Abstract
The hematopoietic- and neurologic-expressed sequence 1 (Hn1) gene encodes a highly conserved protein that is expressed in developing and regenerating tissues. In this study, Hn1 expression was evaluated in human and murine malignant gliomas. Hn1 mRNA and protein were detected in the murine GL261 glioma cell line and in GL261 brain tumors in vivo. HN1 is also expressed in human U118MG and U87MG cell lines. Evaluation of human brain tumors using an anti-Hn1 polyclonal antibody detected strong immunoreactivity in high-grade (WHO III and IV) malignant gliomas. The rate of GL261 cell proliferation in vitro was unaltered by Hn1 depletion using an anti-Hn1 siRNA. However, tumors established from Hn1-depleted GL261 cells formed significantly smaller volumes than those established from control-treated cells. These data suggest a role for Hn1 in the biology of malignant brain tumors.
Keywords: GL261, Tumor, siRNA, Jupiter, AAV, Glioma
Introduction
The rapid growth and invasive characteristics of gliomas can be caused by genetic and external factors that alter intracellular mechanisms responsible for controlling cell proliferation and repair. Identifying the roles of the various genes involved in the aberrant growth properties of tumors is a valuable goal toward uncovering mechanisms of tumorigenesis. The little-studied hematopoietic- and neurologic-expressed sequence 1 (Hn1) gene encodes a small protein that is expressed throughout several biological models of growth. When comparing the expression of genes involved in regenerating and non-regenerating models of injured motoneurons, our lab identified the marked upregulation of Hn1 mRNA and protein in axotomized adult facial motor and vagus nerves, both of which regenerate after injury [1]. The duration of the increased expression correlated to the time required for the neurons to regenerate completely. Moreover, an ortholog of Hn1 in the Japanese common newt is upregulated during dedifferentiation of their regenerating retinal pigment epithelial cells [2]. Numerous tissues, including the brain, spinal cord, heart, thymus, spleen and testis, manifest high levels of Hn1 mRNA during embryonic development [1]. Although the expression of Hn1 throughout the brain of the newborn rat and mouse is diffuse and intense, it becomes restricted to the hippocampus, cortex, and cerebellum, areas that exhibit high plasticity [1]. Furthermore, HN1 is one of four genes that distinguish epithelial ovarian carcinoma cells from normal ovarian surface epithelial cells [3]. While the specific function of Hn1 is not known, this collective information suggests that this gene is involved in processes associated with cell proliferation, repair and/or growth.
The murine Hn1 cDNA was first isolated from embryonic erythroid cells derived from yolk sac blood islands. The naming of this gene followed the realization that it is highly expressed in hematopoietic cells and fetal brain [4]. Hn1 has a unique protein sequence consisting of 154 amino acids and it is conserved among numerous species including humans, rodents, primates, cattle, birds, fish, amphibians, and insects. Murine Hn1 is found on chromo-some 11, which is, in many cases, parallel to human chromosome 17, where the human ortholog is located (17q25.2) [4, 5]. A gene of similar sequence to Hn1, termed Hn1-Like (Hn1L), is also found in several species [5]. The function of Hn1L is also unknown.
Here, we investigated the expression of Hn1 not only in the murine GL261 glioma model, but also in several human gliomas. The murine GL261 model was established in C57BL/6 mice after an intracranial injection of 3-methylcholanthrene [6]. The tumor was originally maintained in vivo by serial transplantation of small tumor pieces onto syngeneic C57BL/6 mice and the GL261 cell line was established thereafter [7, 8]. GL261 cells exhibit rapid in vitro growth rates, lack contact inhibition, and develop an aggressive tumor when injected into their syngeneic host [9, 10]. The GL261 murine glioma model is appropriate for the study of treatments against glioma because it shares numerous characteristics with human gliomas [8, 11–13]. It is invasive but does not metastasize, has a high tumor take rate, both p53 and K-ras are mutated, and c-myc and p53 are upregulated [8]. The results reported here establish that Hn1 is expressed in the GL261 murine glioma model. Moreover, HN1 expression was detected in the human glioma cell lines U118MG and U87MG, as well as high-grade malignant human brain gliomas. In addition, we developed an adeno-associated virus (AAV) engineered to express a recombinant siRNA that targets and degrades murine Hn1 in GL261 cells. The effect of Hn1 depletion on the in vitro and in vivo growth of GL261 cells and tumors was evaluated.
