Abstract
Tumors may be initiated and maintained by a cellular subcomponent that displays stem cell properties. We have utilized the expression of aldehyde dehydrogenase as assessed by the ALDEFLUOR assay to isolate and characterize “cancer stem cell” populations in 33 cell lines derived from normal and malignant mammary tissue. Twenty-three of the 33 cell lines contained an ALDEFLUOR-positive population that displayed stem cell properties in vitro and in NOD/SCID xenografts. Gene expression profiling identified a 413-gene “cancer stem cell” profile that included genes known to play a role in stem cell function as well as genes such as CXCR1/IL8RA not previously known to play such a role. Recombinant IL8 increased mammosphere formation and the ALDEFLUOR-positive population in breast cancer cell lines. Finally, we show that ALDEFLUOR-positive cells are responsible for mediating metastasis. These studies confirm the hierarchical organization of immortalized cell lines, establish techniques that can facilitate the characterization of regulatory pathways of cancer stem cells and identify potential stem cell markers and therapeutical targets.
Keywords: cancer stem cells, cell line, IL8, breast cancer, metastasis
Introduction
The evolution of a normal cell into a fully transformed one requires the deregulation of multiple cellular processes (1, 2). According to classical models of carcinogenesis, these events can occur in any cell. In contrast, the “cancer stem cell hypothesis” holds that the preferential targets of oncogenic transformation are tissue stem or early progenitor cells that have acquired self-renewal potential (3-6). These “tumor-initiating cells” or “cancer stem cells” (CSC), in turn, are characterized by their ability to undergo self-renewal, a process that drives tumorigenesis and differentiation which contributes to tumor cellular heterogeneity. Recent evidence supporting the cancer stem cell hypothesis has been generated utilizing xenografts of primary human tumors. These studies have suggested that tumors are composed of a cellular hierarchy driven by the cancer stem cell component. In addition, recent data suggest that immortalized cell lines derived from both murine and human tissues may also contain a cellular population displaying stem cell properties. Most of these studies have been based on in vitro properties including clonogenic potential, sphere formation and multi-lineage differentiation potential (7-10). More limited studies utilizing functional transplantation of immortalized cell lines in xenografts have also suggested the existence of such a hierarchy. These studies have generally utilized Hoechst dye exclusion to identify the so-called “side population” (SP) (7, 9, 11). In addition, cell surface markers defined using primary tumor xenografts such as CD44 and CD133 have also been utilized to identify similar populations in established cell lines (7, 8). However, the limitations of these techniques have precluded their application across a wide variety of cell lines representing the molecular heterogeneity of tumors such as breast cancer. In addition, a crucial question remains as to whether the stem cell components of cell lines represent a valid model for cancer stem cell biology.
To provide more definitive evidence for the existence of “cancer stem cell populations” within breast cancer cell lines, we have studied the expression of the stem cell marker Aldehyde dehydrogenase (ALDH) in a series of 33 cell lines derived from human breast cancers and non-transformed breast cells. ALDH is a detoxifying enzyme responsible for the oxidation of intracellular aldehydes and is thought to play a role in stem cell differentiation through metabolism of retinal to retinoic acid (12). ALDH activity as assessed by the fluorescent ALDEFLUOR assay has been successfully utilized to isolate cancer stem cells in multiple myeloma and acute myeloid leukemia (AML) as well as from brain tumors (13, 14). We recently demonstrated that ALDH activity can be utilized to isolate a subpopulation of cells that display stem cell properties from normal human breast tissue and breast carcinomas (15). We now demonstrate that the majority of breast cancer cell lines contain an ALDEFLUOR-positive population with a distinct molecular profile that displays cancer stem cell properties. These studies have important implications for the interpretation of data utilizing cell lines and suggest that these lines may be useful for elucidating cancer stem cell regulatory pathways.
Methods
Cell culture
Breast cell lines (BCL) were obtained from the ATCC1 or from collections developed in the laboratories of Drs. S. Ethier2 (SUM44, SUM52, SUM149, SUM159, SUM185, SUM190, SUM225, SUM229), V.J. Möbus (BrCa-MZ-01), and V. Catros (S68). All BCLs tested were derived from carcinomas except MCF10A, which is derived from fibrocystic disease, and the HMEC-derived 184A1, which was derived from normal mammary tissue. The cell lines were grown using the recommended culture conditions (Supplementary Table 1). All experiments were done with subconfluent cells in the exponential phase of growth.
ALDEFLUOR assay and separation of the ALDH-positive population by FACS
ALDH activity was assessed in 33 BCLs representing the main molecular subtypes of human breast cancer. The ALDEFLUOR kit (StemCell technologies, Durham, NC, USA) was used to isolate the population with high ALDH enzymatic activity using a FACStarPLUS (Becton Dickinson)as previously described (15). Briefly, cells were incubated in ALDEFLUOR assay buffer containing ALDH substrate (BAAA, 1 μmol/l per 1×106 cells). In each experiment a sample of cells was stained under identical conditions with 50mmol/L of diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor, as negative control. The sorting gates were established using PI stained cells for viability. Prior to RNA profiling or NOD/SCID mice injection, the purity of sorted populations was checked using double sorting of 10,000 ALDEFLUOR-positive and negative cells in BrCa-MZ-01 and SUM159 cell lines. For both cell lines, sorted ALDEFLUOR-positive populations contained more than 98% of ALDEFLUOR-positive cells and no ALDEFLUOR-positive cells were detected in the ALDEFLUOR-negative population.
Tumorigenicity in NOD/SCID mice
Tumorigenicity of ALDELFUOR-positive, -negative and unseparated SUM159, MDA-MB-453 and BrCa-MZ-01 cells was assessed in NOD/SCID mice. Fat pads were prepared as described (15).
Anchorage-independent culture
ALDEFLUOR-positive, -negative and unseparated cells from 184A1, SUM149 and SUM159 were plated as single cells in ultra-low attachment plates (Corning, Acton, MA) at low density (5000 viable cells/ml). Cells were grown in serum-free mammary epithelial basal medium (Cambrex Bio Science, Walkerville, MD) for 3-7 days, as described (16). The capacity of cells to form spheres was quantified after treatment with different doses of IL8 (GenWay Biotech, San Diego, CA) added to the medium.
RNA extraction
Total RNA was extracted from frozen ALDEFLUOR-positive and -negative cells using DNA/RNA All Prep Maxi Kit, according to the manufacturer's instructions (Qiagen, Sample and Assay technologies, The Netherlands). Eight BCLs were used for transcriptional analysis: 184A1, BrCa-MZ-01, HCC1954, MDA-MB-231, MDA-MB-453, SK-BR-7, SUM149, and SUM159. RNA integrity was controled by denaturing formaldehyde agarose gel electrophoresis and micro-analysis (Agilent Bioanalyzer, Palo Alto, CA).
Gene expression profiling with DNA microarrays
Gene expression analyses used Affymetrix U133 Plus 2.0 human oligonucleotide microarrays containing over 47,000 transcripts and variants including 38,500 well-characterized human genes. Preparation of cRNA, hybridizations, washes and detection were done as recommended by the supplier1. Expression data were analyzed by the RMA (Robust Multichip Average) method in R using Bioconductor and associated packages (17), as described (18). RMA did background adjustment, quantile normalization and summarization of 11 oligonucleotides per gene.
