Abstract
All cellular systems evolve ways to combat predators and genomic parasites. In bacteria and archaea, numerous resistance mechanisms have developed against phage. Our understanding of this defensive repertoire has recently been expanded to include the CRISPR system of Clustered, Regularly Interspaced Short Palindromic Repeats. In this remarkable pathway, short sequence tags from invading genetic elements are actively incorporated into the host's CRISPR locus, to be transcribed and processed into a set of small RNAs that guide the destruction of foreign genetic material. Here, we review the inner workings of this adaptable and heritable immune system and draw comparisons to small RNA-guided defense mechanisms in eukaryotic cells.
Introduction
The battle between predator and prey is perhaps the second-oldest conflict on earth, and phage may represent one of the planet's oldest predators. For bacteria and archaea, phage are a formidable force, being responsible for 4-50% of their destruction (Breitbart and Rohwer, 2005; Rohwer and Thurber, 2009). The predatory challenge is substantial and dynamic; phage outnumber their prey by 10-fold, and benefit from significantly greater genome variability and faster rates of mutation (Hatfull, 2008; Hendrix, 2003).
This diverse and rapidly evolving challenge has prompted the development of multiple layers of resistance mechanisms in bacteria. As a first line of defense, bacteria can disrupt phage adsorption to the cell surface by eliminating or masking the corresponding receptors (Forde and Fitzgerald, 1999). Injection of phage DNA can also be blocked in some cases. Once within the bacterial cells, phage DNA is subject to the well-studied restriction/modification systems that degrade foreign DNA. These rely on differences in methylation status to accomplish self-non-self recognition and block the activity of sequence-specific nucleases toward endogenous DNA, while targeting the invaders (Bickle and Kruger, 1993). Finally, abortive infection systems interfere with various aspects of phage replication and packaging, while leading to death of the host (Chopin et al., 2005). The importance of evading these defenses for the phage is demonstrated by specific adaptations that they have evolved in response (Chibani-Chennoufi et al., 2004; Forde and Fitzgerald, 1999).
The past several years have brought an understanding of an additional bacterial and archaeal defense against exogenous nucleic acids. The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) system is a highly adaptive and heritable resistance mechanism that incorporates sequences derived from the foreign element into a small-RNA-based repertoire. These small RNAs program an enzymatic complex to recognize and destroy the invader. Conceptually, many aspects of the CRISPR system are similar to adaptive mechanisms of small RNA-based defense that protect animal germ cells from mobile genetic elements (Aravin et al., 2007). In this review, we describe the recent, substantial progress toward understanding the CRISPR system and draw parallels to mobile element defense mechanisms in animals. We would also like to point the reader to excellent existing reviews on this topic (Sorek et al., 2008; van der Oost et al., 2009).
Anatomy of a CRISPR locus
The CRISPR story began with the discovery of a peculiar short repeat in the E. coli genome by Ishino and coworkers in the 1980s (Ishino et al., 1987; Nakata et al., 1989). Subsequently, similar repeats were noted in a number of bacteria and archaea (Bult et al., 1996; Groenen et al., 1993; Hermans et al., 1991; Hoe et al., 1999; Kawarabayasi et al., 1999; Kawarabayasi et al., 1998; Klenk et al., 1997; Masepohl et al., 1996; Mojica et al., 1995; Nelson et al., 1999; Sensen et al., 1998; She et al., 1998; She et al., 2001; Smith et al., 1997). Mojica and Jansen and their colleagues unified these observations, coined the CRISPR acronym, and characterized the CRISPR locus (Jansen et al., 2002; Mojica et al., 2000). In prokaryotes, genes which impact similar biological processes often travel through evolution as physically linked units (Galperin and Koonin, 2000; Overbeek et al., 1999). Accordingly, the investigators also characterized the protein coding genes that were often adjacent to the repeat cluster (CRISPR-associated genes or cas genes). We now know that these genes form elemental components of the CRISPR defense pathway. Our understanding of CRISPR loci and cas genes was further refined and expanded as more genomic sequence information became available (Bolotin et al., 2005; Godde and Bickerton, 2006; Grissa et al., 2007; Haft et al., 2005; Kunin et al., 2007; Lillestol et al., 2006; Makarova et al., 2002; Makarova et al., 2006; Pourcel et al., 2005). A wealth of this information on CRISPRs and cas genes is now accessible in the form of online databases and tools (Grissa et al., 2007; Grissa et al., 2008; Oberle et al., 1991). Overall, CRISPR loci have been found in about 40% of bacterial and in most archaeal species with sequenced genomes (Godde and Bickerton, 2006; Kunin et al., 2007).
A CRISPR locus is defined as an array of short direct repeats interspersed with spacer sequences (Fig. 1A). Within a given locus, the repeats are practically identical in length and sequence. The spacers are also uniform in length but have highly variable sequence content. Among different species, repeats vary from 21 to 47bp, being 32bp on average (Godde and Bickerton, 2006; Grissa et al., 2007), and spacers are of a similar size, 20-72bp (Grissa et al., 2007; Mojica et al., 2000). Related species can have similar repeat sequences, but the overall bacterial and archaeal sequence diversity of both spacers and repeats is great (Kunin et al., 2007).
Figure 1. The structure of a CRISPR locus.
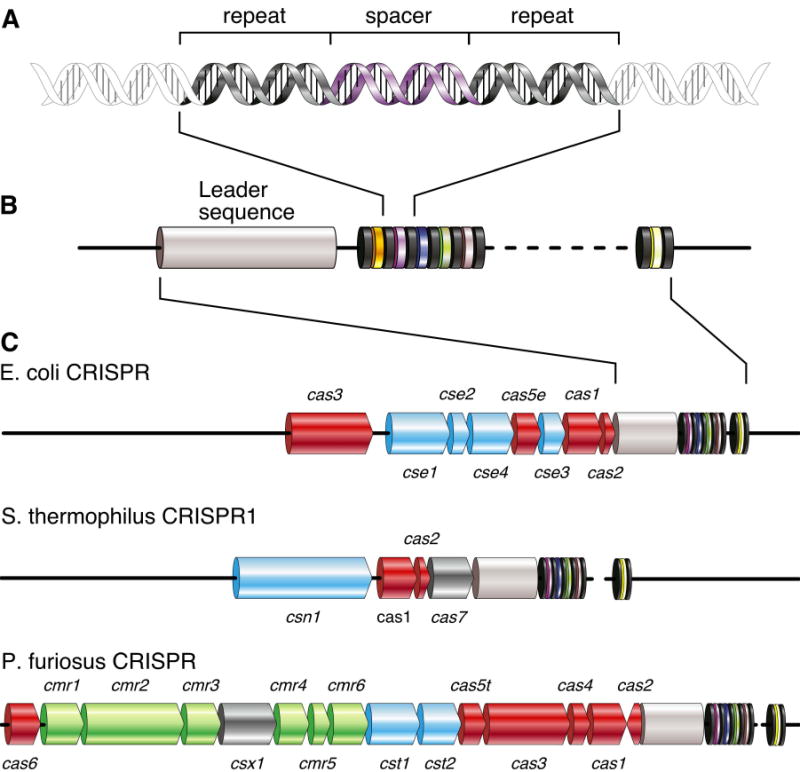
(A) Repeat sequences averaging 32bp are interleaved by variable spacers of approximately the same size. (B) The number of repeat-spacer units varies greatly. A conserved leader sequence (gray) of several hundred base pairs is located on one side of the cluster. (C) CRISPR-associated (cas) genes surround the CRISPR locus. Three examples of well-studied CRISPR loci are shown. Core cas genes are depicted in red, subtype-specific genes in blue, and the RAMP module in green. Unclassified genes are shown in dark grey. Gene names follow the nomenclature of Haft et al., except cas7, which was named by Barrangou et al.