Materials and Methods
Cell Culture
GL261 cells, obtained from the NCI (Frederick, MD) were grown in RPMI (Gibco BRL) containing 10% FBS, 1% penicillin-streptomycin, and 4 mM L-glutamine. B16.F10, HEK293, U118MG and U87MG were obtained from the ATCC (Manassas, Virginia). B16.F10 and HEK293 cells were grown in DMEM (Gibco BRL) containing 10% FBS, 1% penicillin-streptomycin, and 1% sodium pyruvate. The sodium bicarbonate content in DMEM was 3.7 g/L for HEK293 cells and 1.5 g/L for B16.F10 cells. U118MG cells were grown in DMEM with 4 mM L-glutamine, 1.5 g/L sodium bicarbonate, 4.5 g/L glucose and 10% FBS. U87MG cells were grown in Minimum essential medium (Gibco BRL) with 2 mM L-glutamine and Earle's BSS adjusted to contain 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acids, 1.0 mM sodium pyruvate, and 10% FBS. All cells were cultured in an incubator maintained at 37°C with 5% CO2.
Northern Blot Analysis
Total RNA was isolated from GL261 cells using TRIzol Reagent according to the manufacturer's recommended procedure (Life Technologies, Grand Island, NY). RNA (20 μg/lane) was electrophoresed through denaturing 1.2% agarose and subjected to Northern blot analysis [14]. The nylon membrane was hybridized with a 32P-radiolabeled cDNA generated from a 450-bp murine Hn1 protein coding DNA sequence, which was 32P-radiolabeled by the random primer method to a specific activity of 1.2×109 dpm/μg.
Development of Anti-Hn1 Antibody and Western Blot Analysis
Mouse Hn1 was expressed in E. coli using the pET22a and pATH11 expression vectors. The pATH11 vector produces Hn1 fused to the C-terminus of E. coli Trp-E, while pET22a produces a His-tagged protein. The Trp-E fusion protein was purified by excision of the appropriate gel band from a 6 M urea extract of a bacterial inclusion body preparation as described [15]. The His-tagged protein was affinity purified using a nickel column using standard procedures. New Zealand white rabbits were injected with the Trp-E-Hn1 fusion protein and then boosted with His-tagged Hn1. Sera were tested by Western blotting for activity on Trp-E-Hn1 and His-tagged Hn1. Strong reactivity with both proteins indicated good reactivity with Hn1. Serum was collected from rabbits and affinity purified on His-tagged Hn1 coupled to cyanogen bromide activated Sepharose 4B.
Protein samples were subjected to SDS-PAGE, transferred onto a PVDF membrane and probed with the affinity purified polyclonal rabbit anti-Hn1 antibody at a 1/1,000 dilution. The secondary antibody used was an HRP-conjugated goat anti-rabbit (Sigma-Aldrich, St. Louis, MO) and was utilized at a 1/2,000 dilution.
Development of siRNA Against Hn1
Using siDirect (http://design.RNAi.jp), we identified three potential small interfering RNA (siRNA) sequences predicted for maximum Hn1-target specificity to degrade the Hn1 mRNA. These siRNA sequences were initially tested for their ability to inhibit expression of murine Hn1 in a cotransfection paradigm using HEK293 cells, which lack endogenous Hn1. The sequences were individually cloned into a vector (pH1rSC) containing the H1 promoter that allows for packaging into a recombinant self-complementary (double stranded) adeno-associated virus (AAV) [16]. The most efficient of the three Hn1-siRNA-expressing plasmids was generated by cloning the following complimentary oligonucleotides into the AscI and NheI sites of pH1rSC: 5′-CGC GGG GAG AAG GTG ATA TGC ATT TCA AGA CAA TGC ATA TCA CCT TCT CCC TTT TTG GAA A-3′ and 5′-CTA GTT TCC AAA AAG GGA GAA GGT GAT ATG CAT TGT CTT GAA ATG CAT ATC ACC TTC TCC C-3′. Plasmids were packaged into an AAV6, a serotype of AAV that was determined to be the most efficient at transducing GL261 cells in culture by fluorescence microscopy of GL261 cells transduced with different GFP-expressing AAV serotypes. High titer virus was produced and purified according to previously published protocols [17].
GL261 cells were seeded on a 12-well cell culture plate at a density of 300,000 cells per well. Two days later, cells were rinsed with serum-free RPMI and transduced with serum-free media containing either the Hn1-siRNA-AAV6 or the control H1-AAV6. All dishes were incubated for 3 h at 37°C in 5% CO2, after which the volumes were doubled with media containing 20% FBS, 1% pen/strep, and 4 mM L-glutamine. Two days after transduction, protein extracts were analyzed by Western blot analysis.
