the light cycle (6:00 a.m.) as time 0. Gene expression was normalized to 18S ribosomal RNA and expressed relative to gene expression in ad libitum–fed control mice at time 0.
) (n = 10 per group) with the onset of the light cycle (6:00 a.m.) as time 0. Gene expression was normalized to 18S ribosomal RNA and expressed relative to gene expression in ad libitum–fed control mice at time 0.
ATGL and adiponutrin are oppositely regulated by insulin in vitro
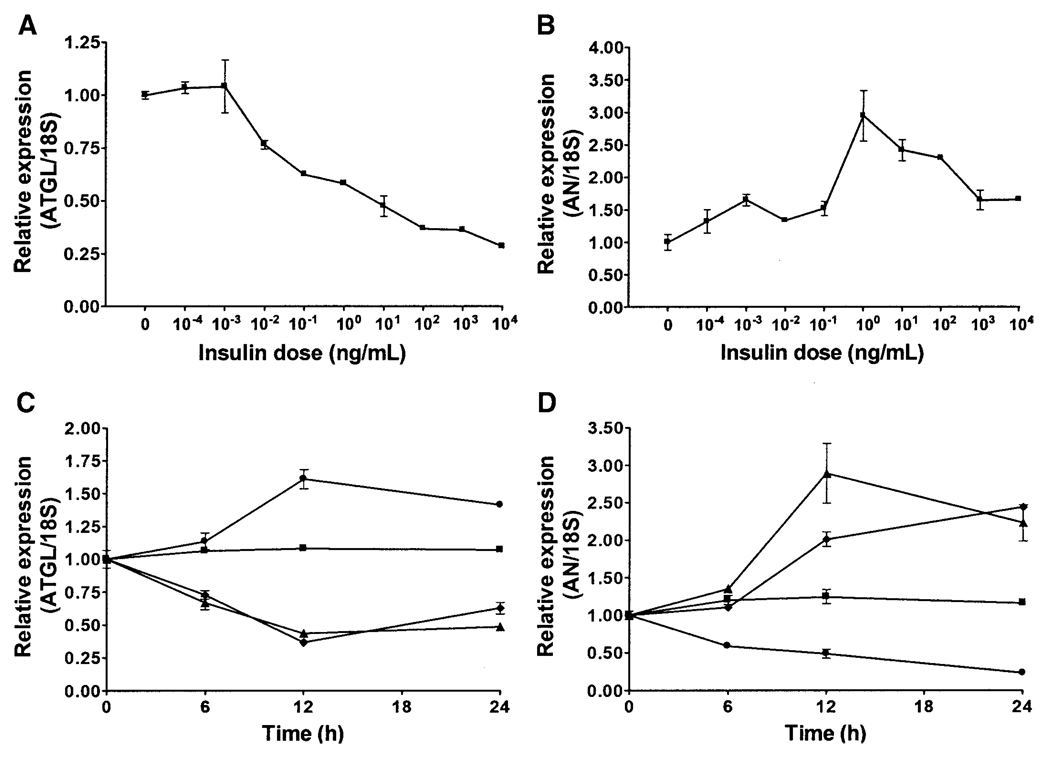
Incubation of fully differentiated 3T3-L1 adipocytes in serum-free DMEM supplemented with 10−4 to 104 ng/ml insulin for 12 h resulted in a dose-dependent decrease in ATGL expression (P < 0.0001) (Fig. 7A). Similar treatment resulted in a biphasic pattern for adiponutrin expression characterized by an initial dose-dependent increase followed by a decrease at higher doses (Fig. 7B). Incubation in serum-free medium, and hence in the absence of insulin as well as other serum factors, resulted in a time-dependent increase in ATGL expression, with a peak at 12 h (P < 0.001) (Fig. 7C). Incubation in serum-free medium supplemented with insulin at 1 ng/ml prevented this increase (data not shown). Incubation in medium supplemented with 1,000 ng/ml insulin resulted in a time-dependent decrease in ATGL expression, with the nadir occurring at 12 h regardless of the presence or absence of serum (P < 0.0001). For adiponutrin, incubation in the serum-free medium resulted in a time-dependent decrease in adiponutrin expression (P < 0.0001) (Fig. 7D). Incubation in medium supplemented with 1 ng/ml of insulin resulted in a time-dependent increase in adiponutrin expression, regardless of the presence or absence of serum (P < 0.0001). However, the increase at 12 h was more pronounced for adipocytes incubated in serum-free medium.
