Abstract
Kisspeptin is a product of the Kiss1 gene and is expressed in the forebrain. Neurons that express Kiss1 play a crucial role in the regulation of pituitary luteinizing hormone secretion and reproduction. These neurons are the direct targets for the action of estradiol-17β (E2), which acts via the estrogen receptor α isoform (ERα) to regulate Kiss1 expression. In the arcuate nucleus (Arc), where the dynorphin gene (Dyn) is expressed in Kiss1 neurons, E2 inhibits the expression of Kiss1 mRNA. However, E2 induces the expression of Kiss1 in the anteroventral periventricular nucleus (AVPV). The mechanism for differential regulation of Kiss1 in the Arc and AVPV by E2 is unknown. ERα signals through multiple pathways, which can be categorized as either classical, involving the estrogen response element (ERE), or nonclassical, involving ERE-independent mechanisms. To elucidate the molecular basis for the action of E2 on Kiss1 and Dyn expression, we studied the effects of E2 on Kiss1 and Dyn mRNAs in the brains of mice bearing targeted alterations in the ERα signaling pathways. We found that stimulation of Kiss1 expression by E2 in the AVPV and inhibition of Dyn in the Arc required an ERE-dependent pathway, whereas the inhibition of Kiss1 expression by E2 in the Arc involved ERE-independent mechanisms. Thus, distinct ERα signaling pathways can differentially regulate the expression of identical genes across different brain regions, and E2 can act within the same neuron through divergent ERα signaling pathways to regulate different neurotransmitter genes.
Introduction
Gonadotropin-releasing hormone (GnRH) neurons represent the final common pathway through which the brain regulates reproduction. The secretion of GnRH is controlled by the negative and positive feedback actions of 17β-estradiol (E2), which in turn coordinate reproductive cycles. The primary effects of E2 on GnRH secretion are mediated by estrogen receptor α (ERα) (Rissman et al., 1997; Shughrue et al., 1997, 2002; Couse and Korach, 1999); however, GnRH neurons do not express ERα (Herbison and Theodosis, 1992). Kisspeptin neurons in the hypothalamus are thought to relay information about estrogen status directly to GnRH neurons (Popa et al., 2008). Kisspeptin, which is coded by the Kiss1 gene, is expressed in the rodent by neurons in the arcuate (Arc) and anteroventral periventricular (AVPV) nuclei that coexpress ERα (Smith et al., 2005). E2 inhibits Kiss1 expression in the Arc (Smith et al., 2005), a nodal point in negative feedback regulation of GnRH/luteinizing hormone (LH) secretion (Nishihara et al., 1986; Thind and Goldsmith, 1988; Goodman, 1996), and conversely, E2 stimulates the expression of Kiss1 in the AVPV (Smith et al., 2005), an essential area for generating the preovulatory GnRH surge through positive feedback (Wiegand et al., 1978; Terasawa et al., 1980; Popolow et al., 1981; Ronnekleiv and Kelly, 1988; Simerly et al., 1990; Gu and Simerly, 1997; Polston and Simerly, 2006). Although the induction and inhibition of Kiss1 expression in the AVPV and Arc both require ERα (Smith et al., 2005), the molecular pathways through which ERα differentially regulates Kiss1 expression in these regions remain a mystery.
ERα exerts its effects through multiple signaling pathways (O'Malley and Tsai, 1992; Glass, 1994; McKenna et al., 1999; Smith and O'Malley, 2004; McDevitt et al., 2008). One pathway involves the translocation of ERα into the nucleus, where it recruits cofactors to estrogen response element (ERE) regulatory sites to alter gene transcription—termed classical signaling (Glass, 1994; Tsai and O'Malley, 1994; McKenna et al., 1999). ERα signaling can also employ ERE-independent genomic pathways that entail interactions with other transcription factors—termed the nonclassical pathway (Gaub et al., 1990; Ray et al., 1994; Stein and Yang, 1995; Webb et al., 1995; Kushner et al., 2000; Jakacka et al., 2001, 2002). Recently, it has been shown that the positive feedback effects of E2 require an ERE-dependent pathway, whereas negative feedback involves an ERE-independent mechanism (Glidewell-Kenney et al., 2007). We argued that if Kiss1 in the AVPV mediates positive feedback, the induction of Kiss1 by E2 in the AVPV would require ERE-dependent signaling. Likewise, if Kiss1 in the Arc mediates negative feedback, the inhibition of Kiss1 and dynorphin [Dyn; which is coexpressed with Kiss1 (Goodman et al., 2007) (V. M. Navarro, D. K Clifton, R. A. Steiner, our unpublished observations)] in the Arc would require an ERE-independent mechanism. To test this hypothesis and dissect the molecular pathways by which E2 regulates Kiss1 and Dyn gene expression in the brain, we studied mice with genetically engineered alterations in the ERα signaling pathways and analyzed the effects of E2 on Kiss1 and Dyn mRNAs (Glidewell-Kenney et al., 2007).
Materials and Methods
Animals.
Animals were housed at Northwestern University and surgeries were conducted there according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animal use procedures were approved by the Northwestern University Animal Care and Use Committee. Nonclassical estrogen receptor knock-in (ERαAA/−) mice were created by crossing mutant mice that have an ERα allele that cannot bind ERE (E207A/G208A; AA) on a 129SvJ background with ERα−/− mice. ERαAA/− mutants used in these experiments are the result of the AA mutant allele crossed 7–11 generations onto the C57BL/6 background. ERα−/− mice (obtained from Pierre Chambon, Collège de France, Illkirch, France) were on a C57BL/6 background. Mice were maintained on a 14 h light:10 h dark cycle with standard chow (7912; Harlan Teklad) and water available ad libitum.
Ovariectomy and estradiol replacement.