GL261 Tumor Preparation
GL261 cells were harvested from plates by trypsinization, washed one time in phosphate buffered saline (PBS), and suspended in PBS at a density of 67,000 cells/μl. Cells (200,000) were implanted intracranially into isoflurane-anesthetized female C57BL/6 mice (7 weeks old) on the right hemisphere of the brain (1 mm posterior and 2 mm lateral to Bregma; 3 mm deep to the dural surface). The mice were monitored carefully for signs of depressed activity and motor instability. Twenty days after GL261 cell implantation, the mice were given an injection of sodium pentobarbital (32 mg/kg) and euthanized by transcardial perfusion with 0.9% saline followed by buffered 4% paraformaldehyde (PFA). The brains were removed, post-fixed for 1 h with 4% PFA, sectioned, cryoprotected overnight with 30% sucrose, and frozen in isopentane cooled by liquid N2. Coronal cryosections (20 μm) of the brainstem and midbrain were prepared at the level of the tumor. Sections were thaw mounted on Superfrost/Plus slides (Fisher Scientific, Pittsburg, PA) and subjected to in situ hybridization or immunohistochemical procedures.
For experiments designed to evaluate the effect of Hn1 depletion on the growth of GL261 tumors, GL261 cells were initially seeded at a density of approximately 2×106 cells per 100 mm plate. Two days later, cells were transduced at an M.O.I. of 5,000 with either the siRNA-AAV6 or the control H1-AAV6 vector under serum-free conditions. Three hours later, the cells were supplemented with medium containing serum to achieve a final 10% FBS content. Two days after transduction, cells were prepared for intracranial implantation as described above.
In situ Hybridization Analysis
Hn1 hybridization probes [1] were used for in situ hybridization (ISH) analysis as previously described [18]. Sections were hybridized separately with antisense and sense riboprobes and then apposed to film and dipped in LM-1 emulsion. After exposure for 1 week, slides were developed, fixed and counterstained with hematoxylin and eosin (H&E).
Immunohistochemistry
The GL261 tumor tissue sections were permeabilized using 0.5% Triton X-100 for 10 min and blocked with 3% BSA for 20 min. The rabbit polyclonal anti-murine Hn1 primary antibody was used at 1:50 dilution in PBS for 1 h at room temperature. Sections were rinsed in PBS three times for 10 min and an anti-mouse Alexa Fluor 594 red fluorescent secondary antibody (Sigma-Aldrich) was added at 1:2,000 in PBS for 1 h at room temperature.
Human brain tumor tissue microarray (TMA) slides obtained from the University of Florida, Department of Pathology, Immunology, and Laboratory Medicine were constructed as previously reported [19]. The final TMA consisted of 28 gliomas, including samples of WHO Grade IV (glioblastoma multiforme) and WHO grade III (anaplastic astrocytoma, anaplastic oligodendroglioma, anaplastic oligoastrocytoma), and seven non-neoplastic control brain samples. Immunohistochemical analysis was performed by incubating with the rabbit polyclonal anti-Hn1 antibody (1:200) overnight. This was followed by incubation with a biotinylated secondary antibody (DakoCytomation, Copenhagen, Denmark). The bound antibodies were visualized with avidin-biotinylated peroxidase complex and diaminobenzidine tetrachloride.
In vitro Cell Proliferation Assay
GL261 cells were seeded on six-well dishes at a density of 10,000 cells per well. Two days later, cells were transduced at an M.O.I. of 5,000 with Hn1-siRNA-AAV6, control H1-AAV6 or no virus. Two days after transduction, triplicate wells of cells were collected by trypsinization and counted manually using a hemacytometer. This procedure was repeated every 24 or 48 h.
Tumor Volume Calculation
Serial sections of each tumor were collected. Tumor volumes were calculated by measuring the longest and shortest diameters on each section, using the formula: Area = (r1)(r2)π and then multiplying each area by the width of the corresponding section. The volumes from all the sections pertaining to a single tumor were summed to obtain the final tumor volume. Paired student's t-test was used for statistical analysis.