Before analysis, a filtering process removed from the dataset genes with low and poorly measured expression as defined by expression value inferior to 100 units in all the 16 samples, retaining 25,285 genes/ESTs. A second filter, based on the intensity of standard deviation (SD), was applied for unsupervised analyses to exclude genes showing low expression variation across the analyses. SD was calculated on log2-transformed data, in which lowest values were first floored to a minimal value of 100 units, i.e. the background intensity, retaining 13,550 genes/ESTs with SD superior to 0.5. An unsupervised analysis was done on 16 ALDEFLUOR-positive, -negative cells on 13,550 genes. Before hierarchical clustering, filtered data were log2-transformed and submitted to the Cluster program (19) using data median-centered on genes, Pearson correlation as similarity metric and centroid linkage clustering. Results were displayed using TreeView program (19). To identify and rank genes discriminating ALDEFLUOR-positive and -negative populations, a Mann and Whitney U test was applied to the 25,285 genes/ESTs and false discovery rate (FDR, was used to correct the multiple testing hypothesis (see Supplementary Table 2 for complete data set). The classification power of the discriminator signature was illustrated by classifying samples by hierarchical clustering. A LOOCV was applied to estimate the accuracy of prediction of the identified molecular signatures and the validity of supervised analysis; each sample was excluded one by one and classified with the linear discriminant analysis (LDA) (20) by using model defined on the non-excluded samples.
Real-time RT-PCR
After ALDEFLUOR-positive and ALDEFLUOR-negative populations from different cell lines were sorted, total RNA was isolated using RNeasy Mini Kit (QIAGEN) and utilized for real-time quantitative RT-PCR (qRT-PCR) assays in a ABI PRISM® 7900HT sequence detection system with 384-well block module and automation accessory (Applied Biosystems). Primers and probes for the Taqman system were selected from the Applied Biosystems website1. The sequences of the PCR primer pairs and fluorogenic probes used are available on the Applied Biosystems website (CXCR1 assay ID: Hs_00174146_mi; FBXO21 assay ID: Hs_00372141_mi, NFYA assay ID: Hs_00953589_mi, NOTCH2 assay ID: Hs_01050719_mi, RAD51L1 assay ID: Hs00172522_mi, TBP assay ID: Hs_00427620_mi). The relative expression mRNA level of CXCR1, FBXO21, NFYA, NOTCH2, RAD51L1 was computed with respect to the internal standard TBP gene to normalize for variations in the quality of RNA and the amount of input cDNA, as described previously (21).
Invasion assay
Assays were done in triplicate in transwell chambers with 8μm pore polycarbonate filter inserts for 12-well plates (Corning, NY). Filters were coated with 30 μl of ice-cold 1:6 basement membrane extract (Matrigel, BD-Bioscience) in DMEM/F12 incubated 1 hour at 37°C. Cells were added to the upper chamber in 200 μl of serum-free medium. For the invasion assay, 5000 cells were seeded on the Matrigel-coated filters and the lower chamber was filled with 600 μl of medium supplemented with 10% human serum (Cambrex) or with 600 μl of serum-free medium supplemented with IL8 (100ng/mL). After 48 hours incubation, the cells on the underside of the filter were counted using light microscopy. Relative invasion was normalized to the unseparated corresponding cell lines under serum condition.
Lentivirus infection
For luciferase gene transduction, 70% confluent cells from HCC1954, MDA-MB-453, and SUM159 were incubated overnight with a 1:3 precipitated mixture of lentiviral supernatants Lenti-LUC-VSVG (Vector Core, Ann Arbor, MI) in culture medium. The following day the cells were harvested by trypsin/EDTA and subcultured at a ratio of 1:6. After 1 week incubation, cells were sorted according to the ALDEFLUOR phenotype and luciferase expression was verified in each sorted population (ALDEFLUOR-positive and ALDEFLUOR-negative) by adding 2 μl D-luciferin 0.0003% (Promega, Madison, WI) in the culture medium and counting photon flux by device camera system (Xenogen, Alameda, CA) (Supplementary Figure 1).
Intracardiac inoculation
Six weeks-old NOD/SCID mice were anesthetized with 2% isofluorane/air mixture and injected in the heart left ventricle with 100,000 cells in 100 μL of sterile Dulbecco's PBS lacking Ca2+ and Mg2+. For each of the three cell lines (HCC1954, MDA-MB-453, SUM159) and for each population (ALDEFLUOR-positive, ALDEFLUOR-negative and unsorted), three animals were injected.
Bioluminescence detection
Baseline bioluminescence was assessed before inoculation and each week thereafter inoculations. Mice were anesthetized with a 2% isofluorane/air mixture and given a single i.p. dose of 150 mg/kg D-luciferin (Promega, Madison, WI) in PBS. For photon flux counting, we used a charge-coupled device camera system (Xenogen, Alameda, CA) with a nose-cone isofluorane delivery system and heated stage for maintaining body temperature. Results were analyzed after 2 to 12 minutes of exposure using Living Image software provided with the Xenogen imaging system. Signal intensity was quantified as the sum of all detected photon flux counts within a uniform region of interest manually placed during data postprocessing. Normalized photon flux represents the ratio of the photon flux detected each week after inoculations and the photon flux detected before inoculation.
Statistical analysis
Results are presented as the mean ±SD for at least three repeated individual experiments for each group. Statistical analyses used the SPSS software (version 10.0.5). Correlations between sample groups and molecular parameters were calculated with the Fisher's exact test or the one-way ANOVA for independent samples. A p-value <0.05 was considered significant.
Results
The majority of breast cell lines contain an ALDEFLUOR-positive population
We utilized the ALDEFLUOR assay (15) to isolate CSC from 33 BCLs representing the diverse molecular subtypes and features of breast cancer (18) (Supplementary Fig. 2). As shown in Fig. 1A, 23 out of the 33 cell lines contained an ALDEFLUOR-positive cell population that ranged from 0.2 to nearly 100%. All 16 basal/mesenchymal BCLs contained an ALDEFLUOR-positive population whereas 7 out of the 12 luminal BCLs did not contain any detectable ALDEFLUOR-positive cells (p=0.0006, Fischer's exact test) (Fig. 1B).
Fig. 1. Global gene expression profiling of 33 breast cell lines analyzed by the ALDEFLUOR assay.
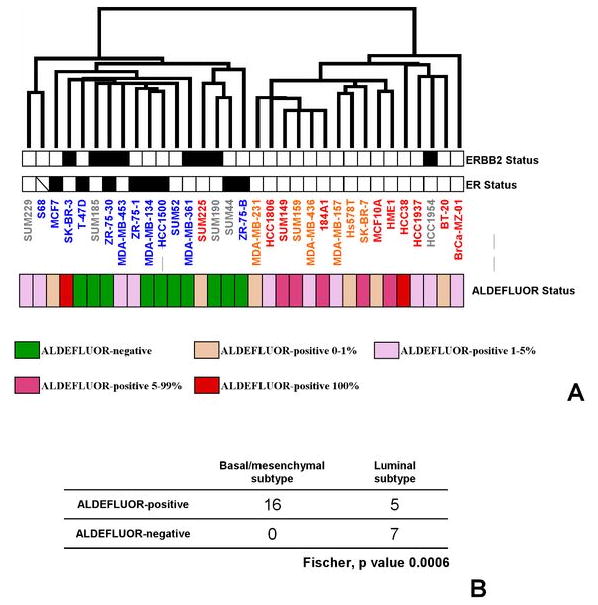
Hierarchical clustering of 33 breast cell lines and 13,550 genes/ESTs based on mRNA expression levels. A. The dendrogram of samples represents overall similarities in gene expression profiles. Two large groups of samples are evidenced by clustering. Name of cell lines is colored as follows: blue for luminal (n=12), red for basal (n=10), brown for mesenchymal (n=6) cell lines according to the correlation of expression profile of each cell line with each Ross and Perou centroid (i.e. molecular subtype). Five cell lines were not attributed any subtype (name in grey). The ER, ERBB2, and ALDEFLUOR status of breast cell lines are represented according to a color ladder (for ER and ERBB2 status: negative, white; positive, black; unavailable, oblique feature; for ALDEFLUOR status: a color scale shown at the bottom of the dendrogram relates the percentage of ALDELFUOR-positive cells found in each breast cell line). B. Comparison of the ALDEFLUOR status with the molecular subtypes of breast cell lines revealed a strong correlation between the basal/mesenchymal subtypes and the presence of ALDEFLUOR-positive cells (p-value=0.0006).