Despite their sequence diversity, a majority of CRISPR repeats have the potential to form hairpin structures (i.e. they are partly palindromic). Conservation of G-U pairs in the stem implies that the structures can form in the transcriptional products of the locus (Kunin et al., 2007). This does not, however, appear to be an absolutely conserved property of all CRISPR repeats (Kunin et al., 2007; Makarova et al., 2006). The number of repeat-spacer units in a locus averages in the mid twenties (Godde and Bickerton, 2006), though this can range from as few as one to as many as several hundred (up to 374 in the current CRISPRdb (Grissa et al., 2007)). While a single CRISPR locus per genome is typical, finding several loci within a single species genome is not uncommon (Jansen et al., 2002). In these cases, similar repeat sequences are often shared among the loci, but there are interesting exceptions.
Sequence analysis of genomes containing multiple CRISPR loci uncovered an additional structural feature directly adjacent to the short repeats (Fig. 1B) (Bult et al., 1996; Jansen et al., 2002; Klenk et al., 1997; Smith et al., 1997). This region of conservation between CRISPR loci, termed the leader sequence, extends several hundred base pairs, lacks coding potential, and is always found on one side of the CRISPR in a fixed orientation. Much like the repeats themselves, leaders are up to 80% identical within a genome, but quite dissimilar among species.
CRISPR loci are surrounded by a cohort of conserved protein-coding genes, which appear in varying orientation and order (see examples in Fig. 1C). Initial homology comparisons by Jansen and colleagues delineated four core CRISPR-associated gene families, cas1-4 (Jansen et al., 2002). Most loci do not contain all four genes. Typically, cas1 and one or more of the others are present, suggesting some functional redundancy among these families. Two independent and concurrent studies expanded the core set to include cas5 and cas6 (Bolotin et al., 2005; Haft et al., 2005). Unfortunately, the nomenclature of these genes can create confusion; cas5 in Bolotin et al. is equivalent to csn1 in Haft et al. (see below), and cas6 in Bolotin et al. (NCBI COG 3512) may be a cas2 variant. Unless otherwise noted, we will follow the Haft nomenclature in this review.
In addition to the core set of cas genes that pervade the entire bacterial and archaeal phylogeny, some families of homologous CRISPR-associated genes are more narrowly conserved. Stereotypic combinations of these families together with the adjacent core genes delineate several cas system subtypes (Fig. 1C). Haft and colleagues defined eight subtypes, each named after a representative organism (for example, cse1 for cas subtype E. coli, Fig. 1C) (Haft et al., 2005). In general agreement, seven subtypes have been classified based on NCBI Clusters of Orthologous Groups (COGs) (Makarova et al., 2006). Members of a cas subtype often maintain the operon organization of core and subtype-specific genes within the CRISPR locus.
RAMPs (Repeat-Associated Mysterious Proteins) round out the cast of characters in the CRISPR pathway (Haft et al., 2005; Makarova et al., 2002; Makarova et al., 2006). This protein family is defined by their presence in CRISPR-containing genomes and loose sequence conservation, characterized by a C-terminal G-rich loop (Makarova et al., 2006). RAMP genes can be located either adjacent to or distant from the repeats themselves. A subset of RAMPs, together with a putative novel polymerase, is found in a well-conserved cluster, termed the “RAMP module” or “polymerase cassette” (Fig. 1C). These six genes (cmr1-6) are associated with several cas system subtypes (Haft et al., 2005). CRISPR-related proteins of known function or activity are summarized in Table 1.
Table 1.
Proteins with known genetic or biochemical association with the CRISPR pathway.
| Name (Haft et al) | Alternative name(s) | Function/activity | Model system |
|---|---|---|---|
| cas1 | Acquisition of new spacers / ss/dsRNA endonuclease | P. aeruginosa | |
| cas2 | ssRNA endonuclease | S. solfataricus and others | |
| cas3 | crRNA-guided degradation of invading NAs | E. coli | |
| cas6 | Endonucleolytic cleavage of pre-crRNA | P. furiosus | |
| csn1 | cas5 | Phage resistance using existing spacers | S. thermophilus |
| - | cas7 | Acquisition of new spacers? | S. thermophilus |
| - | SSO0454 | Specific binding of CRISPR repeat DNA | S. solfataricus |
| Cascade complex: | Endonucleolytic cleavage of pre-crRNA | E. coli | |
| cse1 | casA | ||
| cse2 | casB | ||
| cse3 | casE | Catalytic subunit | |
| cse4 | casC | ||
| cas5e | casD | ||
| RAMP module complex: | crRNA-guided endonucleolytic cleavage of RNA targets | P. furiosus | |
| cmr1 | |||
| cmr2 | |||
| cmr3 | |||
| cmr4 | |||
| cmr5 | Dispensable for activity | ||
| cmr6 | |||
The biological function of the CRISPR-cas system
It is truly a testament to post-genomic era research that the essence of the CRISPR-cas system was first discovered purely by computational sequence analysis and that the hypotheses generated through these efforts only later received remarkable experimental support. Searches for informative identities to the variable spacer regions yielded no matches for most. However, the crucial observation was that some were clearly derived from extrachromosomal DNA elements (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). Mojica and colleagues found that 88 out of 4500 spacers from a broad range of bacteria and archaea matched to known sequences, with most being similar to bacteriophage and plasmids (Mojica et al., 2005). Remarkably, species containing identified spacer elements were immune to the corresponding foreign invaders or had no prophage remnants as evidence of prior infections. In contrast, closely related CRISPR-negative species were susceptible. Similar analyses of spacers in multiple strains of S. thermophilus and Y. pestis also identified a subset with sequence identity to phage and plasmids (Bolotin et al., 2005; Pourcel et al., 2005). In S. thermophilus, there was a negative correlation between the number of such spacers and phage sensitivity (Bolotin et al., 2005). Thus, an intriguing picture was emerging, wherein the CRISPR loci might constitute a host defense against invading, foreign genetic elements, with the spacers providing specificity to the system.
In this model, to acquire resistance, new spacer information must be incorporated into the CRISPR locus. One source of that information might be the elements themselves, leading to the notion that the content of the locus can also serve as a record of infections from which a host had recovered. In accord with this hypothesis, repeat-spacer units show remarkable polymorphism in the number and identity of spacer sequences even among closely related strains (Fabre et al., 2004; Fang et al., 1998; Groenen et al., 1993; Hoe et al., 1999; Jansen et al., 2002; Kamerbeek et al., 1997). In fact, the rapidly evolving nature of CRISPR loci was quickly exploited for strain genotyping. Examination of such spacer content differences revealed a number of characteristics, which hinted to mechanisms of short-term CRISPR evolution (Fig. 2A). The distal end of the cluster contains “older” spacers, those that are shared among strains (Horvath et al., 2008; Lillestol et al., 2006; Pourcel et al., 2005). “Newer,” strain-specific spacers accumulate next to the leader sequence at the proximal end of the cluster. Clusters that lack a leader sequence do not appear to incorporate new spacers, suggesting that they are inactive remnants (Lillestol et al., 2006). This suggests a role for the leader sequence in cluster evolution, adaptation, or function, and points to an orchestrated mechanism of polarized cluster growth. In addition to increases in cluster content, spacers also appeared to be lost by internal deletions of one or more repeat units.
Figure 2. Diversity and acquisition of spacer content.