Results
Hn1 mRNA and Protein are Expressed in GL261 Cells and Tumors
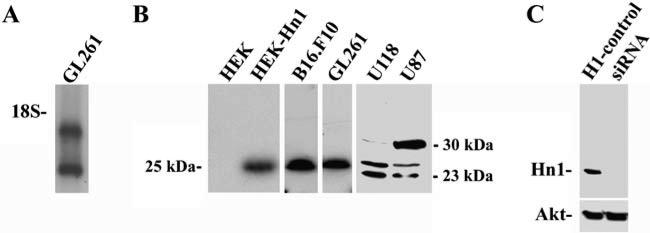
Since Hn1 expression has been reported in biological models involving cellular growth and proliferation, the expression of this gene was explored in GL261 cells. Northern blot analyses identified two mRNA species of 0.7 and 1.4 kb (Fig. 1a). Hn1 protein in GL261 cells cultured in vitro was detected by Western blot analysis using a rabbit polyclonal anti-murine Hn1 antibody. This antibody identified a 25 kDa band in these cells which co-migrated on SDS-PAGE with a recombinant form of Hn1 (Fig. 1b) expressed in HEK293 cells, which do not normally express Hn1 protein. The 25 kDa band was also detected in B16. F10 murine melanoma cells using this antibody. Western blot analysis of the human glioma cell lines U118 and U87 detected three distinct bands with apparent molecular masses of 23, 25, and 30 kDa. The presence of these three bands in human cell lines is consistent with the identification of three variants of HN1 in humans (Genbank accession #s: variant 1: NP057269; variant 2: NP001002032; variant 3: NP001002033). To further validate the specificity of the Hn1 antibody, GL261 cells were treated with a serotype 6 recombinant adeno-associated virus (AAV6) engineered to express a silencing RNA sequence (siRNA) against murine Hn1. Western blot analysis of these samples collected 2 days later revealed that the 25 kDa protein recognized by the anti-Hn1 antibody was no longer present in the samples treated with the anti-Hn1 siRNA-AAV6 (Fig. 1c).
Fig. 1.
Hn1 is expressed in murine and human glioma cell lines. (a) Northern blot analysis of Hn1 mRNA in GL261 cells. The migration of the 18 S rRNA is indicated. (b) Western blot analysis of HEK293 cells (HEK), HEK293 cells transfected with a recombinant murine Hn1 protein (HEK-Hn1), murine B16.F10 and GL261 cells, and the human U118 and U87 glioma cells using the rabbit polyclonal anti-Hn1 antibody. (c) Western blot analysis, using the anti-Hn1 antibody, show Hn1 expression in GL261 cells transduced at an M.O.I. of 5,000 with either Hn1-siRNA AAV6 (siRNA) or control H1-AAV6 (H1-control)
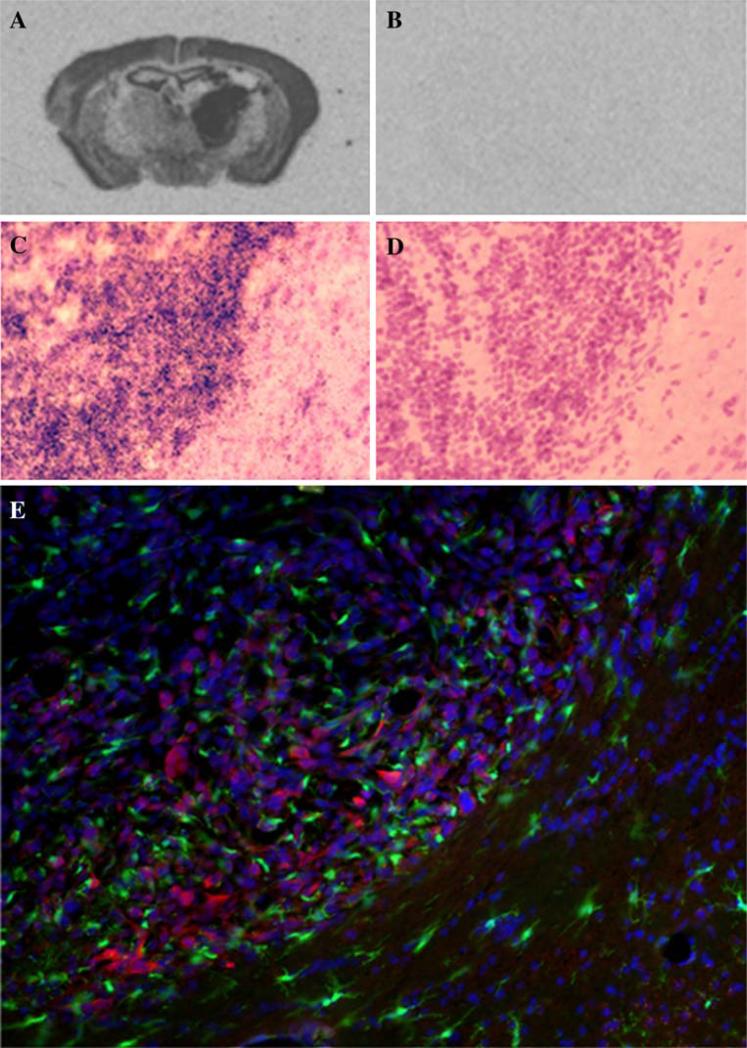
The in vitro findings were complemented by in situ hybridization (ISH) and immunohistochemical analyses of GL261 tumors established in murine (C57BL/6) brain (Fig. 2). Strong hybridization signals were evident within the GL261 intracranial tumor mass, indicating high levels of Hn1 mRNA (Fig. 2a–d). Hn1 protein expression in GL261 tumors was detected by immunohistochemical methods using the anti-Hn1 antibody (Fig. 2e). The tumor was generated in a C57BL/6 mouse that expresses green fluorescent protein (GFP) in all the CX3CR1-expressing cells. In this model, the GFP/CX3CR1-expressing cells represent glioma-infiltrated microglia [20]. Hn1 protein did not co-localize with the GFP/CX3CR1-expressing microglia, consistent with its expression within the tumor cells.