ALDEFLUOR-positive cells have tumorsphere-forming capacity
We have previously reported that mammary epithelial stem and progenitor cells are able to survive and proliferate in anchorage-independent conditions and form floating spherical colonies which we termed mammospheres (16). Data from breast tumors, as well as cell lines, have demonstrated that cancer stem-like cells or cancer-initiating cells can also be isolated and propogated as “tumorspheres” in similar assays (22). All mammosphere-initiating cells in the normal human mammary gland are contained within the ALDEFLUOR-positive population (15). To characterize the ALDEFLUOR-positive population from BCLs, we compared the ability of ALDEFLUOR-positive and -negative populations from 184A1, SUM149 and SUM159 to form tumorspheres. In each cell line, the ALDEFLUOR-positive population showed increased tumorsphere-forming capacity compared to ALDEFLUOR-negative cells (Supplementary Fig. 3).
ALDEFLUOR-positive BCL cells have cancer stem cell properties in vivo
To determine the hierarchical organization of BCL, we analyzed the stem cell properties of the ALDEFLUOR-positive and -negative populations of MDA-MB-453, SUM159, and BrCa-MZ-01 cell lines. The ALDEFLUOR-positive populations of these three BCLs constituted between 3.54±1.73% and 5.49±3.36% of the total cell populations (Supplementary Fig. 4). As shown in Fig. 2A-B the size and latency of tumor formation correlated with the number of ALDEFLUOR-positive cells injected. Remarkably, 500 ALDEFLUOR-positive cells from MDA-MB-453 and 1,000 ALDEFLUOR-positive cells from SUM159 were able to form tumors. The tumor-generating capacity was maintained through serial passages demonstrating the self-renewal capacity of these cells. In contrast, ALDEFLUOR-negative cells failed to generate tumors, although limited growth was produced when 50,000 ALDEFLUOR-negative MDA-MB-453 cells were injected (Supplementary Table 3). H&E staining of the fat pad sections confirmed that tumors formed by ALDEFLUOR-positive cells contained malignant cells whereas only residual Matrigel, apoptotic cells and mouse tissue were seen at the sites of ALDELFUOR-negative cell injections (Fig. 2C-D). Consistent with the ALDEFLUOR-positive population having cancer stem cell characteristics, tumors generated by this population recapitulated the phenotypic heterogeneity of the initial tumor, with a similar ratio of ALDEFLUOR-positive and -negative cells (Supplementary Fig.4). This indicates that ALDEFLUOR-positive cells were able to self-renew, generating ALDEFLUOR-positive cells and were able to differentiate, generating ALDEFLUOR-negative cells.
Fig. 2. The ALDEFLUOR-positive cell populations from breast cancer cell lines (MDA-MB-453, SUM159) have cancer stem cell properties.
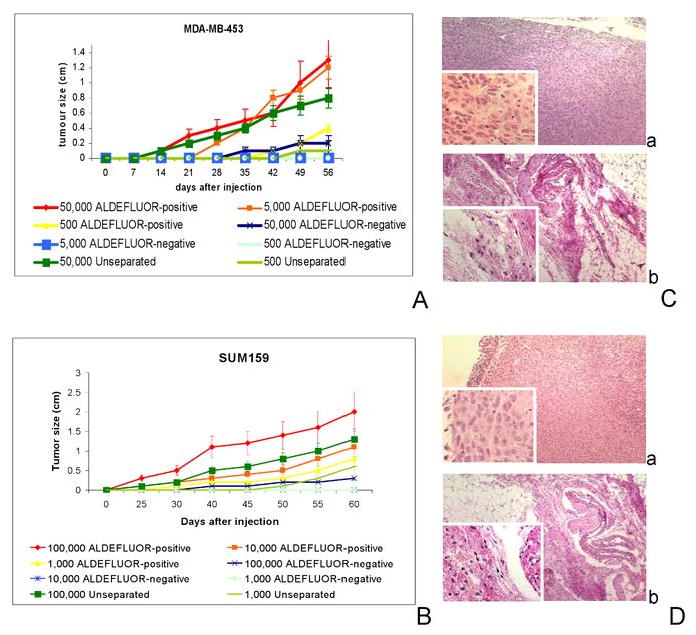
A-B. Tumor growth curves were plotted for different numbers of cells injected (for MDA-MB-453: 50,000 cells, 5,000 cells, and 500 cells and for SUM159: 100,000 cells, 10,000 cells, and 1,000 cells) and for each population (ALDEFLUOR-positive, ALDEFLUOR-negative, unseparated). Tumor growth kinetics correlated with the latency and size of tumor formation and the number of ALDEFLUOR-positive cells. C-D. H&E staining of ALDEFLUOR-positive cells' injection site, revealing presence of tumor cells (Ca: MDA-MB-453 ALDEFLUOR-positive cells' injection site, and Da: SUM59 ALDEFLUOR-positive cells' injection site). Cb, Db. The ALDEFLUOR-negative cells' injection site contained only residual Matrigel, apoptotic cells, and mouse tissue (Cb: MDA-MB-453 ALDEFLUOR-negative cells' injection site, and Db: SUM59 ALDEFLUOR-negative cells' injection site). Data represent mean ± SD.
When BrCa-MZ-01 cells were separated into ALDEFLUOR-positive and -negative components, both were capable of tumor generation. Tumors generated by the ALDEFLUOR-positive population consisted of both ALDEFLUOR-positive and -negative cells recapitulating the phenotypic heterogeneity of the initial tumor (Supplementary Fig. 5). In contrast, tumors generated by ALDEFLUOR-negative cells gave rise to slowly growing tumors containing only ALDEFLUOR-negative cells (Supplementary Fig. 5F, G). In contrast to the ability of ALDEFLUOR-positive cells to be serially transplanted, serial passages of ALDEFLUOR-negative tumors produced decreasing tumor growth with no growth following three passages. This suggests that the ALDEFLUOR-positive component of the BrCa-MZ-01 cells contain cells with stem cell properties, whereas the ALDEFLUOR-negative cells contain progenitor cells able to undergo limited growth but not self-renewal.
Gene expression profiling of ALDELFUOR-positive and -negative cell populations
To determine whether ALDEFLUOR-positive cells isolated from different BCLs expressed a common set of “cancer stem cell” genes, we analyzed ALDEFLUOR-positive and -negative cell populations isolated from eight BCLs (184A1, BrCa-MZ-01, HCC1954, MDA-MB-231, MDA-MB-453, SK-BR-7, SUM49, and SUM159) by using Affymetrix whole-genome oligonucleotide microarrays. Unsupervised hierarchical clustering, applied to the 16 samples and the 13,550 filtered genes/ESTs, did not separate ALDEFLUOR-positive and –negative populations (not shown). Instead, ALDEFLUOR-positive and –negative populations clustered with the parental cell line. This suggests that the differences in mRNA transcripts between clonal cell lines supersede differences between ALDEFLUOR-positive and ALDEFLUOR-negative cells. This further suggests that only a limited number of genes are differentially expressed between putative cancer stem cells and their progeny.