FIG. 7.
Regulation of ATGL and adiponutrin (AN) expression by insulin in 3T3-L1 adipocytes. Dose-response curves for ATGL (A) and adiponutrin (B) expression were generated by incubating fully differentiated 3T3-L1 adipocytes in serum-free DMEM supplemented with insulin at the doses indicated for 12 h (n = 3, representative one of three experiments). Data were normalized to 18S ribosomal RNA and expressed relative to gene expression in control cells incubated without insulin. Time course for ATGL (C) and adiponutrin (D) expression were generated by incubating fully differentiated 3T3-L1 adipocytes in either DMEM containing 10% FBS or serum-free DMEM (serum starved) with and without insulin at 1,000 ng/ml for ATGL or 1 ng/ml for adiponutrin for the time indicated (n = 3, representative one of three experiments). ■, FBS; ●, serum starved; ♦, FBS plus insulin; ▲, serum starved plus insulin. Data were normalized to 18S and expressed relative to gene expression in the control group at time 0. To convert ng/ml to nmol/l, multiply by 0.174.
ATGL and adiponutrin are oppositely regulated by insulin in vivo
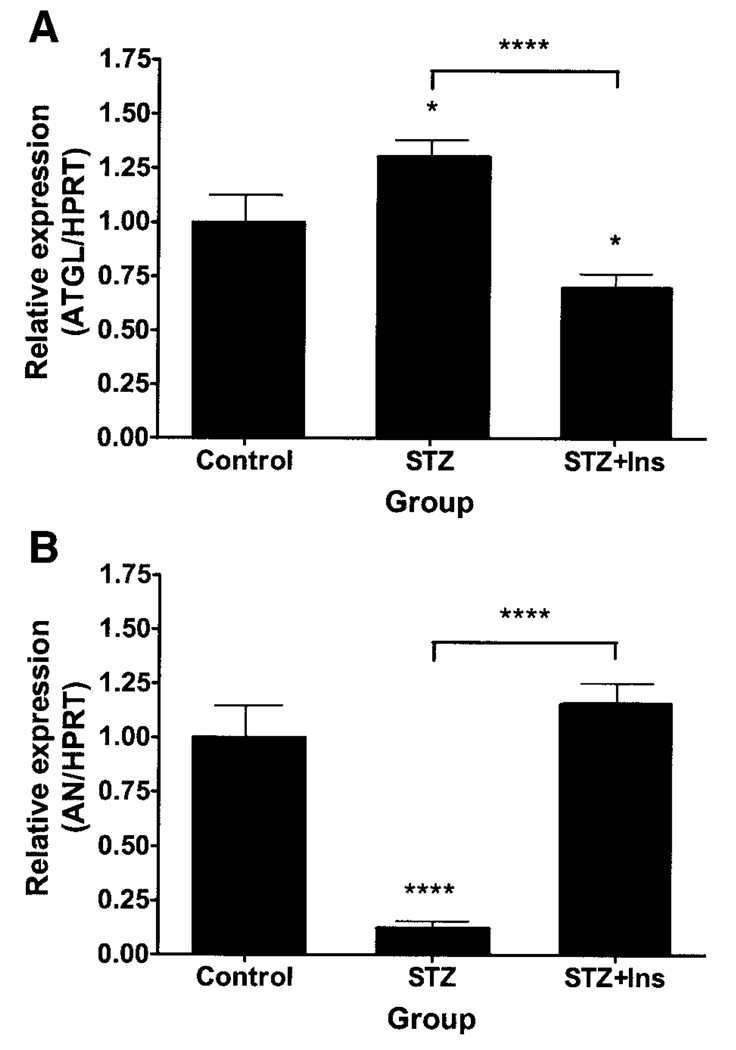
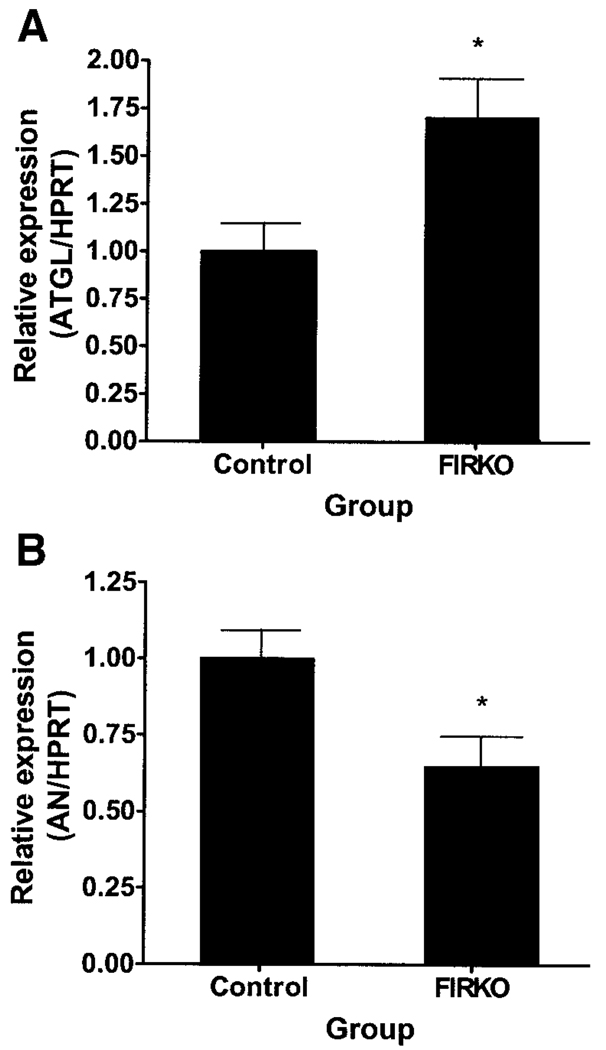
ATGL expression was increased by insulin deficiency due to STZ administration and decreased by insulin replacement (Fig. 8A). Conversely, adiponutrin expression was decreased by insulin-deficiency due to STZ administration and normalized by insulin replacement (Fig. 8B). The differences between ATGL and adiponutrin expression in STZ-administered mice that did and did not receive insulin were highly significant. In the PGAT of FIRKO mice, ATGL expression was increased (Fig. 9A) and adiponutrin expression was decreased (Fig. 9B). HSL expression was unchanged in both of the above models (data not shown).
FIG. 8.