Fig. 2.
Hn1 is expressed in GL261 brain tumors of C57BL/6 mice. Sections were derived from tumor bearing brains 20 days post-GL261 cell implantation. Panels a–b depict low resolution ISH analysis of Hn1 mRNA in GL261 tumors using anti-sense (a) and sense (b) Hn1 riboprobes. Panels (c–d) depict higher resolution silver grain analysis of adjacent sections from a GL261 brain tumor hybridized with anti-sense (c) and sense (d) Hn1 riboprobes. High silver grain density on the left side of the section indicates Hn1 expression within the tumor cells. Sections were counterstained with H&E following ISH. (e) Immunohistochemical analysis using a rabbit polyclonal anti-Hn1 antibody showing Hn1 (red) in the GL261 tumor cells and not the GFP-expressing microglia (green). Sections were counterstained with DAPI nuclear stain (blue)
HN1 Protein Expression in Human Brain Tumors
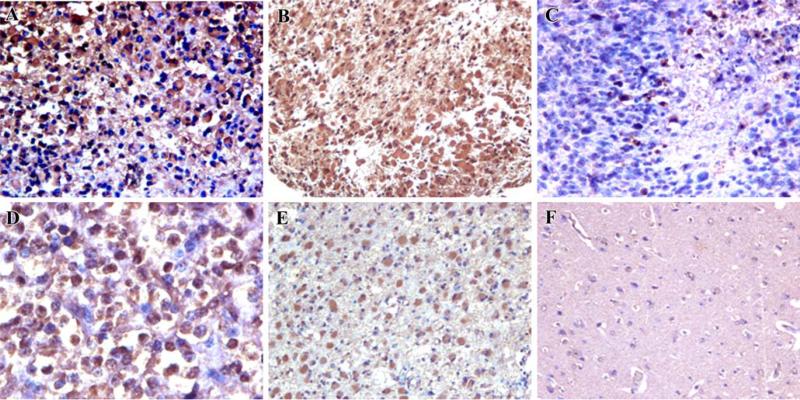
The expression of HN1 protein was evaluated in human brain tumors. A human brain tumor tissue microarray, containing multiple WHO tumor grade III and IV infiltrating gliomas [21], was subjected to immunohistochemical analysis using the polyclonal anti-Hn1 antibody. The antibody detected strong HN1 immunoreactivity in human malignant gliomas including glioblastoma and anaplastic forms of astrocytoma and oligodendroglioma (Fig. 3). The localization of HN1 protein paralleled what was seen in the mouse GL261 tumors, where the tumor cells were the prominent HN1-expressing cell type. HN1 expression in a healthy cortical region of human brain was negative except on two of the tissue samples where weak immunoreactivity was detected in neuronal cells (Table 1). The expression of HN1 in human cortical neurons is consistent with the localization of this gene in murine brain [1]. Of the WHO grade III and IV gliomas, 75% expressed strong anti-Hn1 immunoreactivity while 11% had moderate Hn1 expression (Table 1).
Fig. 3.
HN1 is expressed in multiple types of high-grade human glioma. A human brain tumor tissue microarray containing a variety of tumor samples was subjected to immunohistochemistry using the rabbit polyclonal anti-Hn1 antibody. Strong immunoreactivity was identified in human malignant gliomas including glioblastoma multiforme (a–c), anaplastic oligodendroglioma (d), and anaplastic astrocytoma (e); panel (f) shows a healthy region of brain cortex
Table 1.