Adult female mice (10–13 weeks of age) were ovariectomized on day 0. The animals were anesthetized by isoflurane inhalation. Immediately after ovariectomy (OVX), animals received midscapular subcutaneous SILASTIC capsules (1.5 cm in length that were plugged with silicone adhesive on each end to leave 1 cm to fill with treatment, inner diameter 1.47 mm; outer diameter 1.95 mm) that were either filled with only sesame oil (OVX + vehicle group; n = 5) or capsules containing 1 mg/ml E2 in sesame oil (OVX + E2 group; n = 6) (Miller et al., 1995; Smith et al., 2005; Dungan et al., 2007). Between 9:00 and 10:00 A.M. 7 d after OVX, animals were killed and brains and sera were obtained. Brain tissue was sent to the University of Washington for analysis. Serum measurements for E2 and LH were conducted at Northwestern University.
Serum collection and assays.
Blood was allowed to coagulate for 90 min at room temperature and centrifuged at 2000 × g for 15 min. Serum was transferred to a fresh tube and stored at −20°C. Serum levels of LH and E2 were measured at Northwestern University (Evanston, IL). Reagents for the LH assay were from the National Institutes of Health, the antiserum used was anti-r-LH-S11, and the standard was rLH-RP3. The assay sensitivity was 0.2 ng/ml, and the intra-assay coefficient of variation was 4%. E2 was measured with a double-antibody kit (Diagnostics Production). The assay sensitivity was 2.0 pg/ml, and the intra-assay coefficient of variation was 6%.
Tissue preparation.
Mice were anesthetized with isoflurane and then killed by decapitation. Brains were removed and frozen on dry ice. Sections in the coronal plane (20 μm) were cut on a cryostat, thaw-mounted onto SuperFrost Plus slides (VWR Scientific), and stored at −80°C. Sections were collected from the diagonal band of Broca to the mammillary bodies.
Detection of Kiss1 mRNA.
The Kiss1 probe used for detection of Kiss1 mRNA was previously described by Gottsch et al. (2004). The Kiss1-specific sequence of the probe spans bases 76–486 of the mouse cDNA sequence (GenBank accession no. AF472576). The procedure for in situ hybridization (ISH) is outlined below.
Detection of Dynorphin mRNA.
Total RNA was extracted from mouse brain using an RNAqueous Kit (Ambion). RNA was reverse transcribed into cDNA with a RetroScript kit (Ambion) primed with oligodeoxythymidine (dT) for subsequent PCR. Primers were designed based on the published sequence of the Pro-Dynorphin mouse gene (GenBank accession number NM_018863) with forward primers starting at 132 bp and reverse primers starting at 540 bp for a product of ∼409 bp. Primers were custom synthesized (OPERON). PCRs contained the following in a volume of 25 μl: 2 μl of reverse transcriptase reaction product; 0.2 μm of each primer; 12.5 μl of RediTaq polymerase (Sigma-Aldrich); and 8.5 μl of water. Reactions were performed in a PTC-100 thermal cycler (MJ Research) using the following protocol: cDNA was denatured for 5 min at 94°C, then 35 cycles were performed at 94°C for 1 min, 55°C for 1.5 min, and 72°C for 2 min, with a final 5 min extension at 72°C. After electrophoresis on a 2% agarose (w/v) gel, a single DNA fragment was obtained of approximately the expected size (409 bp) and gel purified with a QiaQuick gel extraction kit (QIAGEN). The PCR product was confirmed to be the mouse Dynorphin probe by sequencing. Clamp polymerase sequences for T7 or T3 polymerase were added for the final primer product sequence of Dyn forward primer: CAGAGATGCAATTAACCCTCAACTAAAGGGAGAGATTTGCTCCCTGGAGTC and Dyn reverse primer: CCAAGCCTTCTAATACGACTCACTATAGGGAGACATCTCGGAACTCCTCTTGG and transcribed for ISH.
In situ hybridization.
Antisense mouse Kiss1 probe was transcribed from linearized pAMP1 plasmid containing the mouse Kiss1 insert with T7 polymerase (Fermentas) (Gottsch et al., 2004), and Dyn sense and antisense probes were transcribed with T7 or T3 polymerase (Fermentas). Radiolabeled probes were synthesized in vitro by inclusion of the following ingredients in a volume of 20 μl: 250 μCi 33P-UTP (Perkin-Elmer Life Sciences); 1 μg of linearized DNA (or 1 μg of PCR product); 0.5 mm each ATP, CTP, GTP; and 40 U of polymerase. Residual DNA was digested with 4 U of DNase (Ambion), and the DNase reaction was terminated by addition of 2 μl of 0.5 m EDTA, pH 8.0. The riboprobes were separated from unincorporated nucleotides with NucAway Spin Columns (Ambion).
Slides with mouse hypothalamic sections from the three experimental groups were processed before hybridization, as previously reported (Cunningham et al., 2002; Gottsch et al., 2004). Briefly, sections were fixed in 4% paraformaldehyde, pretreated with acetic anhydride, rinsed in 2× sodium citrate, sodium chloride (SSC), delipidated in chloroform, dehydrated in graded ethanols, and then allowed to air dry before the hybridization procedure. Radiolabeled antisense Kiss1 riboprobes or antisense and sense Dyn riboprobes were denatured, dissolved in hybridization solution at a concentration of 0.1 pmol/ml along with tRNA (1.9 mg/ml) (Roche Biochemicals) in 0.1 m Tris/0.01 m EDTA, pH 8.0, to produce the probe mix. The probe mix was heat denatured in boiling water for 3 min, then returned to ice for 5 min. The denatured probe mix was added to prewarmed hybridization buffer (60% deionized formamide, 5× hybridization salts, 0.1× Denhardt's buffer, 0.2% SDS), at a ratio of 1:4, and added to each slide (100 μl/slide). The sections were then coverslipped and placed in humidity chambers at 55°C for 16 h. After hybridization, coverslips were removed, and the slides were washed in 4× SSC at room temperature. Slides were then placed into ribonuclease (RNase) [10 mg/ml RNase (Roche Biochemicals) in 0.15 m sodium chloride, 10 mm Tris, 1 mm EDTA, pH 8.0] for 30 min at 37°C, then in RNase buffer, without RNase, at 37°C for another 30 min. After a 30 min wash in 2× SSC at room temperature, slides were washed twice in 0.1× SSC at 62°C, then dehydrated in graded ethanols and air dried. The slides were then dipped in Kodak NTB emulsion (VWR), air dried, and stored at 4°C for 8–9 d. Slides were then developed, dehydrated in graded ethanols, cleared in Citrasol (VWR), and coverslips were applied with Permaslip (Sigma).