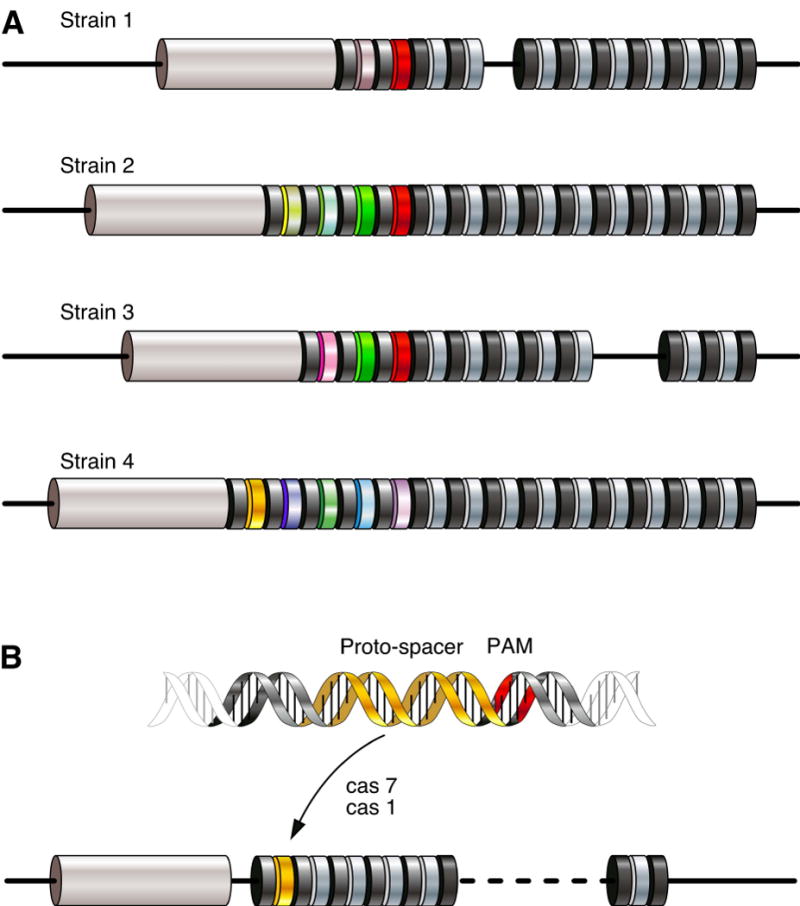
(A) A schematic representation of the CRISPR loci in related bacterial strains. Colored spacers represent sequences that differ among strains. All strains share the same ancestral spacers at the leader-distal end (gray spacers). Deletions after divergence have occurred in some strains. Spacer content becomes more strain-specific near the leader end. Strain ancestry can be traced by the presence of common spacers (green, red). (B) Acquisition of new spacers occurs next to the leader sequence. The spacer sequence is derived from the invading nucleic acid, selected based upon the presence of a proto-spacer adjacent motif.
Overall, computational analysis revealed a rapidly evolvable system, which enabled hosts to recognize invading genetic elements based upon sequence similarity to CRISPR spacers. The most parsimonious mechanism for acquiring resistance would be to steal sequence content from an invading pathogen and to incorporate it into the locus. A transition from computational to experimental approaches yielded the next insights into this remarkable system, specifically how information resident within CRISPRs could be used to combat infection.
Acquisition of CRISPR-directed resistance
Over the last few years, a stream of elegant experimental studies has validated many of the predictions arising from computational analysis and has provided new insights into how the CRISPR system operates. In 2007, Barrangou and colleagues provided the first glimpse of CRISPRs in action (Barrangou et al., 2007). In this study, bacteriophage-insensitive mutants (BIMs) were isolated following a challenge of S. thermophilus with two related phages. Remarkably, all of the resistant mutants had gained from one to four novel spacers with sequence identity to the invading genomes. In each case, as predicted by evolutionary analyses, the insertion(s) occurred proximal to the leader. Not only did the simple presence of a spacer correlate with protection against the phage, but the degree of resistance also correlated with the number of acquired sequences. Critically, resistance could be granted or revoked by experimentally engineered insertion or deletion of spacers corresponding to the challenging phage. The presence of the leader sequence next to the CRISPR locus proved essential, since introduction of unrelated sequence between the two abrogated the resistance. In all cases, CRISPR-mediated protection required csn1 (cas5 in the nomenclature of Bolotin et al.), a large protein containing a McrA/HNH nuclease domain (Gorbalenya and Koonin, 1993) and a RuvC-like nuclease domain (Haft et al., 2005; Makarova et al., 2006). Subsequent studies provided additional observations of the natural plasticity of the CRISPR locus. These demonstrated, for example, iterative addition of spacers after repeated phage challenges, and sporadic internal deletion of older spacer segments (Deveau et al., 2008). Furthermore, expression studies of phage infection in T. Thermophilus detected the upregulation of a number of cas genes, RAMP module proteins and CRISPR loci, indicating an infection-sensing mechanism within the host, with a subset of the genes induced through a cAMP receptor protein (Agari et al., 2009).
Thus, during the acquisition of a defensive repertoire, the CRISPR machinery appears to select sequences from the phage genome and incorporate these as novel spacers (Fig. 2B). The selection is not random. Instead, sequence motifs can be detected in proximity to those regions that ultimately become part of the CRISPR, termed proto-spacers. Analysis of spacers newly added to the CRISPR1 locus of independently selected S. thermophilus phage-insensitive mutants identified a short motif (NNAGAAW) directly downstream of the proto-spacer in the phage genome (Deveau et al., 2008). A similar motif was independently observed by Bolotin and colleagues (Bolotin et al., 2005). Interestingly, spacers from CRISPR3, a locus with some divergence from CRISPR1, showed a different downstream motif, NGGNG, near proto-spacers (Horvath et al., 2008). In S. mutans, one CRISPR locus displayed a preference for a short motif 3′ of the proto-spacer, while another CRISPR locus favored a 5′-adjacent motif (van der Ploeg, 2009). Similarly, three different CRISPR families in the archaeal Sulfolobales have distinct 5′ proto-spacer adjacent motifs (PAMs) (Lillestol et al., 2009). Like the repeat and leader sequences, distinct PAMs correspond to specific CRISPR/cas subtypes (see the section on CRISPR/cas co-evolution below). This suggests that spacer acquisition is driven by recognition of phage sequences by subtype-specific proteins in different species (Mojica et al., 2009). However, it also implies that when an individual species harbors multiple CRISPR loci from different subtypes, these represent distinct and compartmentalized resistance systems. In addition to its suggested role in spacer selection, the PAM also appears to be important at the effector stage of defense, since phage can evade resistance to a particular spacer by mutating this nearby motif (Deveau et al., 2008).
Presently, the precise mechanisms by which information is transferred from phage or plasmids into CRISPR loci are obscure. The original experimental study in S. thermophilus showed a requirement for cas7 to generate bacteriophage-insensitive mutants, but not to mount a response using existing spacers (Barrangou et al., 2007). Although cas7 (str0660) resides in the CRISPR1 locus, it has homology limited to the Streptococcus clade and lacks any classification by comparative sequence analyses (Haft et al., 2005; Makarova et al., 2006). Notably, its place in the mostly syntenic CRISPR3 locus of the same genome is occupied by csn2 (Horvath et al., 2008); however, there is no similarity between the two genes.
Another likely participant in resistance acquisition is the hallmark CRISPR gene, cas1. Cas1 is dispensable for the employment of existing CRISPR spacers in the effector phase of defense (see below) and has been proposed to act in making new spacers, either cleaving foreign DNA or facilitating integration of new sequences into the CRISPR locus. Sequence analysis predicted Cas1 to be a novel nuclease/integrase (Makarova et al., 2002). Recently, Wiedenheft and colleagues validated these predictions, demonstrating a ss/dsDNA endonuclease activity for P. aeruginosa cas1 (Wiedenheft et al., 2009). The DNase activity was not sequence- or methylation-specific and yielded final products of ∼80bp. Interestingly, dsDNA cleavage required Mn2+ or Mg2+, while ssDNA degradation was only supported by Mn2+. Moreover, the authors determined the cas1 crystal structure, which revealed a novel α-helical fold and a single metal ion site. Another study of cas1 from S. solfataricus revealed ss/dsDNA and ss/dsRNA binding and annealing activities, but could not detect any nuclease activity, possibly due to the somewhat unusual Mn2+ dependence (Han et al., 2009).