Immunohistochemical analysis of Hn1 expression in human gliomas
| Tissue type | Number of cases | Relative intensity of Hn1 immunoreactivity | ||
|---|---|---|---|---|
| Non-neoplastic (negative control) | 7 | Neurons | 2/7 weak | |
| 5/7 absent | ||||
| Astrocytes | 7/7 absent | |||
| Oligos | 7/7 absent | |||
| WHO Grade IV Tumor [21] | Glioblastoma Multiforme | 11 | 6 strong | |
| 2 moderate | ||||
| 3 weak | ||||
| WHO Grade III Tumor [21] | Anaplastic Astrocytoma | 5 | 4 strong | |
| 1 moderate | ||||
| Anaplastic Oligoastrocytoma | 1 | 1 weak | ||
| Anaplastic Oligodendroglioma | 11 | 11 strong | ||
| All tumors | 28 | 21 (75%) strong | ||
| 3 (11%) moderate | ||||
| 4 (14%) weak | ||||
Criteria: Strong: over 80% of cells have anti-Hn1 immunoreactivity. Moderate: between 21% to 79% of tumor cells have anti-Hn1 immunoreactivity. Weak: less than 20% of cells have anti-Hn1 immunoreactivity.
Impact of Hn1 Depletion on Growth of GL261 Cells and Tumors
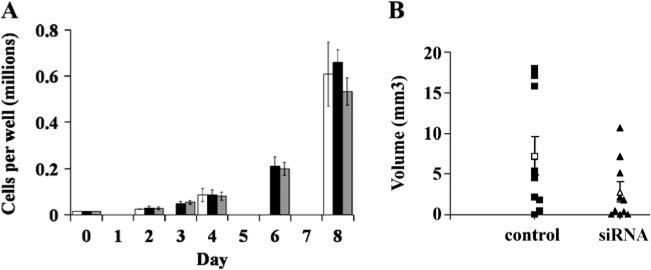
To evaluate the in vitro proliferation rates of cells with and without Hn1 protein, GL261 cells were treated independently with the Hn1-siRNA AAV6, control H1 AAV6 (each at an M.O.I. of 5,000), or no virus. Cell counts were obtained at 2, 3, 4, 6, and 8 days after addition of virus. Protein lysates from each sample were also collected at each time point for Western blot analysis in order to confirm the effect of the siRNA on Hn1 protein expression (data not shown). The in vitro proliferation rate of the Hn1-depleted GL261 cells did not differ from either the control-treated or the untreated cells (Fig. 4a). A parallel experiment was conducted in vivo on C57BL/6 mice. Twenty mice were implanted i.c. with GL261 cells pre-treated with either Hn1-siRNA AAV6 or the control H1 AAV6. The brain tumors were allowed to develop for 20 days, at which time all the mice were euthanized. Excised brains were cryosectioned and the volumes were calculated based on the area of each tumor section (Fig. 4b). Tumor volumes were significantly smaller (p<0.05) in the animals implanted with Hn1-depleted cells.
Fig. 4.
Hn1 depletion affects growth of GL261 tumors in vivo but not in vitro proliferation rates of GL261 cells. (a) In vitro growth rates of GL261 cells transduced with either Hn1-siRNA AAV6 (grey), control H1 AAV6 (black) at an M.O.I. of 5,000, or no virus (white). There was no difference in the in vitro growth rates of GL261 cells with or without Hn1. (b) Comparison of the growth of in vivo GL261 tumors established from cells pretreated with either control or Hn1-siRNA-expressing AAV6. The scatter-plot shows individual tumor volumes as well as means and SEMs. The difference between the mean volumes of tumors derived from Hn1-expressing and Hn1-depleted cells was statistically significant (p<0.05)
Discussion
The main findings reported here include the identification of Hn1 mRNA and protein expression in GL261 cells cultured in vitro as well as in murine intracranial tumors established from these cells. HN1 expression was also detected in the U118 and U87 human glioma cell lines as well as high-grade human gliomas, including glioblastoma and anaplastic forms of astrocytoma and oligodendroglioma. While the in vitro proliferation of cells lacking Hn1 was not altered, tumors derived from Hn1-depleted GL261 cells were significantly smaller than tumors generated from Hn1-expressing cells. Collectively, the data identify Hn1 as a novel protein involved in the biology of malignant gliomas.
Gliomas are very difficult to treat because of the delicate intracerebral environment in which they develop and their highly invasive and immunosuppressive properties. Identifying genes involved in the regulation of tumor growth, as well as the invasive and immunosuppressive properties of gliomas is a valuable goal toward developing novel anti-glioma therapies. Previously published results indicate that Hn1 protein is expressed in numerous tissues during development, in highly regenerating nervous tissue, and in at least one type of cancer, the human ovarian carcinoma [1–3]. These varied Hn1-expressing scenarios are suggestive of a role for Hn1 in mechanisms involved in cell proliferation or in maintaining cells in a dedifferentiated state common to cell repair and development. As such, we explored the expression of Hn1 in a murine model of malignant glioma as well as in high-grade human gliomas and determined that each of these brain tumors express the Hn1 protein. The murine GL261 model of malignant glioma reproduces many aspects of the high-grade human glioblastoma multiforme including the latter's rapid growth, invasive characteristics, and genetic alterations [8, 11–13], and the finding of Hn1 protein expression in both the human and GL261 gliomas add to the existing evidence that this is an appropriate murine model to study gliomas. Uncovering the cellular function of Hn1 in human malignant gliomas will be facilitated by parallel studies in this murine model.