To determine which genes discriminated ALDEFLUOR-positive and -negative populations, the Mann and Whitney U test was applied to all genes but those with low and poorly measured expression, i.e. 25,285 probe sets. This test identified and ranked after FDR correction, 413 genes/ESTs that discriminated the ALDEFLUOR-positive and -negative cell populations. The 28 overexpressed genes corresponding to unique genes and the most frequently underexpressed genes are shown in Table 1 upper, lower. The classification power of this discriminating signature was illustrated by classifying the 16 ALDEFLUOR-positive and -negative samples with the 413 differentially expressed genes/ESTs. Hierarchical clustering ranked 15 out of the 16 samples (Supplementary Fig. 6).
Table 1.
Genes up- and down- regulated in ALDEFLUOR-positive populations
| Category | Symbol | Description | Cytoband | Probe set ID | Function | |
|---|---|---|---|---|---|---|
| UP-REGULATED | Genes previously described to have a role in stem cell biology | TPRXL | tetra-peptide repeat homeobox-like | chr3p25.1 | 239061_at | early embryonic development |
| NOTCH2 | Notch homolog 2 (Drosophila) | chr1p13-p11 | 202443_x_at | Self-renewal program | ||
| RBM15 | RNA binding motif protein 15 | chr1p13 | 1555760_a_at | determination of hematopoietic cell fate | ||
| ST3GAL3 | ST3 beta-galactoside alpha-2,3-sialyltransferase 3 | chr1p34.1 | 1555181_a_at | maintenance of the embryonic antigens SSEA-3 and -4 | ||
| NFYA | nuclear transcription factor Y, alpha | chr6p21.3 | 204107_at | Self-renewal program | ||
| PCNX | pecanex homolog (Drosophila) | chr14q24.2 | 213173_at | determination of neural cell fate of early developing embryo | ||
| Signaling | FBXO21 * | F-box protein 21 | chr12q24.22 | 212231_at | Ubiquitination | |
| WWOX | WW domain containing oxidoreductase | chr16q23.3-q24.1 | 210695_s_at | Protein degradation, transcription, and RNA splicing | ||
| CAMK2B | Calcium/calmodulin-dependent protein kinase (CaM kinase) II beta | chr22q12 | 34846_at | Calcium signaling | ||
| PNPLA2 | patatin-like phospholipase domain containing 2 | chr11p15.5 | 39854_r_at | Triglyceride hydrolysis | ||
| CLIC5 | chloride intracellular channel 5 | chr6p12.1-21.1 | 213317_at | chloride ion transport | ||
| UGCGL1 | UDP-glucose ceramide glucosyltransferase-like 1 | chr2q14.3 | 222569_at | Protein glucosylation | ||
| FBXL18 | F-box and leucine-rich repeat protein 18 | chr7p22.2 | 220896_at | Ubiquitination | ||
| ADRBK1 | adrenergic, beta, receptor kinase 1 | chr11q13 | 38447_at | Phosphorylation of G-protein-coupled receptors | ||
| SLC38A2 | Solute carrier family 38, member 2 | chr12q | 1559924_at | neutral amino acid transporter | ||
| Membrane protein | IL8RA* | interleukin 8 receptor, alpha | chr2q35 | 207094_at | Inflamatory response | |
| TAS2R14 | Taste receptor, type 2, member 14 | chr12p13 | 241997_at | Bitter perception | ||
| CD300LB | CD300 molecule-like family member b | chr17q25.1 | 1554173_at | Immune response | ||
| GIPC3 | GIPC PDZ domain containing family, member 3 | chr19p13.3 | 236730_at | |||
| DNA repair | RAD51L1 | RAD51-like 1 (S. cerevisiae) | chr14q23-q24.2 | 1570166_a_at | homologous recombination repair | |
| Chromatin remodeling | ARID1B | AT rich interactive domain 1B (SWI1-like) | chr6q25.1 | 225181_at | Chromatin remodeling (SWI/SNF complexe) | |
| Cytoskeleton | EPPK1 | epiplakin 1 | chr8q24.3 | 208156_x_at | Maintenance of the keratin intermediate filaments | |
| Extracellular matrix | COL11A2 | collagen, type XI, alpha 2 | chr6p21.3 | 216993_s_at | skeletal morphogenesis | |
| KLK3 | Kallikrein 3, (prostate specific antigen) | chr19q13.41 | 231629_x_at | Protease | ||
| RNA interference | EIF2C2 | Eukaryotic translation initiation factor 2C, 2 | chr8q24 | 213310_at | short-interfering-RNA-mediated gene silencing | |
| Unknown | ZFP41 | zinc finger protein 41 homolog (mouse) | chr8q24.3 | 227898_s_at | Unknown | |
| FAM49B | Family with sequence similarity 49, member B | chr8q24.21 | 243182_at | Unknown | ||
| PSORS1C2 | psoriasis susceptibility 1 candidate 2 | chr6p21.3 | 220635_at | Unknown | ||
| DOWN-REGULATED | Protein synthesis | MRPL42* | mitochondrial ribosomal protein L42 | chr12q22 | 217919_s_at | Protein synthesis within the mitochondrion |
| MRPL54* | mitochondrial ribosomal protein L54 | chr19p13.3 | 225797_at | Protein synthesis within the mitochondrion | ||
| MRPL47* | mitochondrial ribosomal protein L47 | chr3q26.33 | 223480_s_at | Protein synthesis within the mitochondrion | ||
| MRPS23* | mitochondrial ribosomal protein S23 | chr17q22-q23 | 223156_at | Protein synthesis within the mitochondrion | ||
| EIF3S9* | eukaryotic translation initiation factor 3, subunit 9 eta, 116kDa | chr7p22.2 | 236274_at | Initiation of protein synthesis (EIF3 multiprotein complex) | ||
| Signaling | ALG5* | asparagine-linked glycosylation 5 homolog | chr13q13.3 | 218203_at | Protein glucosylation | |
| DNAJC19* | DnaJ (Hsp40) homolog, subfamily C, member 19 | chr3q26.33 | 225359_at | Importation of mithochondrial protein | ||
| HBLD2* | HESB like domain containing 2 | chr9q21.33 | 221425_s_at | iron-sulfur cluster biogenesis | ||
| GART* | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | chr21q22.1 | 230097_at | de novo purine biosynthesis | ||
| NUP37* | nucleoporin 37kDa | chr12q23.2 | 218622_at | Intrcellular protein transport across nuclear membrane | ||
| RNF7* | ring finger protein 7 | chr3q22-q24 | 224439_x_at | subunit of SKP1-cullin/CDC53-F box protein ubiquitin ligases | ||
| DC2* | DC2 protein | chr4q25 | 223001_at | protein glycosylation | ||
| USP15* | ubiquitin specific peptidase 15 | chr12q14 | 210681_s_at | Protein degradation | ||
| COMMD6* | COMM domain containing 6 | --- | 225312_at | Inhibition NF-KappaB Signaling | ||
| UBL5* | Ubiquitin-like 5 | chr19p13.3 | 218011_at | Ubiquitination | ||
| Apoptosis | MRPL41* | mitochondrial ribosomal protein L41 | chr9q34.3 | 225425_s_at | Stabilization of p53 protein, Cell cycle arrest (p21(WAF1/CIP1) and p27(Kip1) dependent) | |
| PDCD10* | programmed cell death 10 | chr3q26.1 | 210907_s_at | Initiation of apoptosis | ||
| PDCD5* | Programmed cell death 5 | chr19q12- | 227751_at | Initiation of apoptosis | ||
| Differentiation program | NACA* | nascent-polypeptide-associated complex alpha polypeptide | chr12q23-q24.1 | 222018_at | Erythroid differentiation | |
| Cell cycle | FAM82B* | family with sequence similarity 82, member B | chr8q21.3 | 218549_s_at | Regulation of microtubule dynamic | |