Kiss1 and Dyn mRNA quantification and analysis.
All Kiss1 or Dyn mRNA-containing sections were analyzed unilaterally. Slides from all of the animals were assigned a random three-letter code, alphabetized, and read under dark-field illumination with custom-designed software designed to count the total number of cells and the number of silver grains (corresponding to radiolabeled Kiss1 or Dyn mRNA) over each cell (Chowen et al., 1990). The system consists of a Scion VG5 video acquisition board (Perceptics) attached to a Power Macintosh G5 computer running custom grain-counting software. Cells were counted as Kiss1 or Dyn mRNA positive when the number of silver grains in a cluster exceeded that of background. Thus, cell counts represent the number of cells that achieved a detectability threshold, and the grains per Kiss1 or Dyn cell reflects a semiquantitative index of mRNA content in those cells that achieve the detectability threshold (Cunningham et al., 2002; Gottsch et al., 2004; Smith et al., 2005). Cells were considered Kiss1 or Dyn positive when the number of silver grains in a cluster exceeded that of background by threefold.
Statistical analysis.
All data are expressed as the mean ± SEM for each group. ANOVA followed with Tukey/Kramer post hoc tests were used to assess variation among experimental groups in each experiment. Significance level was set at p < 0.05. All analyses were performed with Statview 5.0.1 for Macintosh (SAS Institute).
Results
Serum hormone levels
Serum levels of LH in ERα+/+ female OVX + vehicle-treated mice were elevated relative to those of OVX + E2-treated ERα+/+ mice. Treatment of ERα+/+ OVX female mice adults with E2-filled implants significantly reduced LH concentrations (ERα+/+ OVX + vehicle = 2.88 ± 0.55 ng/ml vs ERα+/+ OVX + E2 = 0.23 ± 0.03 ng/ml; p < 0.01), indicating that the implants were functional and able to provide steroidal negative feedback. The LH values of ERα−/− mice were elevated regardless of treatment (ERα−/− OVX + vehicle mice = 3.60 ± 1.05 ng/ml vs ERα−/− OVX + E2 = 1.63 ± 0.27 ng/ml; p = 0.08), and the LH values of OVX + E2-treated ERαAA/− mice were decreased compared with OVX + vehicle-treated ERαAA/− mice (ERαAA/− OVX + vehicle = 5.90 ± 1.13 ng/ml vs ERαAA/− OVX + E2 = 0.46 ng/ml ± 0.27 ng/ml; p < 0.01).
E2 regulation of Kiss1 mRNA in the AVPV
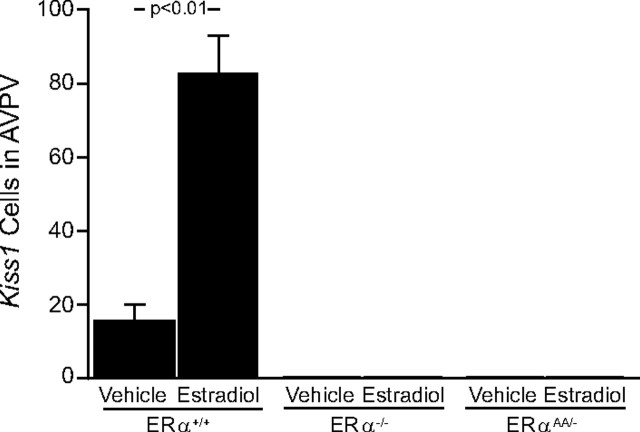
In the AVPV, the results observed in ERα+/+ mice were the opposite of those found in the Arc of ERα+/+ mice. In ERα+/+ that were OVX + vehicle replaced, the number of identifiable Kiss1 mRNA-containing cells was decreased compared with those ERα+/+ mice that were OVX + E2 replaced (ERα+/+ OVX + vehicle mice = 17.5 cells vs ERα+/+ OVX + E2 mice = 83 ± 11 cells; p < 0.01) (Figs. 1, 2), as was the cellular content of Kiss1 mRNA in OVX + vehicle-treated ERα+/+ mice [ERα+/+ OVX + vehicle mice = 39.5 ± 3.5 grains per cell (gpc) vs ERα+/+ OVX + E2 mice = 114.8 ± 27.3 gpc; p < 0.01; data not shown]. In the AVPV of ERα−/− mice, the number of Kiss1 mRNA-expressing cells was not changed with OVX + vehicle treatment or OVX + E2 treatment, as there were essentially no identifiable cells in either group of ERα−/− mice (Fig. 1). The same was true for ERαAA/− mice, in which there were no identifiable Kiss1 cells in the AVPV regardless of treatment (Figs. 1, 2).
Figure 1.
The effects of estradiol on the relative number of cells expressing Kiss1 mRNA in the AVPV of female ERα+/+, ERα−/−, or ERαAA/− mice. There were 5–6 animals per group; bars represent means + SEM.
Figure 2.