Insight into the function of Hn1 in gliomas was obtained from experiments utilizing a recombinant AAV engineered to express an anti-Hn1 siRNA. The Hn1-siRNA efficiently inhibited the expression of Hn1 protein in the GL261 cells within 2 days after addition of the virus, suggesting a relatively short half-life of the protein. Despite a lack of differences in the in vitro proliferation rates of GL261 cells with and without Hn1, Hn1-depleted GL261 cells formed intracranial tumors that were significantly smaller than those established from control-treated cells. As a group, the tumors from Hn1-depleted cells developed tumors with smaller intracranial volumes. In addition, a greater number of animals had no visible tumors after implantation with Hn1-depleted cells. The difference between the in vitro and in vivo growth effects of Hn1 depletion suggests that Hn1 does not play a direct role in regulating the proliferation rate of the GL261 cells and indicates that the function of Hn1 is dependent upon the environment in which the tumor cells grow and invade normal brain tissue. It is noteworthy to mention that Hn1 expression on the GL261 tumors, viewed by immunohistochemistry and in situ hybridization, was particularly higher on the edge of the tumors. This is an area where cells proliferate faster than on the center of the tumor [22] and where the interaction with factors from the surrounding environment, e.g. membrane-type 1 matrix metalloproteinase and matrix metalloproteinase 2 (MT1-MMP/MMP2), is critical for the invasive capabilities of the tumor [23]. The development of the anti-Hn1 antibody as well as the highly efficient anti-Hn1-siRNA AAV6 will facilitate future studies addressing the specific cellular function of Hn1 in murine cancer cells and other biological processes.
Acknowledgements
Financial support for these studies was provided by the Evelyn F. and William L. McKnight Brain Institute of the University of Florida and the National Institutes of Health (AI058256) to J.K.H. We are thankful to Ms. Irene Zolotukhin for manufacturing the adeno-associated viruses utilized in this study. The anti-Hn1 antibody used in this study was generated at EnCor Biotechnology, Inc., Gainesville, Florida. A portion of these data were presented at the 2007 annual meetings of the Society for Neuroscience in San Diego, CA and Society for Neuro-oncology in Dallas, TX.
Contributor Information
Katharine M. Laughlin, Department of Pharmacology & Therapeutics, University of Florida College of Medicine, P.O. Box 100267, 1600 SW Archer Rd, Gainesville, FL 32610-0267, USA
Defang Luo, Department of Pharmacology & Therapeutics, University of Florida College of Medicine, P.O. Box 100267, 1600 SW Archer Rd, Gainesville, FL 32610-0267, USA.
Che Liu, Department of Pharmacology & Therapeutics, University of Florida College of Medicine, P.O. Box 100267, 1600 SW Archer Rd, Gainesville, FL 32610-0267, USA.
Gerry Shaw, Department of Neuroscience, University of Florida College of Medicine, Gainesville, FL 32610-0267, USA.
Kenneth H. Warrington, Jr., Department of Pediatrics, University of Florida College of Medicine, Gainesville, FL 32610-0267, USA
Jingxin Qiu, Department of Pathology, University of Florida College of Medicine, Gainesville, FL 32610-0267, USA.