As a whole, the aforementioned studies strongly support a model in which incorporation of sequences from invading nucleic acids into CRISPR loci allows acquisition of resistance based upon sequence similarity. Thus far, only the barest hints have emerged to how this is accomplished at a mechanistic level. For example, the factors responsible for motif recognition and proto-spacer selection remain unknown, as do the proteins that ensure proper spacer length.
The mechanics of CRISPR-mediated defense
A major step forward in understanding the effector phase of the pathway came with the discovery of processed RNAs from the locus, termed crRNAs (CRISPR RNAs) or psiRNAs (prokaryotic silencing RNAs) (Fig. 3). Transcription of the CRISPR repeats initiates in or near the leader sequence and generates a long pre-crRNA precursor that can span the entire locus (Lillestol et al., 2006; Lillestol et al., 2009). The pre-crRNA is then endonucleolytically processed into fragments corresponding to the interval between repeats, producing mature products and a laddering pattern of intermediates (Brouns et al., 2008; Carte et al., 2008; Tang et al., 2002; Tang et al., 2005).
Figure 3. Processing of CRISPR content into crRNAs.
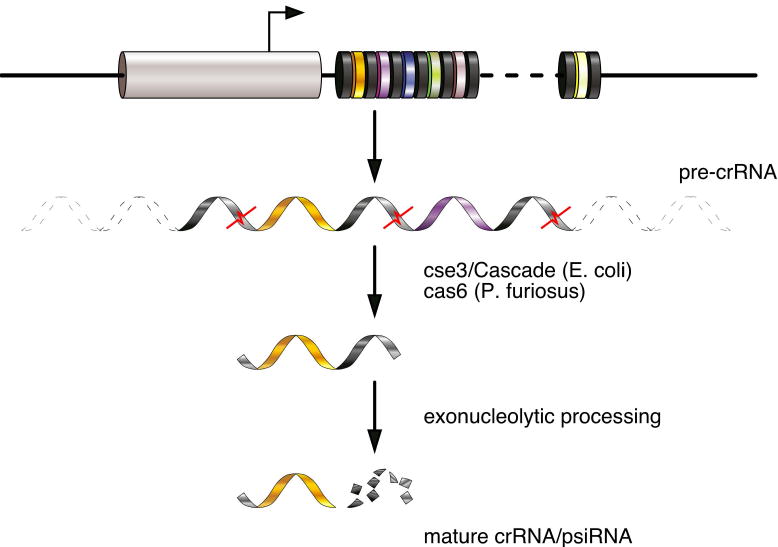
The locus is transcribed from the leader sequence and the RNA is cleaved within the repeat by cse3 or cas6. An additional processing step yields the mature crRNA/psiRNA consisting of an 8nt repeat tag and the spacer sequence.
Irregular patterns of transcripts have also been detected from the opposite strand in S. acidocaldarius (Lillestol et al., 2006; Lillestol et al., 2009). However, no evidence of antisense products was seen in S. epidermidis (Marraffini and Sontheimer, 2008), P. furiosus (Hale et al., 2008), or E. coli (Brouns et al., 2008). Further studies will be needed to resolve this discrepancy and to determine the relevance of these products to phage defense.
In E. coli, a complex termed Cascade produces 57nt units from the multimeric precursor transcript by cleavage within the repeat sequence (Brouns et al., 2008). Cascade is comprised of cse1, cse2, cse4, cas5e, and cse3 (also known as CasA-CasE). Within the complex, Cse3/casE (cas subtype E. coli 3) is necessary and sufficient to define the 5′ end of the product. At least two nucleotides are removed from the 3′ end of cse3 products by unknown mechanisms. Remarkably, the orthologous sequence-specific cleavage activity in P. furiosus is carried out by a different protein, cas6 (Carte et al., 2008). This protein has no homolog within the E. coli cas operon subtype that includes cse3 (Haft et al., 2005). The product of cse3 or cas6 is an RNA consisting of an 8nt repeat sequence “tag” followed by the spacer sequence, followed by the next partial repeat (Fig. 3).
In P. furiosus, an additional processing step was characterized that produces two discrete species of mature psiRNA, 38-45nt and 43-46nt, depending on the spacer length (Hale et al., 2008). This final step is presumed to be exonucleolytic. The resulting RNAs maintain the 5′ repeat tag, but lose the downstream repeat-derived sequence (Fig. 3) (Carte et al., 2008; Hale et al., 2008). In E. coli, potentially similar, shorter species can also be seen on northern blots in addition to the prominent 57mers (see Fig. 2, (Brouns et al., 2008)), but these have not been discussed in the literature. In S. acidocaldarius, CRISPR-derived small RNAs appear as products from 35 to 52 nt, presumably generated by endonucleolytic cleavage of long precursors (Lillestol et al., 2006). Thus, at present, the maturation to a 35-46nt RNA appears to be a conserved processing feature. An examination of the ribonucleoprotein complexes (RNPs) that are assembled on the RNAs revealed that the precursor and mature crRNAs are found in distinct RNPs, providing the first details of the processing/assembly pathway (Hale et al., 2008).
The structures of T. thermophilus cse3 and P. furiosus cas6 explain their common endonucleolytic function. These proteins display similar architectures, despite their lack of sequence homology (Carte et al., 2008; Ebihara et al., 2006; van der Oost et al., 2009). Both enzymes are composed of a duplicated ferredoxin fold, a common domain topology that also underlies the well-known RNA-recognition motif (RRM) domain. However, the conserved sequence signatures of RRMs are absent in cse3 and cas6. The two proteins contain a spatially conserved active site with an essential histidine residue and a G-rich loop (van der Oost et al., 2009). The crystal structure of T. thermophilus cse2/casB, another component of the Cascade complex, reveals a novel α-helical fold with a conserved basic patch that may be involved in binding RNA (Agari et al., 2008).
Accumulating evidence supports a model in which processed crRNAs serve as sequence-specific guides during the effector stage of resistance against invading elements. This was demonstrated by reconstitution of a functioning CRISPR system in E. coli BL21(DE3), which lacks endogenous cas genes (Brouns et al., 2008). These cells were engineered to express the Cascade complex, as well as Cas3 and a modified CRISPR locus in which spacer sequences targeting phage lambda had been incorporated. This was sufficient to create de novo resistance to the phage and allowed an exploration of the properties of cas proteins that were important for mounting an effective response. The catalytic activity of the cse3 nuclease within Cascade proved essential, indicating a crucial role for crRNAs in the overall defense pathway. Cas3 is not required for the generation of crRNAs, as described above, but was necessary for phage resistance in this system. This fact, along with a consideration of the domain structure of cas3, has led to the proposal that it might catalyze crRNA-guided destruction of foreign nucleic acids. Cas3 has an HD nuclease domain fused to a DExD/H helicase module (Makarova et al., 2002). The two domains also exist as separate proteins in the CRISPR loci of some species, indicating some flexibility in this arrangement (Makarova et al., 2002). Interestingly, one such stand-alone HD domain in S. solfataricus was demonstrated to possess nucleolytic activity, being able to use either dsDNA or dsRNA as a substrate (Han and Krauss, 2009). Like Cas1, the only remaining gene in the E. coli CRISPR locus, cas2, was not required for the effector phase, implicating this gene in some other aspect of the response.