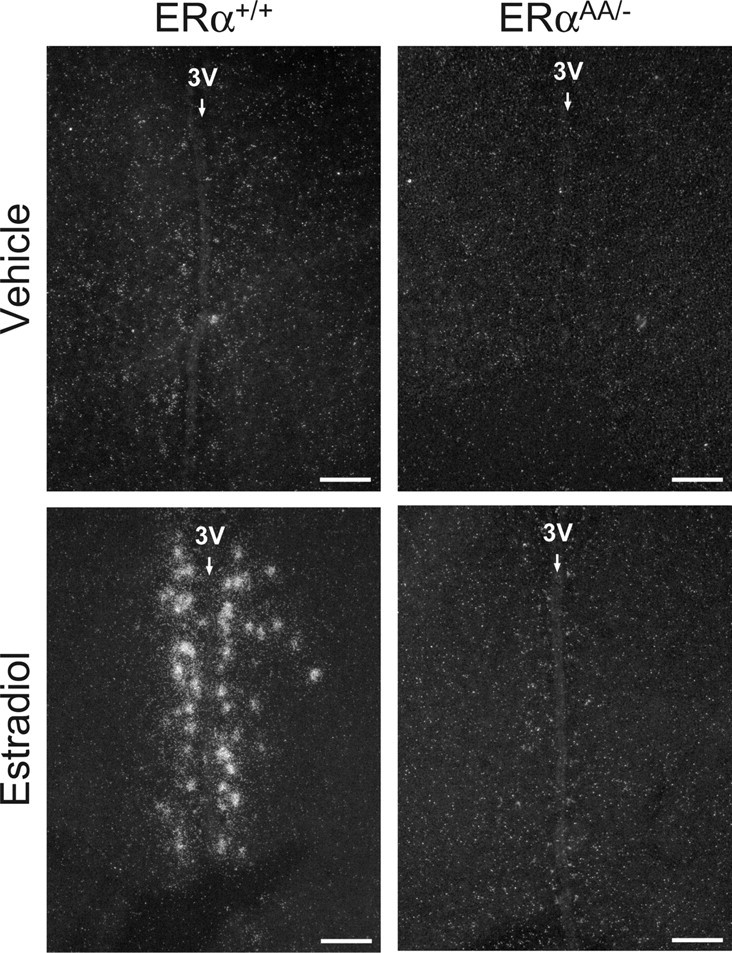
Photomicrographs depicting the effects of estradiol on Kiss1 mRNA expression in the AVPV of female ERα+/+ and ERαAA/− mice. Silver grains (white dots) represent Kiss1 mRNA. Photomicrographs depicting Kiss1 mRNA expression in ERα−/− mice were omitted, as they were the same as ERαAA/− mice. 3V, Third ventricle. Scale bars, 100 μm.
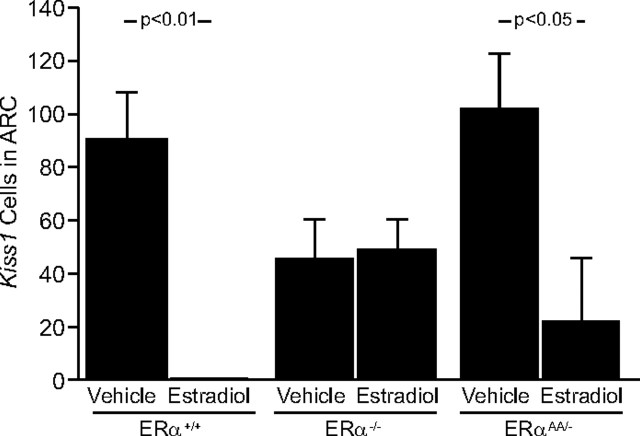
E2 regulation of Kiss1 mRNA expression in the Arc
In the Arc of vehicle-treated ERα+/+ mice, OVX increased the number of cells expressing Kiss1 mRNA (OVX + vehicle = 91 ± 16 cells vs no identifiable Kiss1 cells in E2-treated ERα+/+ mice; p < 0.01) (Figs. 3, 4) and increased the cellular content of Kiss1 mRNA as reflected by gpc (140 ± 7 gpc vs no gpc in E2-replaced ERα+/+ mice; p < 0.01; data not shown). In ERα−/− mice, there was no effect of E2 upon the number of Kiss1 mRNA-expressing cells in the Arc (p > 0.05; OVX + vehicle ERα−/− mice = 43.4 ± 17.4 cells vs OVX + E2 ERα−/− mice = 47 ± 15 cells) (Figs. 3, 4). Nor was there any effect of treatment upon Kiss1 mRNA content in these cells (ERα−/− OVX + vehicle mice = 157 ± 33 gpc vs ERα−/− OVX + E2 mice = 99 ± 14 gpc; p > 0.05; data not shown). In vehicle-treated ERαAA/− mice, the number of Kiss1 mRNA-expressing cells was upregulated in the Arc compared with E2-treated ERαAA/− mice (ERαAA/− OVX + vehicle mice = 101.3 ± 27.5 cells vs ERαAA/− OVX + E2 mice = 20 ± 20 cells; p < 0.05) (Figs. 3, 4), and the cellular content of Kiss1 mRNA was also increased (ERαAA/− OVX + vehicle mice = 130.5 ± 17.7 gpc vs ERαAA/− OVX + E2 mice = 18.5 ± 18.5 gpc; p < 0.01; data not shown).
Figure 3.
The effects of estradiol on the relative number of cells expressing Kiss1 mRNA in the Arc of female ERα+/+, ERα−/−, or ERαAA/− mice. There were 5–6 animals per group; bars represent means + SEM.
Figure 4.
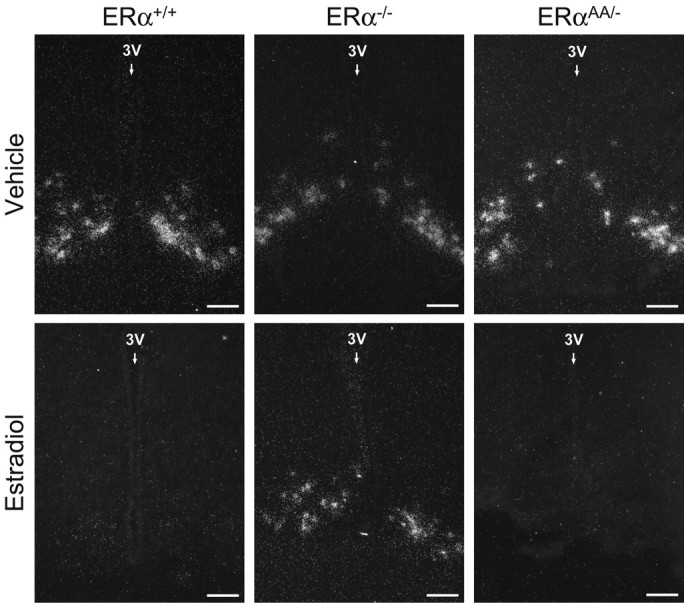
Photomicrographs depicting the effects of estradiol on Kiss1 mRNA expression in the Arc of female ERα+/+, ERα−/−, and ERαAA/− mice. Silver grains (white dots) represent Kiss1 mRNA. 3V, Third ventricle. Scale bars, 100 μm.
