Abstract
Obscurin is a giant structural and signaling protein that participates in the assembly and structural integrity of striated myofibrils. Previous work has examined the physical interactions between obscurin and other cytoskeletal elements but its in vivo role in cell signaling, including the functions of its RhoGTPase Exchange Factor (RhoGEF) domain have not been characterized. In this study, morpholino antisense oligonucleotides were used to create an in-frame deletion of the active site of the obscurin A RhoGEF domain in order to examine its functions in zebrafish development. Cardiac myocytes in the morphant embryos lacked the intercalated disks that were present in controls by 72 hpf and, in the more severely affected embryos, the contractile filaments were not organized into mature sarcomeres. Neural abnormalities included delay or loss of retinal lamination. Rescue of the phenotype with co-injection of mini-obscurin A expression constructs demonstrated that the observed effects were due to the loss of small GTPase activation by obscurin A. The immature phenotype of the cardiac myocytes and the retinal neuroblasts observed in the morphant embryos suggests that obscurin A-mediated small GTPase signaling promotes tissue-specific cellular differentiation. This is the first demonstration of the importance of the obscurin A-mediated RhoGEF signaling in vertebrate organogenesis and highlights the central role of obscurin A in striated muscle and neural development.
INTRODUCTION
The Rho GTPases have very important roles in directing a wide range of cellular events including transcriptional regulation, cell growth and differentiation, and cell migration (Govek et al., 2005; Hall, 2005; Jaffe and Hall, 2005; Luo, 2000; Van Aelst and D’Souza-Schorey, 1997). RhoGTPase activity in turn is regulated by secondary factors that either facilitate the conversion of the GTPase from its inactive to active state [such as G-protein coupled receptors (GPCRs) and guanine nucleotide exchange factors (GEFs)] or inhibit GTPase activation or activity [such as GTPase activating proteins (GAPs) and guanine nucleotide dissociation inhibitors (GDIs)] (Koh, 2007; Schmidt and Hall, 2002). Many of these secondary factors are localized to specific intracellular locations such that the ratio of active to inactive small GTPases can be precisely regulated at that site. Quite commonly these secondary factors are clustered at sites of linkage between the cellular cytoskeleton and transmembrane adhesive complexes where they help to translate extracellular signals into intracellular responses. Each member of the Rho small GTPase family, which includes RhoA, RhoG, TC10, Rac1, and Cdc42, activates a unique set of target proteins and signaling pathways, eliciting distinct patterns of cellular rearrangements [reviewed in (Wennerberg and Der, 2004)].
While much has been learned regarding the importance of activated Rho GTPases and their downstream effectors during development, much less is known about the factors that regulate their activity. Recently, a novel giant structural and signaling protein, obscurin, was identified that has a RhoGEF domain and is highly expressed during vertebrate development (Raeker et al., 2006; Russell et al., 2002; Young et al., 2001). Previous work has determined that there are multiple isoforms of obscurin (Russell et al., 2002) and that the best characterized, obscurin A, localizes to the M band and to a lesser extent, the Z disk of striated myofibrils (Borisov et al., 2004; Kontrogianni-Konstantopoulos and Bloch, 2005; Raeker et al., 2006; Young et al., 2001). It facilitates myosin integration into the sarcomere (Borisov et al., 2004; Borisov et al., 2006; Kontrogianni-Konstantopoulos and Bloch, 2005; Kontrogianni-Konstantopoulos et al., 2004; Kontrogianni-Konstantopoulos et al., 2006; Raeker et al., 2006), organizes the sarcoplasmic reticulum (Bagnato et al., 2003; Kontrogianni-Konstantopoulos et al., 2003; Lange et al., 2009; Raeker et al., 2006) and promotes the lateral alignment of myofibrils (Borisov et al., 2006; Raeker et al., 2006).