ATGL and adiponutrin (AN) expression in adipose tissue of STZ-administered mice. ATGL (A) and adiponutrin (B) expression were determined by Q-PCR in PGAT of ad libitum–fed, 10-week-old male mice given vehicle (control), STZ, or STZ followed by insulin replacement for 24 h (STZ + Ins) (n = 8–10 per group). Data were normalized to HPRT and expressed relative to gene expression in the control group. *P < 0.05; ****P < 0.0001.
FIG. 9.
ATGL and adiponutrin (AN) expression in adipose tissue of FIRKO mice. ATGL (A) and adiponutrin (B) expression were determined by Q-PCR in PGAT of 2-year-old, ad libitum–fed, male FIRKO mice (n = 4 per group). Data were normalized to HPRT and expressed relative to gene expression in the control group. *P < 0.05.
DISCUSSION
Using microarray analysis, we identified a Riken gene 0610039C21 that was markedly induced during adipogenesis and highly expressed in adipose tissue, and we performed studies designed to determine its function and regulation. The identification and characterization of Riken 06010039C21, alternatively designated ATGL or desnutrin, has been reported by other groups using different approaches while our studies were being completed (11,12). The human homolog iPLA2ζ (previously TTS-2.2) and the Drosophila homolog Brummer lipase have also been reported (13,20). ATGL shares considerable sequence and structural homology with several proteins in a variety of organisms, ranging from prokaryotes to humans (21,22), and with functions ranging from plant nutrient storage and host defense to bacterial virulence (23,24). Its NH2-terminal sequence contains a patatin-like domain, characteristic of patatin, a potato protein with lipid acyl hydrolase, acyl transferase, and esterase activity (25–28). It also contains a predicted α/β hydrolase fold, a GXSXG serine hydrolase motif, and a Ser-Asp catalytic dyad, all features commonly found in known lipases. ATGL shares the greatest sequence homology with adiponutrin, a previously reported nutritionally regulated gene that is induced during adipogenesis, enriched in adipose tissue, and upregulated in genetic models of obesity (29–32).