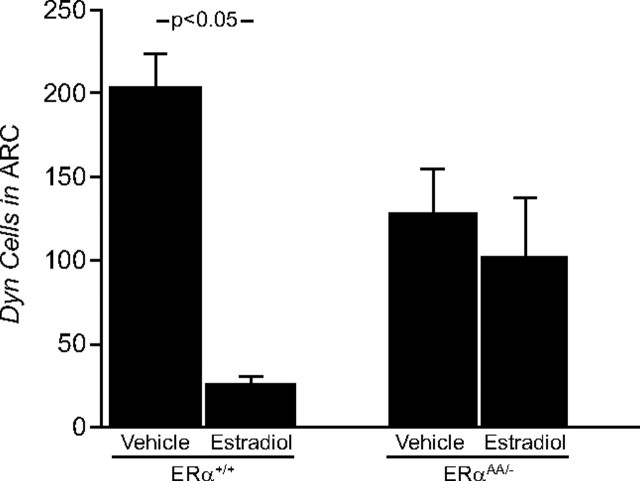
E2 regulation of Dynorphin mRNA expression in the Arc
In the Arc of ERα+/+ mice, OVX + vehicle treatment increased the number of cells expressing Dyn mRNA, as well as the cellular content of Dyn mRNA, compared with ERα+/+ mice that were OVX + E2 treated (ERα+/+ OVX + vehicle mice = 197.6 ± 24.5 cells vs ERα+/+ OVX + E2 mice = 25.3 ± 6.15 cells; p < 0.05) (Figs. 5, 6) (ERα+/+ OVX + vehicle mice = 57.9 ± 8.1 gpc vs ERα+/+ OVX + E2 mice = 23.6 ± 3.9 gpc; p < 0.05; data not shown). In vehicle-treated ERαAA/− mice, the number of Dyn mRNA-expressing cells was not changed in the Arc compared with E2-treated ERαAA/− mice (ERαAA/− vehicle-treated mice = 128.2 ± 28 cells vs ERαAA/− E2-treated mice = 103.2 ± 35.6 cells; p > 0.05) (Figs. 5, 6), nor was the cellular content of Dyn mRNA (ERαAA/− vehicle-treated mice = 46.5 ± 8.6 gpc vs ERαAA/− E2-treated mice = 35.7 ± 5.2 cells; p > 0.05; data not shown). In OVX vehicle-treated and OVX E2-treated ERα−/− female mice, there was no change in Dyn expression (Navarro, Clifton, Steiner, unpublished observations). There were no identifiable Dyn cells on slides that received Dyn sense riboprobe (data not shown).
Figure 5.
The effects of estradiol on the relative number of cells expressing Dyn mRNA in the Arc of female ERα+/+ and ERαAA/− mice. There were 5–6 animals per group; bars represent means + SEM.
Figure 6.
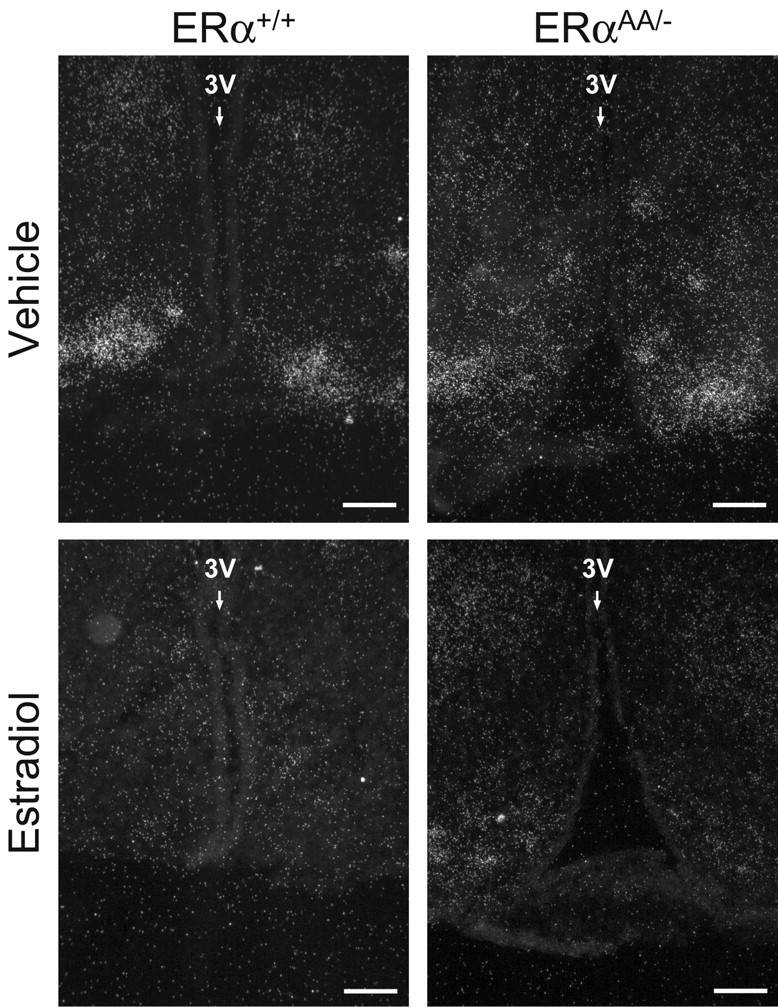
Photomicrographs depicting the effects of estradiol on Dyn mRNA expression in the Arc of female ERα+/+ and ERαAA/− mice. Silver grains (white dots) represent Dyn mRNA. 3V, Third ventricle. Scale bar, 50 μm.