| RNA splicing | CCNL1* | cyclin L1 | chr3q25.32 | 1555411_a_at | Pre-mRNA processing | |
| PRPF39* | PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae) | chr14q21.3 | 220553_s_at | Pre-mRNA processing | ||
| LSM3* | LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | chr3p25.1 | 202209_at | Pre-mRNA processing | ||
| SFRS7* | splicing factor, arginine/serine-rich 7, 35kDa | chr2p22.1 | 213649_at | Regulation of RNA splicing | ||
| PRPF4B* | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | chr6p25.2 | 202127_at | Pre-mRNA processing | ||
| Oxidative phosphorylation | ATP5S* | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s (factor B) | chr14q22.1 | 206992_s_at | subunit of mitochondrial ATP synthase | |
| NDUFA2* | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, | chr5q31 | 209224_s_at | components of the complex I multi-subunit enzyme | ||
| ATP5J2* | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 | chr7q22.1 | 202961_s_at | subunit of mitochondrial ATP synthase | ||
| IMMP1L* | IMP1 inner mitochondrial membrane peptidase-like | chr11p13 | 230556_at | Proteolysis | ||
| Unknown | ASTE1* | asteroid homolog 1 (Drosophila) | chr3q22.1 | 221135_s_at | Unknown | |
| MGC61571* | hypothetical protein MGC61571 | chr3p24.1 | 228283_at | Unknown | ||
| WDR53* | WD repeat domain 53 | chr3q29 | 227814_at | Unknown | ||
| DKFZP686A10121* | hypothetical protein | chr7q21.13 | 234311_s_at | Unknown | ||
| CHCHD8* | coiled-coil-helix-coiled-coil-helix domain containing 8 | chr11q13.4 | 220647_s_at | Unknown | ||
| FLJ32745* | hypothetical protein FLJ32745 | chr2q13 | 235644_at | Unknown | ||
| CHURC1* | churchill domain containing 1 | chr14q23.3 | 233268_s_at | Unknown | ||
| XTP3TPA* | XTP3-transactivated protein A | chr16p11.2 | 218069_at | Unknown | ||
| FLJ37953* | hypothetical protein FLJ37953 | chr2q33.1 | 235181_at | Unknown | ||
| SNORD50A* | small nucleolar RNA, C/D box 50A | chr6q14.3 | 244669_at | Unknown | ||
| LOC644053* | hypothetical protein LOC644053 | chr1q41 | 235466_s_at | Unknown | ||
| TMEM141* | transmembrane protein 141 | chr9q34.3 | 225568_at | Unknown | ||
| C8orf59* | chromosome 8 open reading frame 59 | chr8q21.2 | 226165_at | Unknown |
These tables comprise the 28 up-regulated genes in ALDEFLUOR-positive population (upper part of the table) and the 42 down-regulated genes in ALDEFLUOR-positive population (lower part), common between the 413-gene and the 49-gene signatures (table 2). They are grouped together according to a common biological function and ranked according to their discriminating score (DS). The DS was calculated for each gene as DS=(M1-M2)/(S1+S2) where M1 and S1 respectively represent mean and standard deviation of expression levels of the gene in subgroup 1, and M2 and S2 in subgroup 2. Because of multiple hypothesis testing, confidence levels were estimated by 100 iterative random permutations of samples as previously described with a false positive rate of 1/1.000(50).
follows the name of genes found in the 49-gene signature. Genes underlined are cited in the text (results and discussion sections).
A number of genes known to play a role in stem cell biology were upregulated in the ALDEFLUOR-positive populations (Table 1, upper), including NFYA, NOTCH2, PCNX, RBM15, ST3GAL3, and TPRXL. Other genes encode proteins that have putative or uncharacterized role in stem cell function, such as ARID1B, RAD51L1, and the chemokine receptor CXCR1/IL8RA (23). Genes underexpressed in the ALDEFLUOR-positive population are involved in cell differentiation, apoptosis, RNA splicing, and mitochondrial metabolism.
To increase the stringency of analysis, we raised the threshold of the Mann and Whitney analysis to the 0.5 risk and obtained a list of 49 genes/ESTs that discriminated ALDELFLUOR-positive and -negative populations (genes with asterisk in Table 1). With this list, all of the ALDEFLUOR-positive cells, except from SK-BR-7, clustered together (Supplementary Fig. 7A). Among these 49 genes/ESTS, 45 corresponded to identified unique genes; only 3 of these 45 were overexpressed in the ALDEFLUOR-positive group while 42 were underexpressed. Characterized overexpressed genes code for an F-box protein FBXO21 and CXCR1/IL8RA. Underexpressed genes include those coding for mitochondrial proteins (MRPL41, MRPL42, MRPL47, MRPL54, MRPS23, IMMP1L), and differentiation (NACA) and pre-mRNA splicing factors (LSM3, pre-mRNA processing factor PRPF39 and PRPF4B).
Leave-one-out cross-validation (LOOCV) at 0.5% risk estimated the accuracy of prediction of the identifier molecular signature and 88% of the samples were predicted in the right class with this “cancer stem cell signature” confirming the supervised analysis (Supplementary Fig. 7B-C).
Quantitative RT-PCR assessment confirmed a significant increase of CXCR1 and FBXO21 in ALDEFLUOR-positive cells
We performed guantitative RT–PCR analysis of five discriminator genes overexpressed in ALDEFLUOR-positive populations (CXCR1/IL8RA, FBXO21, NFYA, NOTCH2 and RAD51L1). Three cell lines used in the profiling analysis (BrCa-MZ-01, MDA-MB-453, SUM159) and two additional luminal cell lines (MCF7, S68) were sorted by ALDEFLUOR-assay and ALDEFLUOR-positive and -negative populations were processed separately for quantitative RT-PCR analysis. The quantitative RT-PCR expression level of CXCR1 and FBXO21 are presented in Fig. 3 A and B. Analyses of NFYA, NOTCH2 and RAD51L1 are presented in Supplementary Fig. 8. Gene expression levels measured by quantitative RT-PCR confirmed the results obtained using DNA microarrays with an increase of CXCR1 and FBXO21 mRNA level in the ALDEFLUOR-positive population compared to the ALDEFLUOR-negative population (p<0.05).
Fig. 3. Validation of gene expression results by quantitative RT-PCR.
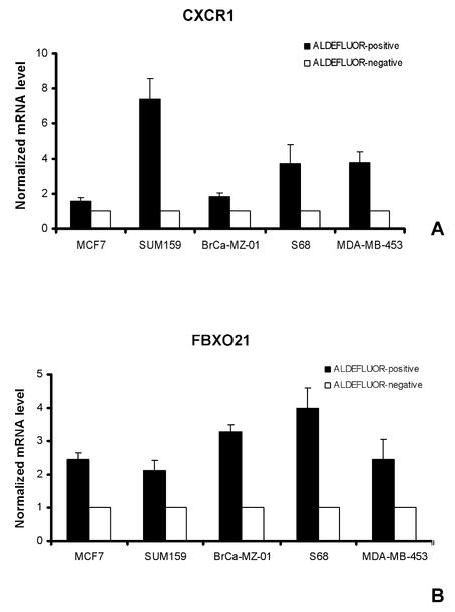
A-B. To confirm our gene expression results, we measured in a set of five breast cancer cell lines sorted for the ALDEFLUOR phenotype the expression of five discriminator genes overexpressed in ALDEFLUOR-positive populations (CXCR1/IL8RA, FBXO21, NFYA, NOTCH2 and RAD51L1) by quantitative RT-PCR. The quantitative RT-PCR expression levels of CXCR1 and FBXO21 are presented in this figure and the ones of NFYA, NOTCH2, and RAD51L1 are presented in Supplementary Fig. 8. Gene expression levels measured by quantitative RT-PCR confirm the results obtained using DNA microarrays with an increase of CXCR1 and FBXO21 mRNA level in the ALDEFLUOR-positive population compared to the ALDEFLUOR-negative population (p<0.05).