ATGL and adiponutrin are induced during adipogenesis in 3T3-L1 adipocytes. However, although adiponutrin is undetectable until day 3, ATGL is expressed at low but detectable levels early in differentiation and begins to increase within the first 24 h of differentiation. This early expression precedes the expression of several adipogenic genes, including C/EBP-α and PPAR-γ, suggesting a potential role for ATGL in the adipogenic process. In addition, both ATGL and adiponutrin are highly enriched in adipose tissue in a depot-specific manner, with higher expression in BAT and PGAT. This differential expression may contribute to the metabolic heterogeneity among adipose tissue depots. Interestingly, ATGL expression exceeds adiponutrin expression in cultured 3T3-L1 adipocytes, whereas the opposite is true in adipose tissue. Furthermore, although adiponutrin expression is confined to adipose tissue and the adrenal gland, ATGL is expressed in a variety of tissues, most notably cardiac and skeletal muscle. Indeed, the expression of ATGL in muscle is comparable with its expression in several white adipose tissue depots, suggesting a potentially important role for ATGL in this metabolically active tissue as well.
Functional analysis of ATGL demonstrates that stable retroviral-mediated overexpression increases while siRNA-mediated knockdown decreases both basal and stimulated glycerol and NEFA release in 3T3-L1 adipocytes. Qualitatively similar effects of overexpression and knockdown of ATGL in 3T3-L1 adipocytes using adenoviral-mediated delivery systems (and antisense RNA) have been reported (11). The quantitatively greater knockdown of ATGL in our study was associated with a comparably greater effect on glycerol and NEFA release. Despite this decrease in absolute basal and stimulated lipolysis after ATGL knockdown, some residual fold-stimulation in lipolysis was observed. This may have been due to residual ATGL expression or to other ATGL-independent mechanisms. It has been previously observed that PKA increases phosphorylation of ATGL only 2-fold compared with 26-fold for HSL but does not significantly affect ATGL activity (11), thereby implicating a PKA-independent mechanism for isoproterenol’s effects on ATGL-mediated lipolysis. Nevertheless, the dramatic effect of experimental manipulation of ATGL expression on absolute glycerol and NEFA release suggests that ATGL-mediated lipolysis has major physiological relevance.
Direct comparison of ATGL and HSL knockdown in fully differentiated 3T3-L1 adipocytes demonstrated a greater effect of ATGL relative to HSL knockdown on both basal and stimulated glycerol and NEFA release. In addition, knockdown of HSL in combination with ATGL did not have an additive effect on basal or stimulated glycerol or NEFA release. Although the measurable functional effects observed after manipulation of ATGL and HSL mRNA expression suggested comparable changes in protein expression, an antibody against ATGL protein is not currently available and hence direct comparison of ATGL and HSL protein expression under the above experimental conditions cannot be made at this time. Nevertheless, the above data suggest that these lipolytic enzymes may work in a serial rather than in a parallel fashion, with ATGL-mediated triglyceride hydrolysis providing diglyceride substrate for HSL, thus driving subsequent steps in the lipolytic pathway. This conclusion is supported by data characterizing the substrate specificity of ATGL, which show that ATGL increases hydrolysis of triglycerides but not of other substrates, whereas HSL has broad substrate specificity and a 10-fold greater activity against diglycerides than triglycerides (11–13). Furthermore, HSL plays a relatively greater role in isoproterenol-stimulated compared with basal lipolysis (7,8,10). Despite HSL’s broad substrate specificity and clear contribution to lipolysis, however, HSL was not able to compensate for the decrease in basal or stimulated lipolysis after siRNA-mediated knockdown of ATGL. More definitive studies in cells and tissues with targeted deletion of these genes, alone and in combination, will be useful in further evaluating the relative contribution of ATGL and HSL to lipolysis.
Although these data support a role for ATGL in triglyceride-specific hydrolysis and net glycerol and NEFA release, the function of the related protein adiponutrin remains less clear. siRNA-mediated knockdown of adiponutrin in fully differentiated 3T3-L1 adipocytes did not have a significant effect on basal or stimulated glycerol or NEFA release, despite a >75% knockdown of adiponutrin mRNA. Furthermore, knockdown of adiponutrin in combination with ATGL did not have an additive effect on ATGL-mediated glycerol or NEFA release. These findings are further supported by those of Zimmerman et al. (11) who parenthetically reported an inability to detect lipid hydrolysis activity for adiponutrin. Nevertheless, Jenkins et al. (13) did detect triglyceride hydrolase as well as acyltransferase activity of the human homolog of adiponutrin, iPLA2ϵ. Because an antibody against adiponutrin protein is not currently available, the possibility that siRNA-mediated knockdown of adiponutrin mRNA may not produce a comparable knockdown of adiponutrin protein cannot be entirely excluded. On the other hand, the above finding may represent an inherent functional difference between the murine and human homologs. Alternatively, adiponutrin may participate primarily in triglyceride/NEFA recycling rather than in net lipolysis per se. As noted above, more definitive studies in cells and tissues with targeted deletion of adiponutrin are required to more definitively determine its contribution to lipolysis.