Discussion
The positive feedback effects of E2 on LH release in the rodent have been extensively investigated over the past 60 years. It is now known that E2 acts via ERα to induce the preovulatory GnRH/LH surge (Wintermantel et al., 2006), and that this process involves classical ERE-dependent signaling (Glidewell-Kenney et al., 2007). Since Kiss1 mRNA expression is induced by E2 in the AVPV—an area thought to be responsible for the generation of the preovulatory LH surge in rodents—we and others have argued that kisspeptin neurons in the AVPV participate in the E2-induced preovulatory surge of GnRH/LH, i.e., reflecting the positive feedback effects of E2 (Smith et al., 2005; Clarkson et al., 2008). If this were true, we would expect that E2 would act through the same pathway to regulate Kiss1 in the AVPV as it does to generate the GnRH/LH surge. We previously reported that E2 acts via ERα in the AVPV to regulate Kiss1 in mice and have confirmed that observation here (Smith et al., 2005). Furthermore, our current observations suggest that E2 acts via a classical ERE-dependent pathway to regulate Kiss1 in the AVPV, the same mechanism by which E2 has been implicated to generate LH surges in mice (Glidewell-Kenney et al., 2007). Thus, our current findings are consistent with the hypothesis that kisspeptin neurons in the AVPV mediate the positive feedback effects of E2 on GnRH/LH secretion.
Our observation that E2 inhibits Kiss1 in the Arc of mice with a mutated ERα that cannot bind to ERE sites (ERαAA/− mice) suggests that regulation of the Kiss1 gene by E2 in the Arc involves an ERE-independent signaling pathway. In a study of ERαAA/− mice, Glidewell-Kenney et al. (2007) previously demonstrated that negative feedback regulation of LH by E2 is mediated by an ERE-independent mechanism. The notion that both GnRH/LH secretion and Kiss1 gene expression in the Arc may be regulated by similar (or identical) ERα signaling pathways is consistent with the proposition that these Kiss1 neurons participate in the negative feedback regulation of gonadotropin secretion. Although it would appear that the regulation of Kiss1 by E2 in the Arc of ERαAA/− mice is remarkably similar to that found in ERα+/+, we cannot rule out some minor (but consequential) effects of classical ERα signaling, ERβ signaling, or other nonclassical ER signaling pathways.
E2 can act through ERα to regulate gene expression in an ERE-independent manner in several ways. E2 bound to ERα may also interact with AP-1, Sp1, or NF-κB, which in turn can associate with target DNA elements to induce transcriptional activation (Gaub et al., 1990; Ray et al., 1994; Webb et al., 1995; Kushner et al., 2000; Safe, 2001; Jakacka et al., 2002; Khan et al., 2003; Safe and Kim, 2004; Fleming et al., 2006). The Kiss1 promoter contains both AP-1 and Sp1 sites, and in vitro studies examining the Kiss1 promoter have revealed that Sp1 elements within the first 200 bp of the Kiss1 promoter can regulate Kiss1 gene expression in certain cell lines (Li et al., 2007; Mitchell et al., 2007). Whether this is the case for the in vivo regulation of Kiss1 by E2 in the Arc of the female mouse remains to be determined. However, in this case we have shown that nonclassical ERα signaling pathways are sufficient to regulate Kiss1 gene expression in the Arc in the absence of ERE-dependent ERα signaling pathways. The molecular elements that signal through this nonclassical pathway to regulate Kiss1 expression in the Arc have yet to be identified.
Dyn is coexpressed with Kiss1 in the Arc of mice (Navarro, Clilfton, Steiner, unpublished observations), as has been previously shown in the sheep by Goodman et al. (2007). We were surprised to find that E2 regulates Dyn mRNA through ERE-dependent ERα signaling, whereas in the same cells, Kiss1 is regulated in an ERE-independent manner. Whether this regulation of Dyn and Kiss1 expression by two different ER signaling pathways occurs in all species is unknown. However, the observations reported here imply that ERα can regulate gene expression within individual neurons through multiple pathways in the same cell population. Although genes within the same neuron may be regulated in a parallel manner by E2 acting through the same ER subtype (i.e., either induced or suppressed), different molecular pathways may be involved in the regulation of each gene. Furthermore, since E2 regulates the Dyn gene in an ERE-dependent manner, its regulation does not appear to play an important role in the negative feedback control of LH secretion. This is because E2 effectively suppresses LH secretion in animals with a mutated ERα allele that cannot bind ERE sites, yet it does not change Dyn gene expression (cf. Glidewell-Kenney et al., 2008). Thus, despite the compelling case for kisspeptin in the Arc mediating the negative feedback effects of E2 on GnRH/LH secretion, the function of dynorphin in these same Kiss1 neurons remains a mystery.
There is some concern that the ERαAA/− mouse model might mask a truly ERE-independent signaling mechanism. For example, if the development of Kiss1 neurons in the AVPV required an intact ERE-dependent ERα signaling pathway, there would be no Kiss1 neurons in the AVPV that respond to E2 in ERαAA/− mice. If this were the case, even if an ERE-independent pathway normally regulated the expression of Kiss1, E2 may be incapable of inducing Kiss1 in such an animal. Our observation that the AVPV of ERα−/− and ERαAA/− is virtually devoid of Kiss1 neurons is consistent with this alternative hypothesis. Indeed, this is not what one would expect, since the congenital lack of ERα signaling should in theory yield a feminized AVPV (Bodo et al., 2006; McDevitt et al., 2008). Thus, it is plausible that the presence of an ERE-dependent ERα signaling pathway is a prerequisite for the appropriate development of Kiss1 neurons in the AVPV of females. Another problem with interpreting the finding in the ERαAA/− mouse is that it should not be assumed that only a single pathway is involved. For example, if ERE-dependent and -independent mechanisms were required to produce an effect, the ERαAA/− results would be interpreted to indicate that only an ERE-dependent mechanism was involved. This would not be a major difficulty if both pathways worked in parallel to produce an effect, which would result in a blockade of the effect in the ERα−/− mice and an attenuated response in the ERαAA/− mice. A recent report suggests that this may be the case with respect to negative feedback regulation of LH, where E2 had no effect on LH in ERα−/− mice, and was ∼70% as effective in ERαAA/− mice compared with ERα+/+ (Glidewell-Kenney et al., 2007). We observed a similar trend in the regulation of Kiss1 expression in the Arc, although the differences in the effect of E2 on levels Kiss1 mRNA between ERα+/+ and ERαAA/− mice were not statistically significant. Despite these limitations and concerns, the ERαAA/− mouse can be a useful tool to help unravel the pathways involved in ERα-mediated signaling.