Obvious analogies to eukaryotic RNAi-related pathways provoked an initial model in which a crRNA-guided complex would target mRNAs derived from the invader (Makarova et al., 2006). However, multiple lines of evidence point to the direct recognition of foreign DNA by the core CRISPR machinery. To date, sequence analyses have only detected spacers from phage with DNA genomes (Mojica et al., 2009; Wiedenheft et al., 2009). However, any conclusions based upon this observation must be tempered by the relative scarcity of RNA phage sequences. Detailed analyses in S. thermophilus (Bolotin et al., 2005), and more broadly in bacteria and archaea (Makarova et al., 2006; Shah et al., 2009), show that spacers encode crRNAs corresponding to both the coding and template strands of the phage, without a preference for any particular region. Similar conclusions can be reached by examination of spacers arising in experimentally induced phage-resistant mutants of S. thermophilus (Barrangou et al., 2007). Here, some bias toward the coding strand was observed, but this may be explained by the higher occurrence of the proto-spacer adjacent motif on that strand (Deveau et al., 2008). Direct support for DNA rather than mRNA targeting comes from E. coli, where the use of engineered spacers demonstrated that effective crRNAs could be produced from either the coding or template strand of lambda phage (Brouns et al., 2008).
Additional support for DNA targeting comes from a recent study of CRISPR activity in a clinical isolate of S. epidermidis, RP62a (Marraffini and Sontheimer, 2008). Here, the CRISPR locus contains a spacer against the nickase gene of staphylococcal conjugative plasmids. Since nickase activity is required for conjugation only in donor cells, targeting of its mRNA would ablate RP62a's function as a donor but not as a recipient. The spacer negated both donor and recipient activity, as predicted by a DNA targeting model. Insertion of the proto-spacer into a non-conjugative plasmid prevented that plasmid from being transformed into RP62a, demonstrating that resistance was not linked to the mode of plasmid entry. The DNA-targeting model was supported by the observation that the targeted region was equally effective in either orientation within the plasmid. As an additional test of the model, Marraffini and Sontheimer cleverly designed a variant of the plasmid in which the nickase proto-spacer was interrupted by a self-splicing intron. This split the targeted sequence in the plasmid but reformed it in the encoded mRNA. This construct was capable of conjugation into RP62a, indicating that the CRISPR defense was circumvented when the DNA target was disrupted, but the mRNA target remained as a potential substrate.
The above evidence notwithstanding, very recent results demonstrate a capacity for RNA targeting in CRISPR systems containing the RAMP module. In P. furiosus (Fig. 1C), Hale and colleagues identified the six RAMP module proteins in a ribonucleoprotein complex containing the mature 39nt and 45nt psiRNAs with the shared 8nt 5′ repeat tag (Hale et al., 2009). Remarkably, the complex possessed endonucleolytic activity toward RNA targets with sequence complementarity to endogenous psiRNAs. The same activity was shown in a reconstituted, recombinant complex programmed by either 39nt or 45nt psiRNAs, or both, with only cmr5 being dispensable for the cleavage. The cleavage site is positioned 14nt from the 3′ end of the psiRNA, leading to different products for the 39nt and 45nt guides. While the exact nuclease within the complex is not known, the activity leaves a 3′ or 2′-3′ cyclic phosphate and requires divalent ions. Thus, for CRISPR systems that encode a RAMP module, the effector stage appears to include a mode of targeting the RNA components of the phage's life cycle. Alternatively to targeting phage, the RAMP module of the CRISPR system may be co-opted to impact endogenous cellular processes. Further investigation into the in vivo function of this mode will undoubtedly provide intriguing insights into the question.
It remains to be clarified how the newly discovered RNA cleavage activity relates to DNA targeting. The E. coli, S. thermophilus and S. epidermidis systems that were used to demonstrate DNA targeting do not possess a RAMP module (Haft et al., 2005). However, the RAMP-module-containing CRISPR systems, such as that of P. furiosus, may still retain an ability to affect the phage DNA directly. Two such systems in S. solfataricus and B. halodurans (Haft et al., 2005) contain spacers that are both sense and antisense to extrachromosomal elements, suggesting that DNA targeting is active in these organisms (Makarova et al., 2006).
On the front lines – virus-bacterium population dynamics
Of all the analysed spacer sequences, only a fraction maps to annotated foreign genetic elements, while the origin of the majority remains a mystery. One reason, undoubtedly, is the current under-representation of the overall phage diversity in the available sequence data, which limits our bioinformatic search space (Edwards and Rohwer, 2005) A more interesting contributing factor is the phage's exceedingly high rate of mutation and recombination (Hatfull, 2008). This consideration invokes a dynamic view of the interplay between the phage and the host, where the presence of a resistance-conferring spacer may pressure the targeted phage sequence to become unrecognizable within a very short time span. Remarkably, such adaptations were directly observed in the studies of bacteriophage-insensitive mutants of S. thermophilus. Here, the researchers identified small populations of phage that were pathogenic even in resistant strains. One-upping the bacteria that had gained new spacers, these phage evaded the CRISPR immune system by mutating their proto-spacer or the nearby PAM (Barrangou et al., 2007; Deveau et al., 2008).
Sequence analysis of environmental microbial samples provides a population-wide view of the dynamics between phage and resistance mechanisms within their hosts (Heidelberg et al., 2009; Tyson and Banfield, 2008). In one metagenomic study, two separate biofilm community samples were sequenced at high coverage (Tyson and Banfield, 2008). For each sample, assembly of the sequence data produced a composite genome of the predominant Leptospirillum species. In contrast to the near-clonality of most of the genome, the CRISPR locus showed extensive polymorphism. As would be predicted based upon comparison of sequenced strains and upon analysis of acquired phage resistance, the two communities shared spacer sequences at leader-distal end of their CRISPR loci. These correspond to the “older” parts of the clusters. The middle of the loci contained spacers that were common within each community but not shared between the two. The leader proximal end of the loci contained spacers that were not only unique to each community but also polymorphic within each community.
These studies support a model in which highly plastic CRISPR loci continuously respond to challenge by a rapidly evolving pool of phage. The presence of common, older spacers indicate that periodic selection drives some specific CRISPR loci to fixation, while increasing polymorphism toward the leader proximal end provides support that the CRISPR system is an actively evolving and functioning phage defense in natural populations. Notably, internal deletions of repeat-spacer units were also common. Presumably, this reflects selective pressure to prevent indefinite growth of CRISPR loci, which is accomplished by pruning of unused spacer elements that have lost their protective potency.
The dynamics of the perpetual arms race between phage and bacteria came into even sharper focus with population-wide sequence analysis of the phage camp. Again, the Banfield group reconstructed bacterial and archaeal genomes from deep sequencing data of two biofilm samples (Andersson and Banfield, 2008). The spacers from their CRISPR loci were then used to identify other spacer-containing non-CRISPR reads (potentially corresponding to targeted phage sequences), which were in turn assembled with other reads into families of composite phage genomes. By matching CRISPR-derived spacers with phage proto-spacers, the authors noted clustering into broad phage-host pairs, with virus families having preference for particular bacterial or archaeal groups within the population. In this snapshot of coexisting virus and host populations, up to 40% of spacers of a given CRISPR locus (derived from a single cell) matched viral sequences, a striking contrast to conclusions drawn from snapshot analyses of individual bacterial species in isolation. This finding indicates that most cells are simultaneously defending against several different viruses.
The viral populations were also clearly evolving rapidly. Individual phage families showed significant nucleotide variation. Interestingly, their genomes appear to be constantly shuffled by homologous recombination in chunks of 25 nucleotides or less. Notably, this avoids conservation of blocks targeted by CRISPR loci, which average around 30 nt. Thus, analysis of these natural populations reveals a constant thrust and parry relationship between predator and prey in which each rapidly evolves on unexpectedly short time scales. The high polymorphism of CRISPR loci in natural populations, coupled with their correspondence to viral sequences in the same sample, suggests that the CRISPR system is a primary means of phage defense in bacteria and archaea. The fact that resistance selected in experimental settings is conferred by the CRISPR system provides additional support for this notion.