Despite the above-noted differences in function, both ATGL and adiponutrin are strongly regulated by nutritional status. ATGL expression is increased by fasting and decreased by re-feeding, whereas adiponutrin shows the opposite pattern. This nutritional regulation has been previously reported for adiponutrin (29) and subsequently for ATGL (12). In contrast, HSL is not significantly regulated at the transcriptional level by nutritional status in rodents (33). In addition, the expression of ATGL and adiponutrin are temporally related to the feeding cycle in ad libitum–fed mice, suggesting that transcriptional regulation of these genes may play a metabolic role under normal physiological conditions as well. Although glucocorticoids have been implicated in the nutritional regulation of ATGL (12), several other factors and hormones, notably insulin, may also contribute to this nutritional regulation.
To further explore this possibility, we evaluated the ability of insulin to regulate ATGL and adiponutrin in vitro and in vivo. Treatment of fully differentiated 3T3-L1 adipocytes with insulin produces a dose-dependent decrease in ATGL expression with a half-maximal response near the reported Kd for insulin with its receptor in fully differentiated 3T3-L1 adipocytes (34). Although adiponutrin shows the opposite pattern at lower insulin doses, its expression subsequently decreases at higher doses, underscoring the complex regulation of adiponutrin. A small but significant effect of insulin on adiponutrin expression has been previously reported at an insulin dose of 170 nmol/l (~1,000 ng/ml) (29). However, our results suggest a relatively greater contribution of insulin to adiponutrin expression at insulin doses closer to the physiological range, particularly under serum-starved conditions. Using insulin doses specifically selected for their maximal effect on ATGL and adiponutrin expression, insulin decreased ATGL and increased adiponutrin expression in a time-dependent manner. This time- and dose-dependent regulation of both ATGL and adiponutrin expression by insulin strongly supports a role for insulin in the transcriptional regulation of these genes.
As further support for this hypothesis, adipose tissue expression of both ATGL and adiponutrin are regulated at the transcriptional level in two in vivo models of altered insulin action: the STZ model of systemic insulin deficiency and the FIRKO model of adipocyte-specific insulin receptor deficiency. The STZ model is characterized by activation of lipolysis in adipose tissue, resulting in hydrolysis of stored triglycerides and release of large amounts of NEFAs into the plasma. STZ administration in rats is associated with increased HSL mRNA expression in isolated adipocytes, whereas insulin replacement produces no change in HSL mRNA expression (35). We likewise found no change in HSL expression in response to insulin treatment, but we were also unable to detect a significant change after STZ administration. The discrepancy may have been due to a difference in species (rat versus mice) or adipose substrate (isolated adipocytes versus whole adipose tissue). In contrast to our observations with HSL, adipose tissue expression of ATGL was clearly increased by insulin deficiency and decreased by insulin replacement in STZ-administered mice, while adiponutrin showed the opposite pattern. Similarly, ATGL was increased, adiponutrin was decreased, and HSL was unchanged in adipose tissue of FIRKO mice, which have normal serum triglycerides and NEFAs but increased basal lipolysis and lipogenesis in a subset of “large” adipocytes (19). Although these data support a potential role for insulin in the transcriptional regulation of ATGL and adiponutrin, more specific studies evaluating transcription per se are required to better characterize the specific effect of insulin on mRNA of these genes.