In summary, we have used the ERαAA/− mouse model to demonstrate that the inhibition of Kiss1 gene expression by E2 in the Arc appears to be mediated by an ERE-independent ERα signaling mechanism, whereas the inhibition of Dyn expression in the Arc and the induction of Kiss1 in the AVPV is likely to occur through ERE-dependent ERα signaling pathways. Our results investigating the regulation of Kiss1 by ERα parallel an earlier report demonstrating that negative feedback regulation of LH is mediated by ERE-independent mechanisms, whereas ERE-dependent ERα signaling mechanisms mediate the positive feedback regulation of LH and ovulation. These observations provide additional evidence that Kiss1 neurons in the Arc participate in negative feedback regulation of GnRH/LH secretion by E2 and that Kiss1 neurons in the AVPV are involved in the positive feedback action of E2 that leads to the generation of the preovulatory GnRH/LH surge.
Footnotes
This research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)–National Institutes of Health (NIH) through cooperative agreements U54 HD12629 (to the University of Washington Center for Research in Reproduction and Contraception), NICHD–NIH Grant R01 HD27142, NICHD–NIH Grant P01 HD21921, and Office of Research on Women's Health–NICHD–NIH Specialized Center of Research Grant P50HD44405. We are grateful for the technical assistance provided by Janessa Lawhorn and Maile Parker, at the University of Washington in Seattle, WA, and Brigitte Mann, at Northwestern University in Chicago, IL.
References
- Bodo C, Kudwa AE, Rissman EF. Both estrogen receptor-alpha and -beta are required for sexual differentiation of the anteroventral periventricular area in mice. Endocrinology. 2006;147:415–420. doi: 10.1210/en.2005-0834. [DOI] [PubMed] [Google Scholar]
- Chowen JA, Argente J, Vician L, Clifton DK, Steiner RA. Pro-opiomelanocortin messenger RNA in hypothalamic neurons is increased by testosterone through aromatization to estradiol. Neuroendocrinology. 1990;52:581–588. doi: 10.1159/000125647. [DOI] [PubMed] [Google Scholar]
- Clarkson J, d'Anglemont de Tassigny X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. J Neurosci. 2008;28:8691–8697. doi: 10.1523/JNEUROSCI.1775-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse JF, Korach KS. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- Cunningham MJ, Scarlett JM, Steiner RA. Cloning and distribution of galanin-like peptide mRNA in the hypothalamus and pituitary of the macaque. Endocrinology. 2002;143:755–763. doi: 10.1210/endo.143.3.8661. [DOI] [PubMed] [Google Scholar]
- Dungan HM, Gottsch ML, Zeng H, Gragerov A, Bergmann JE, Vassilatis DK, Clifton DK, Steiner RA. The role of kisspeptin-GPR54 signaling in the tonic regulation and surge release of gonadotropin-releasing hormone/luteinizing hormone. J Neurosci. 2007;27:12088–12095. doi: 10.1523/JNEUROSCI.2748-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JG, Spencer TE, Safe SH, Bazer FW. Estrogen regulates transcription of the ovine oxytocin receptor gene through GC-rich SP1 promoter elements. Endocrinology. 2006;147:899–911. doi: 10.1210/en.2005-1120. [DOI] [PubMed] [Google Scholar]
- Gaub MP, Bellard M, Scheuer I, Chambon P, Sassone-Corsi P. Activation of the ovalbumin gene by the estrogen receptor involves the fos-jun complex. Cell. 1990;63:1267–1276. doi: 10.1016/0092-8674(90)90422-b. [DOI] [PubMed] [Google Scholar]
- Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- Glidewell-Kenney C, Hurley LA, Pfaff L, Weiss J, Levine JE, Jameson JL. Nonclassical estrogen receptor alpha signaling mediates negative feedback in the female mouse reproductive axis. Proc Natl Acad Sci U S A. 2007;104:8173–8177. doi: 10.1073/pnas.0611514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glidewell-Kenney C, Weiss J, Hurley LA, Levine JE, Jameson JL. Estrogen receptor alpha signaling pathways differentially regulate gonadotropin subunit gene expression and serum follicle-stimulating hormone in the female mouse. Endocrinology. 2008;149:4168–4176. doi: 10.1210/en.2007-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RL. Neural systems mediating the negative feedback actions of estradiol and progesterone in the ewe. Acta Neurobiol Exp (Wars) 1996;56:727–741. doi: 10.55782/ane-1996-1178. [DOI] [PubMed] [Google Scholar]
- Goodman RL, Lehman MN, Smith JT, Coolen LM, de Oliveira CV, Jafarzadehshirazi MR, Pereira A, Iqbal J, Caraty A, Ciofi P, Clarke IJ. Kisspeptin neurons in the arcuate nucleus of the ewe express both dynorphin A and neurokinin B. Endocrinology. 2007;148:5752–5760. doi: 10.1210/en.2007-0961. [DOI] [PubMed] [Google Scholar]
- Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- Gu GB, Simerly RB. Projections of the sexually dimorphic anteroventral periventricular nucleus in the female rat. J Comp Neurol. 1997;384:142–164. [PubMed] [Google Scholar]
- Herbison AE, Theodosis DT. Localization of oestrogen receptors in preoptic neurons containing neurotensin but not tyrosine hydroxylase, cholecystokinin or luteinizing hormone-releasing hormone in the male and female rat. Neuroscience. 1992;50:283–298. doi: 10.1016/0306-4522(92)90423-y. [DOI] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Martinson F, Ishikawa T, Lee EJ, Jameson JL. An estrogen receptor (ER)alpha deoxyribonucleic acid-binding domain knock-in mutation provides evidence for nonclassical ER pathway signaling in vivo. Mol Endocrinol. 2002;16:2188–2201. doi: 10.1210/me.2001-0174. [DOI] [PubMed] [Google Scholar]