Co-evolution and horizontal transfer of CRISPR systems
In contrast to the highly variable genomic “Wild West” view of CRISPR spacers that emerges from sequencing of microbial communities, analysis of the CRISPR repeats, leaders and cas genes reveals a more conserved picture. Furthermore, several features indicate their functional association. The phylogeny of cas1, the most conserved CRISPR-associated gene, recapitulates the phylogeny of cas subtypes and their operon organization, suggesting co-evolution of these components (Haft et al., 2005; Makarova et al., 2006). A similar pattern of clustering according to cas subtypes emerges if one examines sequence similarity among repeats (Kunin et al., 2007) or the periodicity of repeat units (Haft et al., 2005). Finally, leader sequences exhibit correlations with their corresponding repeats, as exemplified by studies of CRISPRs within the Sulfolobus genus (Lillestol et al., 2009; Shah et al., 2009). These results are as expected if the cas genes, leader sequences, and CRISPR loci have co-evolved as a single functional unit that is vertically inherited through speciation events. Undoubtedly, the cas genes are under pressure not only to maintain interactions amongst their protein products but also with the non-coding components of the system, the repeats and leader sequences.
Interestingly, the pattern of co-evolution between core cas genes, subtype-specific cas genes and the non-coding components does not apply to the RAMP module. The phylogenetic tree for the module's signature polymerase gene bears no similarity to the core cas1 gene tree (Makarova et al., 2006), indicating that the RAMP module, with its associated RNA-targeting effector activity, traveled a somewhat independent evolutionary path by horizontal transfer without the rest of the CRISPR system, predominantly in thermophilic organisms (Haft et al., 2005; Makarova et al., 2006).
Aside from its conventional vertical inheritance, the CRISPR system appears to propagate extensively by horizontal gene transfer as well. It has been widely noted that the phylogenetic tree of CRISPR systems does not agree with the established bacterial/archaeal taxonomy. Closely related species sometimes have different CRISPR content, and divergent species occasionally harbor similar CRISPR systems (Godde and Bickerton, 2006; Haft et al., 2005; Makarova et al., 2002). An example proposed to represent horizontal transfer is the CRISPR locus of the bacterium T. maritima, which is found on a gene island of archaeal origin (Nelson et al., 1999). Plasmids can also harbor CRISPR loci. These often represent subtypes with high similarity to genomic CRISPRs in species closely related to the plasmid host (Godde and Bickerton, 2006; Haft et al., 2005; She et al., 1998). CRISPR loci can even be carried on phage and mobile elements. For example, the genome of C. difficule contains multiple CRISPR loci, with two residing on prophages and one lying within a skin element, a prophage-like, excisable sigK intervening sequence that participates in sporulation (Haraldsen and Sonenshein, 2003; Sebaihia et al., 2006). In these cases, it is unclear what impact the presence of this cargo might have on its carrier, perhaps conferring competitive advantage against other invading elements, but these instances do highlight the many mechanisms by which CRISPR loci and their corresponding resistance phenotypes can move between species.
Loose ends
Though the past few years have seen an explosion in our understanding of the CRISPR-cas pathway, many issues remain unresolved. A number of cas proteins have been biochemically or structurally characterized, but still lack a functional assignment within the CRISPR mechanism. Among these are the Cas2 proteins, which demonstrate a ssRNA-specific endonuclease activity that is conserved across several organisms (Beloglazova et al., 2008). The enzyme is Mg2+-dependent and leaves a 5′ phosphate at the cleavage site. However, the low sequence specificity (some preference for U-rich regions) prevents its placement in the CRISPR pathway. The crystal structure of S. solfataricus cas2 (Beloglazova et al., 2008), as well as two homologs solved by structural genomics projects (PDB codes 1zpw and 2i0x) shows a single ferredoxin-like domain which forms homodimers in solution and in the crystal. This is reminiscent of the duplicated ferredoxin fold in the cse3 and cas6 endonucleases (Carte et al., 2008; Ebihara et al., 2006). While the secondary structure elements of the cas2 and cse3/cas6 structures can be superimposed, it appears that this common structural platform was harnessed to evolve different enzymatic activities. cse3 and cas6 employ an entirely different reaction mechanism and use a distinct set of active site residues than does cas2. Unlike cas2, cse3 and cas6 do not require divalent ions and leave a 2′,3′ cyclic phosphate following cleavage.
Cmr5, a component of the RAMP module, has been structurally examined (Sakamoto et al., 2009). Cmr5 has a single α-helical domain that assembles into an interesting trimeric ring structure. A large, conserved basic patch on one face of the ring suggests an RNA-binding surface. The crystal structure of another ancillary cas protein, csx1 (COG1517) from Vibrio cholerae, has been solved in a structural genomics effort (PDB code 1xmx, unpublished). The protein contains a Rossman-like fold (possibly involved in nucleotide binding), a helix-turn-helix-containing domain, and a restriction endonuclease domain (Makarova et al., 2006). Thus, it may be hypothesized to act in spacer generation/integration, or in degradation of target DNA.
A DNA binding protein with sequence specificity for the CRISPR repeat was purified in the archaeon S. solfataricus P2 by affinity to a biotinylated repeat sequence (Peng et al., 2003). The 18kDa protein, SSO0454, recognizes a single copy of the repeat and induces a conformational change in the DNA upon binding. Consistent with its DNA-binding role, SSO0454 has a tripartite internal repeat structure with helix-turn-helix motifs. However, the gene is distant to any CRISPR loci, and does not belong to any of the classified CRISPR-associated families. Interestingly, most homologs of this protein are found in bacterial prophages, suggesting a possible function in thwarting of the CRISPR system by phages (Sorek et al., 2008).
Additional functions of CRISPRs
Aside from its defensive roles, the CRISPR locus appears to have other functions in some systems. The regular nature of the repeats presents an opportunity for homology-driven genome rearrangements. Accordingly, many large inversions/translocations identified in two related Thermotoga species occur between CRISPR hotspots, rivaling those that occur at tRNA genes (DeBoy et al., 2006). Perhaps even more interesting is the involvement of the CRISPR system in regulating endogenous cellular processes. In the bacterium P. aeruginosa, there is a link to biofilm formation and swarming motility, two group behaviors exhibited by the organism. Lysogeny of P. aeruginosa by DMS3 phage results in loss of the behaviors, possibly as a self-quarantine mechanism to protect the rest of the community. The CRISPR locus is essential for this loss, since disruption of the CRISPR or several cas genes restores biofilm formation and swarming of the lysogens (Zegans et al., 2009). A different group behavior in M. xanthus, fruiting body development upon starvation, involves the devTRS locus. These genes are actually part of a CRISPR locus that is co-transcribed with surrounding cas genes and the repeats (Viswanathan et al., 2007). DevR is a subtype-specific cas gene, cst2, and devS belongs to the cas5 family (Haft et al., 2005). These examples indicate that the existing CRISPR-cas pathway can be adapted to additional functions, though the precise mechanisms by which control of cellular behavior is exerted and if and how it is integrated with defensive roles remains to be determined.
Summary and perspectives
The CRISPR system is an elegant, effective, and fluid mechanism of defense against foreign genetic elements (Fig. 4). It is rightly described as an adaptive immune system, which evolved long before its famed namesake. Interestingly, CRISPR's ability to acquire a resistance phenotype and pass it to progeny could be construed an example of a soft, or Lamarckian, mode of inheritance. One could also view this from a conventional Darwinian perspective, where pressure exerted by the environment simply selects the fittest. However, armed with knowledge of the molecular basis of this response, CRISPR-cas does seem to fit more firmly with a Lamarckian paradigm, in essence because increases in fitness do not rely on random mutations but on a much more specific acquisition of genetic information from environmental sources.