IL8 promotes cancer stem cell self-renewal
Our profiling studies suggested that the IL8 receptor CXCR1/IL8RA was consistently expressed in the ALDEFLUOR-positive cell population. To confirm this association we measured the protein expression of CXCR1/IL8RA by flow cytometry in ALDEFLUOR-positive and -negative populations. The ALDEFLUOR-positive and -negative populations from four different cell lines were isolated by FACS, fixed, and stained with a CXCR1 monoclonal antibody labeled with phycoerythrin. ALDEFLUOR-positive cells were highly enriched in CXCR1-positive cells compared to the ALDEFLUOR-negative populations (Supplementary Fig. 9).
To determine whether IL8 signaling is important in stem cell function, we treated four BCLs with human recombinant IL8 and determined its effect on the cancer stem cell population as measured by the formation of tumorspheres and by ALDH enzymatic activity. As shown in Fig. 4A, addition of IL8 increased the formation of primary and secondary tumorspheres in a dose-dependent manner. Furthermore, IL8 increased the ALDEFLUOR-positive population in a dose-dependent manner in each of the four BCLs analyzed (Fig. 4B). This illustrates the power of the “CSC signature” to identify pathways that may play a role in stem cell function.
Fig. 4. Role of the IL8/CXCR1 axis in the regulation of breast cancer stem cells.
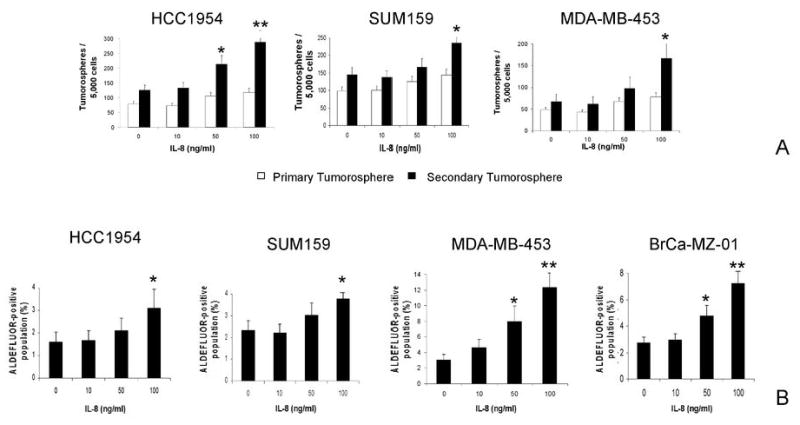
A. Effect of IL8 treatment on tumorosphere formation of three different cell lines (HCC1954, SUM159, MDA-MB-453). IL8 treatment increased the formation of primary and secondary tumorospheres in a dose-dependent manner. B. Effect of IL8 treatment on the ALDEFLUOR-positive population of four different cell lines cultured in adherent conditions. IL8 increased the ALDEFLUOR-positive population in a dose-dependent manner in each of the four cell lines analyzed (* p<0.05/ ** p<0.01, statistically significant differences from the control group).
The IL8/CXCR1 axis is involved in cancer stem cell invasion
The IL8/CXCR1 axis has been reported to play a role in cancer stem cell invasion (24, 25). We first utilized a Matrigel invasion assay, using serum as attractant, to examine the ability of ALDEFLUOR-positive and -negative cell populations from three different cell lines (HCC1954, MDA-MB-453, SUM159) to invade. As shown in Fig. 5A, ALDEFLUOR-positive cells demonstrated 6- to 20-fold higher invasion through Matrigel than the ALDEFLUOR-negative population (p<0.01). When used as a chemo-attractant IL8 (100 ng/ml) increased invasion of the ALDEFLUOR-positive cells (p<0.05) (Fig. 5A). In contrast to its effects on ALDEFLUOR-positive cells, IL8 did not have any effect on the invasive capacity of ALDELFLUOR-negative cells. These results indicate that cancer stem cells exhibited invasive behavior and furthermore that IL8 facilitates this process.
Fig. 5. ALDEFLUOR-positive cells display increased metastatic potential.
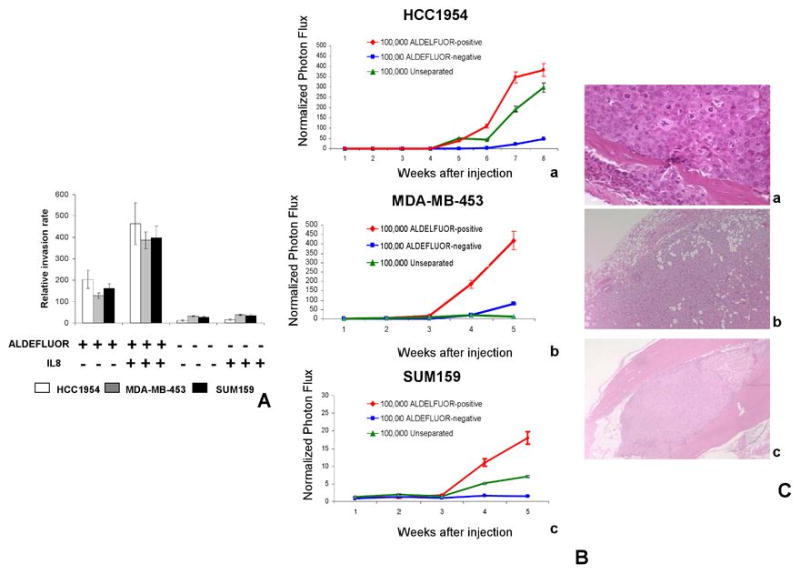
A. The IL8/CXCR1 axis is involved in cancer stem cell invasion. The role of the IL8/CXCR1 axis in invasion was assessed by a Matrigel invasion assay using serum or IL8 as attractant for three different cell lines (HCC1954, MDA-MB-453, SUM159). ALDEFLUOR-positive cells were 6- to 20-fold more invasive than ALDEFLUOR-negative cells (p<0.01). When using IL8 (100 ng/ml) as attractant, we observed a significant increase of ALDEFLUOR-positive cells invading through Matrigel compared to serum as attractant (p<0.05). In contrast IL8 had no effect on the invasive capacity of the ALDEFLUOR-negative population. B-C. The ALDEFLUOR-positive population displayed increased metastatic potential. Ba-c. Quantification of the normalized photon flux measured at weekly intervals following inoculation of 100,000 luciferase infected cells from each group (ALDEFLUOR-positive, ALDEFLUOR-negative, unseparated). Mice inoculated with ALDEFLUOR-positive cells developed several metastasis localized at different sites (bone, muscle, lung, soft tissue) and displayed a higher photon flux emission than mice inoculated with unseparated cells, which developed no more than one metastasis per mouse. In contrast, mice inoculated with ALDEFLUOR-negative cells developed only an occasional small metastasis, which was limited to lymph nodes. C. Histologic confirmation, by H&E staining, of metastasis in bone (Ca), soft tissue (Cb) and muscle (Cc) resulting from injection of ALDEFLUOR-positive cells.