- Khan S, Abdelrahim M, Samudio I, Safe S. Estrogen receptor/Sp1 complexes are required for induction of cad gene expression by 17beta-estradiol in breast cancer cells. Endocrinology. 2003;144:2325–2335. doi: 10.1210/en.2002-0149. [DOI] [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Li D, Mitchell D, Luo J, Yi Z, Cho SG, Guo J, Li X, Ning G, Wu X, Liu M. Estrogen regulates KiSS1 gene expression through estrogen receptor alpha and SP protein complexes. Endocrinology. 2007;148:4821–4828. doi: 10.1210/en.2007-0154. [DOI] [PubMed] [Google Scholar]
- McDevitt MA, Glidewell-Kenney C, Jimenez MA, Ahearn PC, Weiss J, Jameson JL, Levine JE. New insights into the classical and non-classical actions of estrogen: evidence from estrogen receptor knock-out and knock-in mice. Mol Cell Endocrinol. 2008;290:24–30. doi: 10.1016/j.mce.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Miller GM, Silverman AJ, Rogers MC, Gibson MJ. Neuromodulation of transplanted gonadotropin-releasing hormone neurons in male and female hypogonadal mice with preoptic area brain grafts. Biol Reprod. 1995;52:572–583. doi: 10.1095/biolreprod52.3.572. [DOI] [PubMed] [Google Scholar]
- Mitchell DC, Stafford LJ, Li D, Bar-Eli M, Liu M. Transcriptional regulation of KiSS-1 gene expression in metastatic melanoma by specificity protein-1 and its coactivator DRIP-130. Oncogene. 2007;26:1739–1747. doi: 10.1038/sj.onc.1209963. [DOI] [PubMed] [Google Scholar]
- Nishihara M, Matsukawa T, Kimura F. Responses of arcuate neurons to some putative neurotransmitters in perfused rat hypothalamic slices: effects of in vivo and in vitro estrogen treatments. Jpn J Physiol. 1986;36:683–697. doi: 10.2170/jjphysiol.36.683. [DOI] [PubMed] [Google Scholar]
- O'Malley BW, Tsai MJ. Molecular pathways of steroid receptor action. Biol Reprod. 1992;46:163–167. doi: 10.1095/biolreprod46.2.163. [DOI] [PubMed] [Google Scholar]
- Polston EK, Simerly RB. Ontogeny of the projections from the anteroventral periventricular nucleus of the hypothalamus in the female rat. J Comp Neurol. 2006;495:122–132. doi: 10.1002/cne.20874. [DOI] [PubMed] [Google Scholar]
- Popa SM, Clifton DK, Steiner RA. The role of kisspeptins and GPR54 in the neuroendocrine regulation of reproduction. Annu Rev Physiol. 2008;70:213–238. doi: 10.1146/annurev.physiol.70.113006.100540. [DOI] [PubMed] [Google Scholar]
- Popolow HB, King JC, Gerall AA. Rostral medial preoptic area lesions' influence on female estrous processes and LHRH distribution. Physiol Behav. 1981;27:855–861. doi: 10.1016/0031-9384(81)90053-6. [DOI] [PubMed] [Google Scholar]
- Ray A, Prefontaine KE, Ray P. Down-modulation of interleukin-6 gene expression by 17 beta-estradiol in the absence of high affinity DNA binding by the estrogen receptor. J Biol Chem. 1994;269:12940–12946. [PubMed] [Google Scholar]
- Rissman EF, Wersinger SR, Taylor JA, Lubahn DB. Estrogen receptor function as revealed by knockout studies: neuroendocrine and behavioral aspects. Horm Behav. 1997;31:232–243. doi: 10.1006/hbeh.1997.1390. [DOI] [PubMed] [Google Scholar]
- Ronnekleiv OK, Kelly MJ. Plasma prolactin and luteinizing hormone profiles during the estrous cycle of the female rat: effects of surgically induced persistent estrus. Neuroendocrinology. 1988;47:133–141. doi: 10.1159/000124903. [DOI] [PubMed] [Google Scholar]
- Safe S. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- Safe S, Kim K. Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog Nucleic Acid Res Mol Biol. 2004;77:1–36. doi: 10.1016/S0079-6603(04)77001-4. [DOI] [PubMed] [Google Scholar]
- Shughrue PJ, Lubahn DB, Negro-Vilar A, Korach KS, Merchenthaler I. Responses in the brain of estrogen receptor alpha-disrupted mice. Proc Natl Acad Sci U S A. 1997;94:11008–11012. doi: 10.1073/pnas.94.20.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shughrue PJ, Askew GR, Dellovade TL, Merchenthaler I. Estrogen-binding sites and their functional capacity in estrogen receptor double knockout mouse brain. Endocrinology. 2002;143:1643–1650. doi: 10.1210/endo.143.5.8772. [DOI] [PubMed] [Google Scholar]
- Simerly RB, Chang C, Muramatsu M, Swanson LW. Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: an in situ hybridization study. J Comp Neurol. 1990;294:76–95. doi: 10.1002/cne.902940107. [DOI] [PubMed] [Google Scholar]
- Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- Stein B, Yang MX. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa E, Wiegand SJ, Bridson WE. A role for medial preoptic nucleus on afternoon of proestrus in female rats. Am J Physiol. 1980;238:E533–E539. doi: 10.1152/ajpendo.1980.238.6.E533. [DOI] [PubMed] [Google Scholar]
- Thind KK, Goldsmith PC. Infundibular gonadotropin-releasing hormone neurons are inhibited by direct opioid and autoregulatory synapses in juvenile monkeys. Neuroendocrinology. 1988;47:203–216. doi: 10.1159/000124914. [DOI] [PubMed] [Google Scholar]
- Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Webb P, Lopez GN, Uht RM, Kushner PJ. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- Wiegand SJ, Terasawa E, Bridson WE. Persistent estrus and blockade of progesterone-induced LH release follows lesions which do not damage the suprachiasmatic nucleus. Endocrinology. 1978;102:1645–1648. doi: 10.1210/endo-102-5-1645. [DOI] [PubMed] [Google Scholar]
- Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne HJ, Todman MG, Korach KS, Greiner E, Pérez CA, Schütz G, Herbison AE. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron. 2006;52:271–280. doi: 10.1016/j.neuron.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]