In conclusion, ATGL and adiponutrin are induced during adipogenesis and are highly enriched in adipose tissue. Adipose tissue expression of ATGL and adiponutrin are reciprocally regulated by nutritional status and by insulin in vitro and in vivo. The identification and characterization of these murine genes and their homologs in other species have now been reported by several groups (11–13,20). Taken together, these data suggest that ATGL participates in triglyceride-specific hydrolysis and hence, like HSL, may play an important role in lipid metabolism. However, although HSL acutely affects lipolysis, primarily via post-translational mechanisms, ATGL may mediate more long-term effects on lipolysis via transcriptional mechanisms. Furthermore, despite sequence and regulatory similarities, the contribution of adiponutrin to lipid metabolism remains unclear. Additional studies, including in vivo gain and loss of function studies, are required to further characterize the role of ATGL and adiponutrin in whole-body metabolism and to assess their potential contribution to disorders such as obesity and diabetes.
ACKNOWLEDGMENTS
J.S.F. has received research support from Takeda Pharmaceuticals.
This research was supported by National Institutes of Health (NIH) Grants 5K08DK065833-02 and 5P30DK046200-13 (to E.E.K.), NIH Grant 4R37DK028082-24 (to J.S.F.), Takeda Chemical Industries Grant (to J.S.F.), and NIH Grant 2P30DK057521-069006 (to the Beth Israel Deaconess Medical Center Metabolic Physiology Core Facility).
We are grateful to Michael Czech and Qionglin Zhou from the University of Massachusetts Medical Center for sharing expertise and advice on using siRNA and C. Ronald Kahn from the Joslin Diabetes Center for providing adipose tissue RNA from FIRKO mice.
Glossary
- ATGL
adipose triglyceride lipase
- BAT
brown adipose tissue
- FBS
fetal bovine serum
- FIRKO
fat-specific insulin receptor knockout
- HPRT
hypoxanthine guanine phosphoribosyl transferase
- HSL
hormone-sensitive lipase
- iPLA2
calcium-independent phospholipase A2
- NEFA
nonesterified fatty acid
- PGAT
perigonadal adipose tissue
- PK
protein kinase
- Q-PCR
quantitative real-time PCR
- STZ
streptozotocin
REFERENCES
- 1.Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans. 2003;31:1120–1124. doi: 10.1042/bst0311120. [DOI] [PubMed] [Google Scholar]
- 2.Kraemer FB, Shen WJ. Hormone-sensitive lipase: control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J Lipid Res. 2002;43:1585–1594. doi: 10.1194/jlr.r200009-jlr200. [DOI] [PubMed] [Google Scholar]
- 3.Yeaman SJ. Hormone-sensitive lipase: new roles for an old enzyme. Biochem J. 2004;379:11–22. doi: 10.1042/BJ20031811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterlund T. Structure-function relationships of hormone-sensitive lipase. Eur J Biochem. 2001;268:1899–1907. doi: 10.1046/j.1432-1327.2001.02097.x. [DOI] [PubMed] [Google Scholar]
- 5.Shakur Y, Holst LS, Landstrom TR, Movsesian M, Degerman E, Manganiello V. Regulation and function of the cyclic nucleotide phosphodiesterase (PDE3) gene family. Prog Nucleic Acid Res Mol Biol. 2001;66:241–277. doi: 10.1016/s0079-6603(00)66031-2. [DOI] [PubMed] [Google Scholar]
- 6.Londos C, Honnor RC, Dhillon GS. cAMP-dependent protein kinase and lipolysis in rat adipocytes. III. Multiple modes of insulin regulation of lipolysis and regulation of insulin responses by adenylate cyclase regulators. J Biol Chem. 1985;260:15139–15145. [PubMed] [Google Scholar]
- 7.Osuga J, Ishibashi S, Oka T, Yagyu H, Tozawa R, Fujimoto A, Shionoiri F, Yahagi N, Kraemer FB, Tsutsumi O, Yamada N. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci U S A. 2000;97:787–792. doi: 10.1073/pnas.97.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang SP, Laurin N, Himms-Hagen J, Rudnicki MA, Levy E, Robert MF, Pan L, Oligny L, Mitchell GA. The adipose tissue phenotype of hormone-sensitive lipase deficiency in mice. Obes Res. 2001;9:119–128. doi: 10.1038/oby.2001.15. [DOI] [PubMed] [Google Scholar]
- 9.Haemmerle G, Zimmermann R, Hayn M, Theussl C, Waeg G, Wagner E, Sattler W, Magin TM, Wagner EF, Zechner R. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- 10.Okazaki H, Osuga J, Tamura Y, Yahagi N, Tomita S, Shionoiri F, Iizuka Y, Ohashi K, Harada K, Kimura S, Gotoda T, Shimano H, Yamada N, Ishibashi S. Lipolysis in the absence of hormone-sensitive lipase: evidence for a common mechanism regulating distinct lipases. Diabetes. 2002;51:3368–3375. doi: 10.2337/diabetes.51.12.3368. [DOI] [PubMed] [Google Scholar]