Figure 4. An overall model of CRISPR/cas activity.
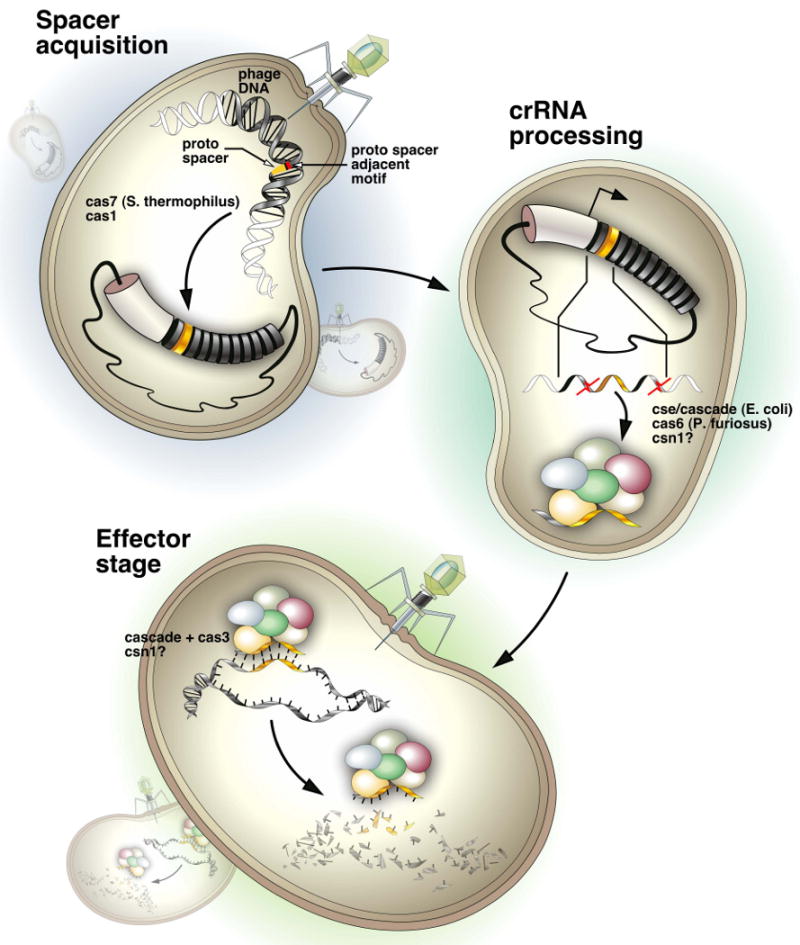
During spacer acquisition, sequence elements from invading nucleic acids become incorporated at the leader-proximal end of the CRISPR locus. In the processing stage, the locus is transcribed and processed into mature crRNA/psiRNAs containing an 8nt repeat tag and a single spacer unit. During the effector stage, the mature crRNAs in complex with associated cas proteins leads to degradation of complementary nucleic acids. See text for details.
The CRISPR-cas system bears many conceptual similarities to eukaryotic RNA interference systems, particularly the piRNA system that acts to combat mobile genetic elements (Aravin et al., 2007), which could perhaps be considered the endogenous analog of phages (Fig. 4 and 5). At the core of both systems is the ability to discriminate self from non-self nucleic acids and selectively inactive the latter. Additionally, both systems must cope with a diversity of elements that show little similarity at the primary sequence level and that, as classes, have the ability to evolve quickly. Both systems must also react to new elements that jump species barriers. In the CRISPR system, this is accomplished by pirating nucleic acids from incoming pathogens and incorporating them into a programmable silencing locus (Fig. 4). This initial step involves cas7 and, likely, cas1. Potential spacer sequences are selected by the presence of a short proto-spacer adjacent motif, PAM. Precisely how the CRISPR system distinguishes foreign from domestic nucleic acids is unclear, but it seems unlikely to depend upon the selective presence of PAMs in invader versus host genomes. In the piRNA pathway there exist similar programmable silencing loci, termed piRNA clusters (Fig. 5). These clusters also acquire additional capacity by the insertion of “foreign” nucleic acids. In this case, no specific active mechanism, analogous to polarized growth of CRISPR repeats, has been tied to building of piRNA clusters. Instead, they seem to rely on the innate mobility of transposons to acquire new content, which can then become fixed by evolutionary selection. In fact, we have proposed that the mobility of transposons is, per se, the self vs. non-self recognition mechanism, since in the absence of a general cellular mechanism to incorporate information into silencing loci, only transposons and not endogenous genes can move into these sites.
Figure 5. Conserved themes in small RNA-guided defense.
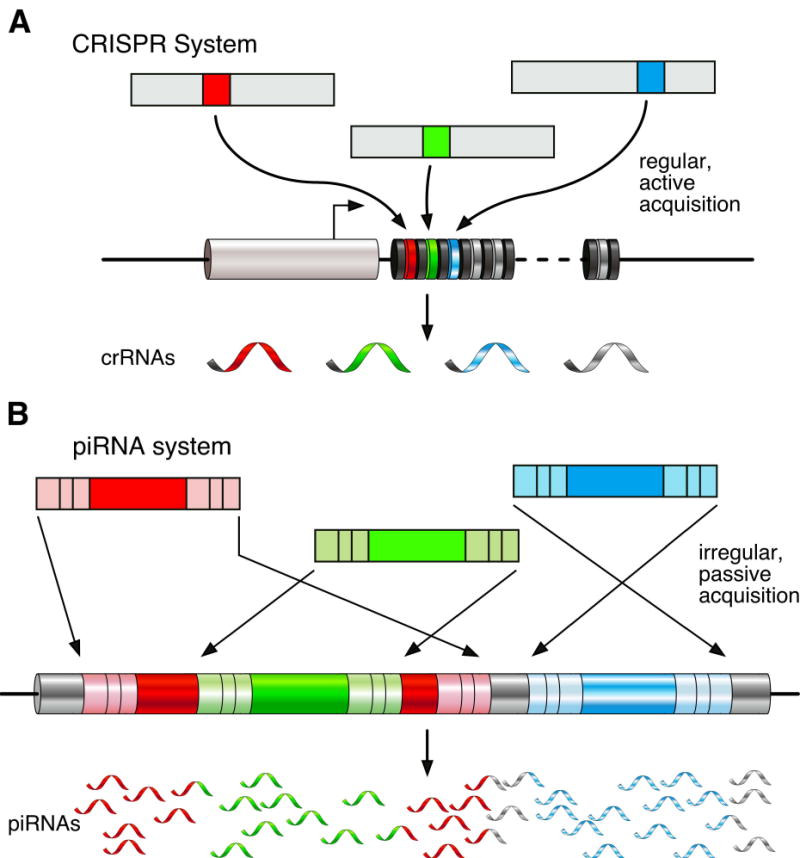
(A) The CRISPR system actively incorporates short sequence tags from invading nucleic acids into the CRISPR locus of regularly interspaced repeats. (B) In the piRNA system, genomic piRNA clusters passively acquire new content by movement of transposons into the locus. In both cases, the locus is transcribed and processed into small RNAs that guide the destruction of exogenous or parasitic sequences.
In both the CRISPR and piRNA systems, the next step in the pathway is also highly analogous. Both the CRISPR repeat and piRNA clusters appear to be transcribed as a continuous unit and later parsed into small RNAs. In the case of the piRNA pathway, the RNAs are 24-30 nt in length, whereas crRNAs can reach 57 nucleotides in length. While little is known of how the initial transcripts of piRNA loci are processed into individual RNAs, cse3/casE in E. coli and cas6 in P. furiosus appear to carry out critical processing steps for CRISPR. In both cases, small RNAs are loaded into specific effector complexes, Piwi/RISC for piRNAs and a complex that likely contains cas3 for crRNAs.