In previous studies in our laboratory, we used the zebrafish as a model to examine the functions of obscurin A (Raeker et al., 2006). Due to the duplication of the zebrafish genome, there are two copies of the obscurin gene, one of which encodes for obscurin A and related isoforms lacking the carboxy terminal kinase domains and one of which encodes for isoforms containing the kinase domains including the isoform obscurin B. This genetic duplication allows for isoform-specific evaluations of obscurin function. Given the proposed role of the obscurin A isoform in myofibril assembly, this isoform was specifically targeted for depletion using antisense morpholinos in developing zebrafish embryos resulting in defects in skeletal muscle, cardiac, and central nervous system development (Raeker et al., 2006). In skeletal muscle, the myofibrils were disorganized and elongated, often extending past where the vertical myoseptum, the zebrafish equivalent of the myotendinous junction, should have been. Cardiac manifestations included ventricular hypoplasia with reduced contractility and pericardial edema indicative of congestive heart failure. More severely affected embryos demonstrated abnormalities of central nervous system development. The manifestations of obscurin A depletion were likely due at least in part to disruption of its physical interactions with other structural elements including titin (Bang et al., 2001; Fukuzawa et al., 2008; Young et al., 2001), myomesin (Fukuzawa et al., 2008) and ankyrin (Bagnato et al., 2003; Kontrogianni-Konstantopoulos et al., 2003). However, the functions of obscurin A’s signaling domains and the contribution of their loss to the observed obscurin depletion phenotype remained to be determined.
In this study, we examined the effects of disruption of obscurin A RhoGEF signaling on zebrafish development. Intron-exon and exon-intron targeted antisense morpholino constructs were used to induce exon skipping, resulting in the in-frame deletion of an exon encoding for part of the active site of the obscurin A RhoGEF domain. As noted with obscurin A depletion, specific inhibition of the RhoGEF domain resulted in abnormalities of central nervous system, cardiac and skeletal muscle development. However, at the cellular level there were significant differences in the obscurin and obscurin RhoGEF morphant phenotypes, suggesting important roles for obscurin RhoGEF signaling in vertebrate neural and striated muscle development. Specifically, inhibition of obscurin RhoGEF signaling delayed advanced differentiation of lineage-committed but immature cardiac myocytes and neuroblasts.
MATERIALS AND METHODS
Zebrafish Maintenance and Breeding
Wild-type adult and cmlc2-EGFP transgenic zebrafish were maintained as described previously (Raeker et al., 2006).
Morpholino Oligonucleotide Microinjection
Two independent morpholino antisense oligonucleotides (ObscRhoMOs) targeting the intron-exon and exon-intron junctions of an exon encoding for the active site of the obscurin RhoGEF domain were designed by GENE TOOLS, LLC (Philomath, OR) [intron-exon MO, 5′-CTGAATAATCCTGATGGATAAAAGG-3′; exon-intron MO, 5′-TTAAACTACCTTCAGTAGAGCCCTG-3′]. ObscRhoMOs were co-injected at the following concentrations: 0.1 mM each for a total of 0.2 mM (1.5 ng/nl) or 0.2 mM each for a total of 0.4 mM (3.0 ng/nl). A standard control (Gene Tools, Inc.) MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was used at 0.4 mM concentration for control injections. The obscurin A antisense morpholino (MO2 from Raeker et al., 2006) was prepared and all injections were performed as previously described. A Leica MZ16F dissecting microscope equipped with DFC340 FX camera was used to visualize the embryos.