ALDEFLUOR-positive cells have increased metastatic potential
It has been proposed that CSCs play a crucial role in cancer metastasis (26, 27). The above experiments demonstrated that ALDEFLUOR-positive cells have increased invasive capacity compared to ALDEFLUOR-negative cells. To determine the relationship between ALDEFLUOR-positivity and metastatic capacity, we infected HCC1954, MDA-MB-453, and SUM159 with a luciferase lentivirus reporter system. Luciferase-infected cells were sorted using the ALDEFLUOR assay and introduced into NOD/SCID mice by intracardiac injection. A suspension of 100,000 cells from each population was injected and metastasis was assessed by bioluminescent imaging. Mice inoculated with ALDEFLUOR-positive cells developed metastases at different sites and displayed a higher photon flux emission than mice inoculated with unseparated cells, which developed no more than one metastasis per mouse, or mice inoculated with ALDEFLUOR-negative cells, which developed only occasional metastases limited to lymph nodes (Fig. 5B-C and Supplementary Fig. 10). Histologic sections confirmed the presence of metastases at these sites (Fig. 5C). Thus, the metastatic capacity of BCLs is predominantly mediated by CSCs contained in the ALDEFLUOR-positive population.
Discussion
The hypothesis that tumors are organized in a cellular hierarchy driven by CSCs has fundamental implications for cancer biology as well as clinical implications for the early detection, prevention and treatment of cancer. Evidence for CSCs has largely relied on primary and early passage xenograft models (28, 29). However, the success of establishing breast tumor xenograft has been low particularly for certain molecular subtypes. In contrast to primary tumors, cell lines are available in unlimited quantities and provide only carcinomatous populations for molecular analysis without normal tissue and stroma. In breast cancer, a large number of immortalized cell lines have been produced which represent the different molecular subtypes found in primary human breast cancers (2, 18). However, a fundamental question remains as to how closely these cell lines are able to recapitulate the biology of human breast cancer.
In vivo evidence for stem cells in cell lines
Recent studies have suggested that although cell lines may be clonally derived, they contain a cellular hierarchy representing different stages of cellular differentiation. Several studies have utilized markers such as CD44+/CD24- to identify CSC within breast cancer cell lines. However, their utility is limited by the observation that frequently a large percentage of cells within a cell line express these putative stem cell markers. For example, greater than 90% of cells in basal breast cancer cell lines display the CD44+/CD24- phenotype. Indeed, the CD44+/CD24- phenotype did not isolate the tumorigenic population of these cell lines (8). An alternative approach has been to use the SP from cell lines. However, functional studies utilizing Hoechst staining are limited by the toxicity of this agent (30). There is also evidence that the functional stem cell activity is not contained within the SP(31).
ALDH activity assessed by the ALDEFLUOR assay isolates cells with stem cell properties from various cancers (13). We demonstrate here that 23 out of 33 BCLs (predominantly basal cell lines) contain an ALDEFLUOR-positive population. Lack of an ALDEFLUOR-positive population in some luminal BCLs may indicate that these luminal BCLs are derived from ALDEFLUOR-negative progenitor cells.
We utilized in vivo assays in NOD/SCID mice to demonstrate the stem cell properties of the ALDEFLUOR-positive populations. Self-renewal was demonstrated by serial passage in NOD/SCID mice and differentiation was demonstrated by the ability of ALDEFLUOR-positive but not ALDEFLUOR-negative cells to regenerate the cellular heterogeneity of the initial tumor.
A breast cancer stem cell signature
Utilizing eight breast cell lines, we identified 413 genes whose expression discriminates ALDEFLUOR-positive and -negative cells. This signature contained a number of genes known to play a role in stem cell biology. Genes overexpressed in the ALDEFLUOR-positive population include Notch homolog 2 (NOTCH2), which regulates self-renewal and differentiation of mammary stem cells (16, 32), NFYA, known to regulate self-renewal and differentiation of stem cells. (33), pecanex homolog PCNX, RBM15/OTT, which plays a pleiotropic role in hematopoietic stem cells (34) and affects myeloid differentiation via NOTCH signaling (35), homeobox-like factor TPRXL involved in embryonic development, ST3GAL3, which codes for a stage-specific embryonic antigen-4 synthase, associated with fetal development and renal and gastric carcinogenesis (36). Notably, stage-specific embryonic antigen-4 protein (SSEA-4) is expressed in stem cell populations such as CXCR4+/CD133+/CD34+/lin- stem cells in human cord blood and quiescent mammary stem cells (37).
Genes underexpressed in the ALDEFLUOR-positive population are involved in cell differentiation, apoptosis, and mitochondrial oxidation. They include genes coding for nascent polypeptide-associated complex alpha subunit NACA, programmed death proteins PDCD5 and PDCD10, mitochondrial ribosomal protein L41 (MRPL41), which induces apoptosis through P53-dependent and independent manner via BCL2 and caspases, and proteins involved in mitochondrial processes such as oxidative phosphorylation (NDUFA2, ATP5J2, IMMP1L) and protein synthesis in the mitochondrion (MRPL42, MRPL47, MRPL54, MRPS23). Downregulation of apoptotic genes in CSCs may play a role in the resistance of these cells to radiation and chemotherapy (38). ALDH1A1 was not identified as a differentially-expressed gene in the ALDEFLUOR-positive signature. However, examination of gene expression profile of individual BCLs revealed that although some showed differential expression of ALDH1A1 in the ALDEFLUOR-positive population, others showed differential expression of ALDH1A3, a different ALDH isoform in this population. This suggests that the expression of different ALDH isoforms could contribute to the ALDEFLUOR-positive phenotype.
From chemokines to “stemokines”
The expression of CXCR1, a receptor for IL8, is increased in a variety of cancers (39, 40). Although IL8 expression is associated with ER-negative breast cancer (41), this chemokine has not previously been reported to play a role in stem cell function. Its implication in the regulation of growth and metastasis is well-established in androgen-independent prostate cancer (42). Futhermore, the expression level of IL8 is associated with tumorigenicity and metastasis through VEGF production and angiogenesis (43, 44). We validated the gene expression data in three ways. First, quantitative RT-PCR analysis confirmed a significant increase of CXCR1 mRNA in ALDEFLUOR-positive population from cell lines both included and not included in profiling analysis. Second, we demonstrated using flow cytometry that CXCR1-containing cells were found exclusively within the ALDEFLUOR-positive population. Third, recombinant IL8 increased mammosphere formation and the percent of ALDEFLUOR-positive cells in BCLs. The IL8/CXCR1 axis may thus regulate mammary stem cell proliferation or self-renewal. Since endothelial and stromal cells secrete IL8 this chemokine may play a role in mediating interactions between tumor stem cells and the tumor microenvironment.
Recent studies have suggested a role for interleukines/chemokines in the regulation of CSCs (45, 46). This includes a role for IL6 in breast CSCs and IL4 in mediating chemoresistance of colon CSCs (46-48). These factors may be involved in the association between inflammation and cancer. This also includes a role for CCL5 (RANTES), a chemokine secreted by mesenchymal stem cells, which acts as a paracrine factor and enhance breast cancer cells motility, invasion and metastasis(45).