The effector mechanisms of the CRISPR and piRNA pathways differ in their specific substrates. CRISPR inactivates the genomes of incoming phage and plasmids, apparently recognizing and targeting destruction of DNA. The piRNA pathway has been most strongly implicated in targeting RNA destruction, but it must be noted that many of the elements that are controlled by piRNAs in animals have RNA genomes (e.g., retrotransposons).
Despite their superficial similarities, the CRISPR and piRNA systems do not share a single protein or non-coding component. One might also argue that strategies used to build a silencing repertoire are fundamentally different: active recognition in the case of CRISPR and passive acquisition by chance transposition in the case of piRNA loci. The CRISPR system is also missing a signature element of the piRNA pathway, the adaptive amplification loop that uses abundant transposon transcripts along with transcripts from the piRNA clusters themselves to shape steady-state small RNA populations. However, in somatic follicle cells of Drosophila, a variant of the piRNA pathway exists that lacks the ampification loop (Malone et al., 2009; Saito et al., 2009) and that, therefore, appears quite similar in its overall construction to CRISPR (Fig. 5). Perhaps the most important difference is that CRISPR seems dedicated to protection against exogenous invaders, whereas the piRNA pathway is tasked to recognize endogenous parasites. While two transposon targeting CRISPR spacers have been detected (Mojica et al., 2009), such sequences are rare amongst the vast number of spacers examined. One might imagine that this reflects the greater danger posed by exogenous pathogens in the microbial world. Alternatively, it might indicate a self-non-self recognition mechanism within the CRISPR pathway that is ill suited to detect transposons selectively and to incorporate their content as spacers.
While the piRNA pathway provides many interesting analogies to the CRISPR-cas system, it must be appreciated that roles for small RNAs in combating pathogenic and parasitic nucleic acids are nearly universal. Within the plant kingdom, small RNAs are essential both for the control of transposons and for antiviral responses (Zilberman and Henikoff, 2005). Similar functions for siRNAs are also common throughout invertebrates (Sijen and Plasterk, 2003). Small RNAs manage repeat and transposon content in the somatic nuclei of protozoa (Mochizuki and Gorovsky, 2004). Considering the evolutionary distance that separates these cited examples, it does seem striking that small RNAs would form a common thread running through all of these distinctly constructed resistance mechanisms. Perhaps a deeper understanding of the biochemical mechanisms and evolution of small RNA pathways throughout the kingdoms of life may provide clues to the reasons underlying this curious convergence.
Acknowledgments
The authors would like to thank P. Rajesh Kumar for help with structural alignments and Xavier Roca for comments on the manuscript. We are greatly indebted to James Duffy for assistance with figures. FVK is supported by a postdoctoral fellowship from the American Cancer Society, PF-07–058-01-GMC. This work was supported by grants from the NIH and by a kind gift from Kathryn W. Davis. GJH is a professor of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription Profile of Thermus thermophilus CRISPR Systems after Phage Infection. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- Agari Y, Yokoyama S, Kuramitsu S, Shinkai A. X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermophilus HB8. Proteins. 2008;73:1063–1067. doi: 10.1002/prot.22224. [DOI] [PubMed] [Google Scholar]
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. Phage-host interaction: an ecological perspective. J Bacteriol. 2004;186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- DeBoy RT, Mongodin EF, Emerson JB, Nelson KE. Chromosome evolution in the Thermotogales: large-scale inversions and strain diversification of CRISPR sequences. J Bacteriol. 2006;188:2364–2374. doi: 10.1128/JB.188.7.2364-2374.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara A, Yao M, Masui R, Tanaka I, Yokoyama S, Kuramitsu S. Crystal structure of hypothetical protein TTHB192 from Thermus thermophilus HB8 reveals a new protein family with an RNA recognition motif-like domain. Protein Sci. 2006;15:1494–1499. doi: 10.1110/ps.062131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Fabre M, Koeck JL, Le Fleche P, Simon F, Herve V, Vergnaud G, Pourcel C. High genetic diversity revealed by variable-number tandem repeat genotyping and analysis of hsp65 gene polymorphism in a large collection of “Mycobacterium canettii” strains indicates that the M. tuberculosis complex is a recently emerged clone of “M. canettii”. J Clin Microbiol. 2004;42:3248–3255. doi: 10.1128/JCM.42.7.3248-3255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Morrison N, Watt B, Doig C, Forbes KJ. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde A, Fitzgerald GF. Bacteriophage defence systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:89–113. [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases - Amino-Acid-Sequence Comparisons and Structure-Function-Relationships. Current Opinion in Structural Biology. 1993;3:419–429. [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2008;36:W145–148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen PM, Bunschoten AE, van Soolingen D, van Embden JD. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. Rna. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Krauss G. Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett. 2009;583:771–776. doi: 10.1016/j.febslet.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Han D, Lehmann K, Krauss G. SSO1450--a CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 2009;583:1928–1932. doi: 10.1016/j.febslet.2009.04.047. [DOI] [PubMed] [Google Scholar]
- Haraldsen JD, Sonenshein AL. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol Microbiol. 2003;48:811–821. doi: 10.1046/j.1365-2958.2003.03471.x. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447–453. doi: 10.1016/j.mib.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hermans PW, van Soolingen D, Bik EM, de Haas PE, Dale JW, van Embden JD. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe N, Nakashima K, Grigsby D, Pan X, Dou SJ, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, Musser JM. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg Infect Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, van Embden JD, Gaastra W, Schouls LM. Identification of a novel family of sequence repeats among prokaryotes. Omics. 2002;6:23–33. doi: 10.1089/15362310252780816. [DOI] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101. 145–152. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestol RK, Redder P, Garrett RA, Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestol RK, Shah SA, Brugger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–272. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masepohl B, Gorlitz K, Bohme H. Long tandemly repeated repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120. Biochim Biophys Acta. 1996;1307:26–30. doi: 10.1016/0167-4781(96)00040-1. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Ferrer C, Juez G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- Nakata A, Amemura M, Makino K. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol. 1989;171:3553–3556. doi: 10.1128/jb.171.6.3553-3556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Brugger K, Shen B, Chen L, She Q, Garrett RA. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J Bacteriol. 2003;185:2410–2417. doi: 10.1128/JB.185.8.2410-2417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Agari Y, Agari K, Yokoyama S, Kuramitsu S, Shinkai A. X-ray crystal structure of a CRISPR-associated RAMP superfamily protein, Cmr5, from Thermus thermophilus HB8. Proteins. 2009;75:528–532. doi: 10.1002/prot.22358. [DOI] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- Sensen CW, Charlebois RL, Chow C, Clausen IG, Curtis B, Doolittle WF, Duguet M, Erauso G, Gaasterland T, Garrett RA, et al. Completing the sequence of the Sulfolobus solfataricus P2 genome. Extremophiles. 1998;2:305–312. doi: 10.1007/s007920050073. [DOI] [PubMed] [Google Scholar]
- Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]
- She Q, Phan H, Garrett RA, Albers SV, Stedman KM, Zillig W. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles. 1998;2:417–425. doi: 10.1007/s007920050087. [DOI] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- van der Ploeg JR. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology. 2009;155:1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol. 2007;189:3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Epigenetic inheritance in Arabidopsis: selective silence. Curr Opin Genet Dev. 2005;15:557–562. doi: 10.1016/j.gde.2005.07.002. [DOI] [PubMed] [Google Scholar]