RNA Isolation and RT-PCR
To determine the efficiency of the obscurin A RhoGEF MOs, total RNA was isolated from uninjected and injected embryos at 48 hours post fertilization stage (hpf) using SV Total RNA isolation System (Promega, Madison, WI). A total of 50 ng RNA was used in a 25-μl reaction volume to assess mRNA expression using the iScript One-Step RT-PCR kit with SYBR green (Bio-Rad Laboratories, Hercules, CA, USA). A 350 bp cDNA that spanned the targeted exon was amplified by PCR (forward primer, 5′-GCCTGACCTGCGCGATTGTGA; reverse primer, 5′-GAGCGCTGGGAGAGGGAGGAC). To estimate the efficiency of exon skipping, primers amplifying the targeted exon (forward primer, 5′-ATTATTCAGTCTCGGAGTTGG; reverse primer, 5′-GTAGAGCCCTGTACTTTTGAAT) and coding sequence 5′ (forward primer, 5′-AACCATTTTCCGTAACATC; reverse primer, 5′-TTTGCCAACTCCGAGACTGAATA) and 3′ (forward primer, 5′-TGCGGCACAGCTGGTCACTCC; reverse primer, 5′-TCCTGGCCATGGTTCGTCTCTGTC) of the targeted exon were used to amplify these domains from 50 ng of total RNA from the 48 hpf control-injected and RhoGEF MO-injected embryos. A 3-step amplification (Denature: 95.0 °C; Anneal: 48.3 °C for targeted exon, 49.2 °C for coding sequence 5′, 56.0 °C for coding sequence 3′; Extend: 72 °C) and mRNA quantification was performed using the BioRad iCycler. Zebrafish β-Actin (forward primer, 5′-CCGTGACATCAAGGAGAAGCT; reverse primer, 5′-TCGTGGATACCGCAAGATTCC) was used as an internal control.
mRNA Rescue Injection
A DNA fragment encoding zebrafish Rho domain (RhoGEF/Ank) was cloned into pcDNA-DEST53 vector and sequenced. The plasmid was linearized with XmaI and capped mRNA was generated using the mMESSAGE mMACHINE T7 kit (Ambion, Austin, TX, USA). 2250 pg of transcribed RNA was co-injected along with 0.2mM ObscRhoMOs into 1 to 4-cell-stage embryos. ObscRhoGEF RNA in rescued embryos at 4 and 24 hpf were detected by RT-PCR (forward primer, 5′-CATGGCCAGCAAAGGAGAAGAA; reverse primer, 5′-GTTGTGGCGAATTTTGAAGTTAGC). Expression of the RhoGEF/Ank-GFP fusion protein (~135 KD) was confirmed by western analysis using an anti-GFP antibody (Zymed Laboratories, Inc.) according to the manufacturer’s instructions.
Site-directed Mutagenesis
Site-specific mutations were introduced into RhoGEF/Ank using QuikChange XL Site-directed Mutagenesis (Stratagene, La Jolla, CA). The following mutations were created and verified by sequencing: (1) A single base, C, was inserted near the start site of obscurin RhoGEF sequence to shift the reading frame (Non-sense), (2) Four bases of the targeted exon of the RhoGEF domain were changed to silence signaling activity (RhoGEF-m/Ank), (3) A stop codon (TAA) was inserted after the PH domain (RhoGEF) (see Fig. 2G). Primers for the QuikChange mutations are given in Table I. All constructs were injected at 2250 pg/embryo. Efficiency of rescue was assessed by an investigator blinded to the treatment group.
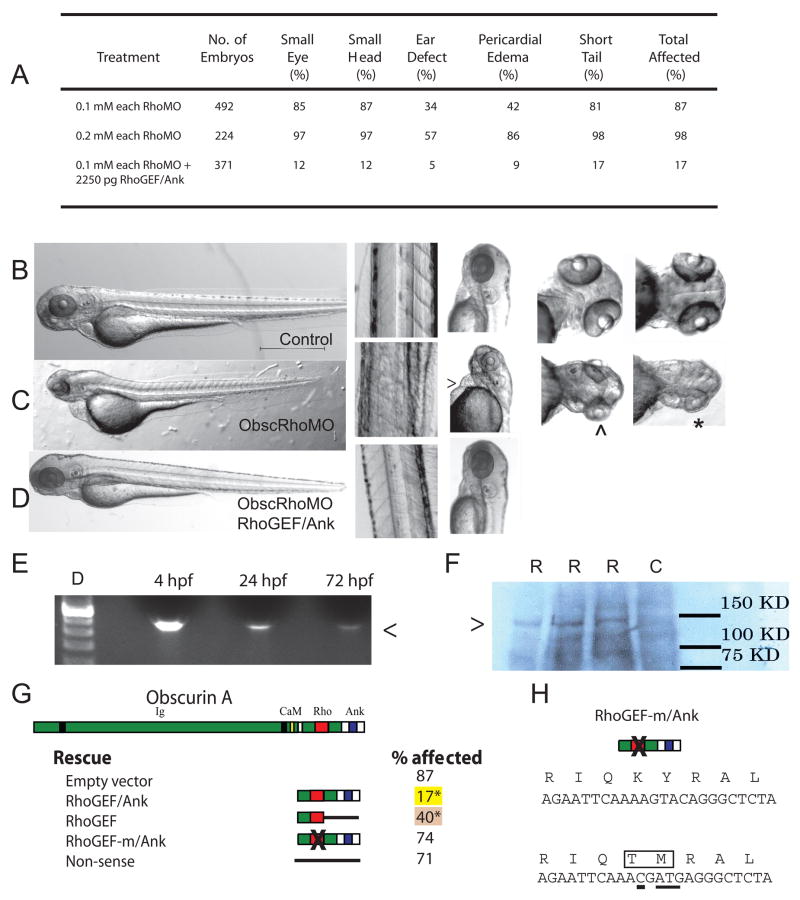
Fig 2.