Anthony T. Yachnis, Department of Pathology, University of Florida College of Medicine, Gainesville, FL 32610-0267, USA
Jeffrey K. Harrison, Department of Pharmacology & Therapeutics, University of Florida College of Medicine, P.O. Box 100267, 1600 SW Archer Rd, Gainesville, FL 32610-0267, USA
References
- 1.Zujovic V, Luo D, Baker HV, Lopez MC, Miller KR, Streit WJ, Harrison JK. The facial motor nucleus transcriptional program in response to peripheral nerve injury identifies Hn1 as a regeneration-associated gene. J Neurosci Res. 2005;82:581–591. doi: 10.1002/jnr.20676. [DOI] [PubMed] [Google Scholar]
- 2.Goto T, Hisatomi O, Kotoura M, Tokunaga F. Induced expression of hematopoietic- and neurologic-expressed sequence 1 in retinal pigment epithelial cells during newt retina regeneration. Exp Eye Res. 2006;83:972–980. doi: 10.1016/j.exer.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, Smith D, Hartmann L, Fishman D, Berchuck A, Schmandt R, Whitaker R, Gershenson DM, Mills GB, Bast RC., Jr. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 4.Tang W, Lai YH, Han XD, Wong PM, Peters LL, Chui DH. Murine Hn1 on chromosome 11 is expressed in hemopoietic and brain tissues. Mamm Genome. 1997;8:695–696. doi: 10.1007/s003359900540. [DOI] [PubMed] [Google Scholar]
- 5.Zhou G, Wang J, Zhang Y, Zhong C, Ni J, Wang L, Guo J, Zhang K, Yu L, Zhao S. Cloning, expression and subcellular localization of HN1 and HN1L genes, as well as characterization of their orthologs, defining an evolutionarily conserved gene family. Gene. 2004;331:115–123. doi: 10.1016/j.gene.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman HM, Arnold H. Experimental brain tumors. I. Tumors produced with methylcholanthrene. Cancer Res. 1941:919–924. [Google Scholar]
- 7.Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30:2394–2400. [PubMed] [Google Scholar]
- 8.Szatmari T, Lumniczky K, Desaknai S, Trajcevski S, Hidvegi EJ, Hamada H, Safrany G. Detailed characterization of the mouse glioma 261 tumor model for experimental glioblastoma therapy. Cancer Sci. 2006;97:546–553. doi: 10.1111/j.1349-7006.2006.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.San-Galli F, Vrignaud P, Robert J, Coindre JM, Cohadon F. Assessment of the experimental model of transplanted C6 glioblastoma in Wistar rats. J Neurooncol. 1989;7:299–304. doi: 10.1007/BF00172924. [DOI] [PubMed] [Google Scholar]
- 10.Weiner NE, Pyles RB, Chalk CL, Balko MG, Miller MA, Dyer CA, Warnick RE, Parysek LM. A syngeneic mouse glioma model for study of glioblastoma therapy. J Neuropathol Exp Neurol. 1999;58:54–60. doi: 10.1097/00005072-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Ishii N, Tada M, Hamou MF, Janzer RC, Meagher-Villemure K, Wiestler OD, Tribolet N, Van Meir EG. Cells with TP53 mutations in low grade astrocytic tumors evolve clonally to malignancy and are an unfavorable prognostic factor. Oncogene. 1999;18:5870–5878. doi: 10.1038/sj.onc.1203241. [DOI] [PubMed] [Google Scholar]
- 12.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 13.Sidransky D, Mikkelsen T, Schwechheimer K, Rosenblum ML, Cavanee W, Vogelstein B. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355:846–847. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- 14.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris J, Ayyub C, Shaw G. A molecular dissection of the carboxyterminal tails of the major neurofilament subunits NF-M and NF-H. J Neurosci Res. 1991;30:47–62. doi: 10.1002/jnr.490300107. [DOI] [PubMed] [Google Scholar]
- 16.McCarty DM, Monahan PE, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 17.Zolotukhin S, Potter M, Zolotukhin I, Sakai Y, Loiler S, Fraites TJ, Jr., Chiodo VA, Phillipsberg T, Muzyczka N, Hauswirth WW, Flotte TR, Byrne BJ, Snyder RO. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. 2002;28:158–167. doi: 10.1016/s1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]
- 18.Harrison JK, Luo D, Streit WJ. In situ hybridization analysis of chemokines and chemokine receptors in the central nervous system. Methods. 2003;29:312–318. doi: 10.1016/s1046-2023(02)00354-7. [DOI] [PubMed] [Google Scholar]
- 19.Qiu J, Ai L, Ramachandran C, Yao B, Gopalakrishnan S, Fields CR, Delmas AL, Dyer LM, Melnick SJ, Yachnis AT, Schwartz PH, Fine HA, Brown KD, Robertson KD. Invasion suppressor cystatin E/M (CST6): high-level cell type-specific expression in normal brain and epigenetic silencing in gliomas. Lab Invest. 2008;88:910–925. doi: 10.1038/labinvest.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Luo D, Streit WJ, Harrison JK. CX3CL1 and CX3CR1 in the GL261 murine model of glioma: CX3CR1 deficiency does not impact tumor growth or infiltration of microglia and lymphocytes. J Neuroimmunol. 2008;198:98–105. doi: 10.1016/j.jneuroim.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pescarmona GP, Scalerandi M, Delsanto PP, Condat CA. Non-linear model of cancer growth and metastasis: a limiting nutrient as a major determinant of tumor shape and diffusion. Med Hypotheses. 1999;53:497–503. doi: 10.1054/mehy.1999.0798. [DOI] [PubMed] [Google Scholar]
- 23.Seiki M. Membrane-type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett. 2003;194:1–11. doi: 10.1016/s0304-3835(02)00699-7. [DOI] [PubMed] [Google Scholar]