The roots of metastasis
CSCs may be responsible for mediating tumor metastasis. A link between CSC and metastasis was first suggested with the identification of stem cell genes in an 11-gene signature generated using comparative profile of metastatic and primary tumors in transgenic mouse model of prostate cancer and cancer patients (49). This signature was also a powerful predictor of disease recurrence, death after therapy and distant metastasis in a variety of cancer types. We have demonstrated that ALDEFLUOR-positive cells are more metastatic than ALDEFLUOR-negative cells and that IL8, previously reported to play a role in tumor metastasis, promotes the invasion and chemotaxis of cancer stem cells which preferentially express the IL8 receptor CXCR1. The ability to isolate metastatic cancer stem cell from cell lines should facilitate studies of the molecular mechanisms by which cancer stem cells mediate tumor metastasis.
Supplementary Material
Acknowledgments
Thanks are due to the University of Michigan Cancer Center Flow Cytometry core. This work was supported by NIH grants CA66233 and CA101860, the University of Michigan Cancer Center NIH Support Grant 5 P 30 CA46592, Inserm, Institut Paoli-Calmettes, Taubman Institute and grants from Ministries of Health and Research (PHRC 2006, PHRC2007), Institut national du Cancer-Institut Lilly (AO 2006), Institut National du Cancer (PL2005, ACI2007), Ligue Nationale Contre le Cancer (Label DB).
Footnotes
Note: Max S. Wicha has financial holdings and is a scientific advisor for OncoMed Pharmaceuticals.
References List
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 4.Glinsky GV. Stem cell origin of death-from-cancer phenotypes of human prostate and breast cancers. Stem Cell Rev. 2007;3:79–93. doi: 10.1007/s12015-007-0011-9. [DOI] [PubMed] [Google Scholar]
- 5.Jaiswal S, Traver D, Miyamoto T, Akashi K, Lagasse E, Weissman IL. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci USA. 2003;100:10002–7. doi: 10.1073/pnas.1633833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 7.Christgen M, Ballmaier M, Bruchhardt H, von Wasielewski R, Kreipe H, Lehmann U. Identification of a distinct side population of cancer cells in the Cal-51 human breast carcinoma cell line. Mol Cell Biochem. 2007;306:201–12. doi: 10.1007/s11010-007-9570-y. [DOI] [PubMed] [Google Scholar]
- 8.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101:781–6. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Setoguchi T, Taga T, Kondo T. Cancer stem cells persist in many cancer cell lines. Cell Cycle. 2004;3:414–5. doi: 10.4161/cc.3.4.799. [DOI] [PubMed] [Google Scholar]
- 11.Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and A. Cancer Res. 2005;65:6207–19. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 12.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–12. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung AM, Wan TS, Leung JC, et al. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007;21:1423–30. doi: 10.1038/sj.leu.2404721. [DOI] [PubMed] [Google Scholar]
- 14.Corti S, Locatelli F, Papadimitriou D, et al. Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24:975–85. doi: 10.1634/stemcells.2005-0217. [DOI] [PubMed] [Google Scholar]
- 15.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–84. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua J, Balagurunathan Y, Chen Y, et al. Normalization benefits microarray-based classification. EURASIP J Bioinform Syst Biol. 2006;43056 doi: 10.1155/BSB/2006/43056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginestier C, Cervera N, Finetti P, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12:4533–44. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 22.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–11. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 23.Ringe J, Strassburg S, Neumann K, et al. Towards in situ tissue repair: human mesenchymal stem cells express chemokine receptors CXCR1, CXCR2 and CCR2, and migrate upon stimulation with CXCL8 but not CCL2. J Cell Biochem. 2007;101:135–46. doi: 10.1002/jcb.21172. [DOI] [PubMed] [Google Scholar]
- 24.Hughes L, Malone C, Chumsri S, Burger AM, McDonnell S. Characterisation of breast cancer cell lines and establishment of a novel isogenic subclone to study migration, invasion and tumourigenicity. Clin Exp Metastasis. 2008;25:549–57. doi: 10.1007/s10585-008-9169-z. [DOI] [PubMed] [Google Scholar]
- 25.Itoh Y, Joh T, Tanida S, et al. IL-8 promotes cell proliferation and migration through metalloproteinase-cleavage proHB-EGF in human colon carcinoma cells. Cytokine. 2005;29:275–82. doi: 10.1016/j.cyto.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Gupta GP, Perk J, Acharyya S, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104:19506–11. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Tiede B, Massague J, Kang Y. Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res. 2007;17:3–14. doi: 10.1038/sj.cr.7310118. [DOI] [PubMed] [Google Scholar]
- 28.Al Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 30.Montanaro F, Liadaki K, Schienda J, Flint A, Gussoni E, Kunkel LM. Demystifying SP cell purification: viability, yield, and phenotype are defined by isolation parameters. Exp Cell Res. 2004;298:144–54. doi: 10.1016/j.yexcr.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 32.Farnie G, Clarke RB. Mammary stem cells and breast cancer--role of Notch signalling. Stem Cell Rev. 2007;3:169–75. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci U S A. 2005;102:11728–33. doi: 10.1073/pnas.0503405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raffel GD, Mercher T, Shigematsu H, et al. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:6001–6. doi: 10.1073/pnas.0609041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma X, Renda MJ, Wang L, et al. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27:3056–64. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peiffer I, Eid P, Barbet R, et al. A sub-population of high proliferative potential-quiescent human mesenchymal stem cells is under the reversible control of interferon alpha/beta. Leukemia. 2007;21:714–24. doi: 10.1038/sj.leu.2404589. [DOI] [PubMed] [Google Scholar]
- 37.Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, et al. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol. 2007;177:87–101. doi: 10.1083/jcb.200611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hambardzumyan D, Becher OJ, Holland EC. Cancer stem cells and survival pathways. Cell Cycle. 2008;7:1371–8. doi: 10.4161/cc.7.10.5954. [DOI] [PubMed] [Google Scholar]
- 39.Maxwell PJ, Gallagher R, Seaton A, et al. HIF-1 and NF-kappaB-mediated upregulation of CXCR1 and CXCR2 expression promotes cell survival in hypoxic prostate cancer cells. Oncogene. 2007;26:7333–45. doi: 10.1038/sj.onc.1210536. [DOI] [PubMed] [Google Scholar]
- 40.Murphy C, McGurk M, Pettigrew J, et al. Nonapical and cytoplasmic expression of interleukin-8, CXCR1, and CXCR2 correlates with cell proliferation and microvessel density in prostate cancer. Clin Cancer Res. 2005;11:4117–27. doi: 10.1158/1078-0432.CCR-04-1518. [DOI] [PubMed] [Google Scholar]
- 41.Freund A, Chauveau C, Brouillet JP. IL-8 expression and its possible relationship with estrogen-receptor-negative status of breast cancer cells. Oncogene. 2003;22:256–65. doi: 10.1038/sj.onc.1206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue K, Slaton JW, Eve BY, et al. Interleukin 8 expression regulates tumorigenicity and metastases in androgen-independent prostate cancer. Clin Cancer Res. 2000;6:2104–19. [PubMed] [Google Scholar]
- 43.Balbay MD, Pettaway CA, Kuniyasu H, et al. Highly metastatic human prostate cancer growing within the prostate of athymic mice overexpresses vascular endothelial growth factor. Clin Cancer Res. 1999;5:783–9. [PubMed] [Google Scholar]
- 44.Kim SJ, Uehara H, Karashima T, Mccarty M, Shih N, Fidler IJ. Expression of interleukin-8 correlates with angiogenesis, tumorigenicity, and metastasis of human prostate cancer cells implanted orthotopically in nude mice. Neoplasia. 2001;3:33–42. doi: 10.1038/sj.neo.7900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 46.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todaro M, Alea MP, Di Stefano AB, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Sansone P, Storci G, Tavolari S, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–21. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