Obscurin A RhoGEF morphant phenotype. (A) Morphant embryos were assessed at 72 hpf and scored for head size (small head), eye morphology (small eye, poorly developed optic cup), ear morphology (one or no otoliths), cardiac structure and function (ventricular hypoplasia or dilatation, pericardial edema) and skeletal muscle appearance (reduced tail length and width). (B-D) Gross morphology of control (B), obscurin A RhoGEF morphant (C) (0.1 mM of each morpholino), and rescued obscurin A RhoGEF morphant embryos (D) (0.1 mM of each morpholino + 2250 pg RhoGEF/Ank expression construct). The obscurin A RhoGEF morphant embryos demonstrate a high incidence of pericardial edema (>), moderate (^) to severe (*) eye hypoplasia, and smaller heads than controls. These defects were rescued or markedly diminished by co-injection of an obscurin A expression construct that included the RhoGEF and ankyrin binding domains (RhoGEF/Ank). (E) RT-PCR of mRNA extracted from 4, 24 and 72 hpf embryos injected with the expression construct demonstrating correct amplification of the expression tag (<). (F) Western analysis (Ab: anti-EGFP) of 72 hpf whole embryo lysates demonstrating expression of a fusion protein (>) of the expected size only in injected embryos (R) and not in control (C). (G) Percentage of obscurin A RhoGEF morphant embryos demonstrating one or more of the phenotypic abnormalities upon co-injection with the indicated rescue construct (*: p < 0.05 compared to empty vector control). Note the high efficiency of rescue with the RhoGEF/Ank and the absence of rescue with the non-sense and RhoGEF-m/Ank constructs. (H) Diagram of the amino acid sequence change introduced into the RhoGEF-m/Ank construct that replaces a conserved KY (Lysine-Tyrosine) motif with TM (Threonine-Methionine). Underlined nucleotides were changed using site-directed mutagenesis.
Table I.
Primers for Site-Directed Mutagenesis of ObscRhoGEF cDNA
| Frameshift (Non-sense) | F | 5′-CTCGCAAATCACAGAAATGCGTATCTGCCAAGATCACAC-3′ |
| R | 5′-GTGTGATCTTGGCAGATACGCATTTCTGTGATTTGCGAG-3′ | |
| Silence RhoGEF (RhoGEF/Ank-m) | F | 5′-CTTCAGAGACCACCTGAGAGAATTCAAACGATGAGGGCTCTACTGAAGG-3′ |
| R | 5′-CCTTCAGTAGAGCCCTCATCGTTTGAATTCTCTCAGGTGGTCTCTGAAG-3′ | |
| Stop after RhoGEF (RhoGEF) | F | 5′-CTTACAGCAGAGCTCGGATAACAGACTGTAAAGCTTGCC-3′ |
| R | 5′-GGCAAGCTTTACAGTCTGTTATCCGAGCTCTGCTGTAAG-3′ |
Wholemount Immunostaining
Paraformaldehyde fixed embryos were labeled with monoclonal anti-RhoA (Santa Cruz Biotechnology, Inc), or monoclonal anti-α-actinin (Sigma-Aldrich Inc., #EA-53) antibodies at 1:20 dilutions. The obscurin antibody (a generous gift of Dr. Robert Bloch, University of Maryland) was used as previously described (Raeker et al., 2006). TRITC-conjugated goat antimouse IgG, FITC-conjugated goat anti-mouse IgG, and FITC-conjugated goat anti-rabbit IgG, Texas Red-conjugated goat anti-rabbit (Jackson Immunoresearch Laboratories) were diluted to 1:100. An Olympus FV-500 confocal microscope was used to visualize the embryos.