- 11.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 12.Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem. 2004;279:47066–47075. doi: 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamm JK, Park BH, Farmer SR. A role for C/EBP in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3–L1 preadipocytes. J Biol Chem. 2001;276:18464–18471. doi: 10.1074/jbc.M100797200. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 18.Jiang ZY, Zhou QL, Coleman KA, Chouinard M, Boese Q, Czech MP. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc Natl Acad Sci U S A. 2003;100:7569–7574. doi: 10.1073/pnas.1332633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bluher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, Kahn CR. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 20.Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metabolism. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Mignery GA, Pikaard CS, Park WD. Molecular characterization of the patatin multigene family of potato. Gene. 1988;62:27–44. doi: 10.1016/0378-1119(88)90577-x. [DOI] [PubMed] [Google Scholar]
- 22.Banerji S, Flieger A. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology. 2004;150:522–525. doi: 10.1099/mic.0.26957-0. [DOI] [PubMed] [Google Scholar]
- 23.Paiva E, Lister RM, Park WD. Induction and accumulation of major tuber proteins of potato stems and petioles. Plant Physiol. 1983;71:161–168. doi: 10.1104/pp.71.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato H, Frank DW, Hillard CJ, Feix JB, Pankhaniya RR, Moriyama K, Finck-Barbancon V, Buchaklian A, Lei M, Long RM, Wiener-Kronish J, Sawa T. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003;22:2959–2969. doi: 10.1093/emboj/cdg290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mignery GA, Pikaard CS, Hannapel DJ, Park WD. Isolation and sequence analysis of cDNAs for the major potato tuber protein, patatin. Nucleic Acid Res. 1984;12:7987–8000. doi: 10.1093/nar/12.21.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews DL, Beames B, Summers MD, Park WD. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem J. 1988;252:199–206. doi: 10.1042/bj2520199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirschberg HJ, Simons JW, Dekker N, Egmond MR. Cloning, expression, purification and characterization of patatin, a novel phospholipase A. Eur J Biochem. 2001;268:5037–5044. doi: 10.1046/j.0014-2956.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 28.Galliard T. The enzymic deacylation of phospholipids and galactolipids in plants: purification and properties of a lipolytic acyl-hydrolase from potato tubers. Biochem J. 1971;121:379–390. doi: 10.1042/bj1210379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baulande S, Lasnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001;276:33336–33344. doi: 10.1074/jbc.M105193200. [DOI] [PubMed] [Google Scholar]
- 30.Polson DA, Thompson MP. Macronutrient composition of the diet differentially affects leptin and adiponutrin mRNA expression in response to meal feeding. J Nutr Biochem. 2004;15:242–246. doi: 10.1016/j.jnutbio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Polson DA, Thompson MP. Adiponutrin mRNA expression in white adipose tissue is rapidly induced by meal-feeding a high-sucrose diet. Biochem Biophys Res Commun. 2003;301:261–266. doi: 10.1016/s0006-291x(02)03027-9. [DOI] [PubMed] [Google Scholar]
- 32.Polson D, Thompson M. Adiponutrin gene expression in 3T3-L1 adipocytes is downregulated by troglitazone. Horm Metab Res. 2003;35:508–510. doi: 10.1055/s-2003-41811. [DOI] [PubMed] [Google Scholar]
- 33.Sztalryd C, Kraemer FB. Regulation of hormone-sensitive lipase during fasting. Am J Physiol. 1994;266:E179–E185. doi: 10.1152/ajpendo.1994.266.2.E179. [DOI] [PubMed] [Google Scholar]
- 34.Chang TH, Polakis SE. Differentiation of 3T3–L1 fibroblasts to adipocytes. Effect of insulin and indomethacin on the levels of insulin receptors. J Biol Chem. 1978;253:4693–4696. [PubMed] [Google Scholar]
- 35.Sztalryd C, Kraemer FB. Regulation of hormone-sensitive lipase in streptozotocin-induced diabetic rats. Metabolism. 1995;44:1391–1396. doi: 10.1016/0026-0495(95)90135-3. [DOI] [PubMed] [Google Scholar]