Electron Microscopy
Electron microscopy was performed as previously described (Raeker et al., 2006). Briefly, embryos at 72 hpf were fixed with 2.5% glutaraldehyde and 2.0% paraformaldehyde and post fixed in 1% osmium tetroxide. After washing, they were stained with saturated uranyl acetate, and embedded in Epon 812. Ultra-thin sections were prepared and stained with saturated uranyl acetate and lead citrate. The sections were examined with a Philips CM100 transmission electron microscope at an accelerating voltage of 60 kV.
Embryo Imaging
Digital movies were obtained on unsedated zebrafish embryos at 48 and 72 hpf using a Leica MZFLIII dissecting microscope with attached Nikon digital camera. Ten second loops were recorded with the embryo right side up. Ventricular diameter was measured both along the axis of flow (Maj) and perpendicular to the axis of flow (Min) at both end systole and end diastole. Ventricular volumes were calculated assuming an ellipsoid shape [V= 4/3π(Maj/2)(Min/2)2]. Ejection fractions were calculated using standard methods. Retinal area was calculated assuming an elliptical shape of the retina. The right eye of the embryo was imaged with the embryo right side up and diameters measured along its long axis and perpendicular short axis [A=π(Maj/2)(Min/2)].
RESULTS
Targeted deletion of the exon encoding the active site of the obscurin RhoGEF domain
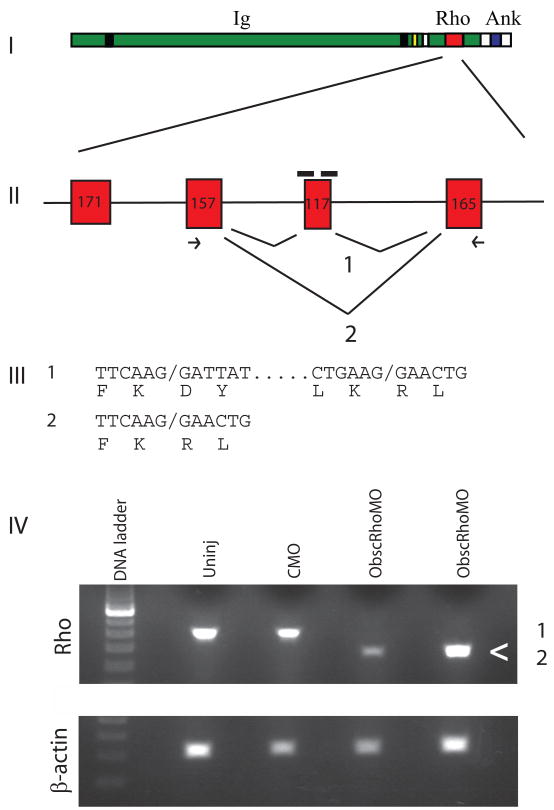
Antisense morpholinos complimentary to the intron-exon and exon-intron boundary of a 117 bp exon encoding for part of the obscurin RhoGEF GTPase activation site (predicted AA 5747–5785) were injected into 1–4 cell stage zebrafish embryos. RT-PCR of RNA isolates from whole 48 hours post-fertilization (hpf) embryos revealed that injection of both morpholinos commonly resulted in skipping of the targeted exon. Sequencing of the RT-PCR products confirmed that the reading frame was maintained in the alternatively-spliced product (Fig. 1). RT-PCR demonstrated preserved expression of the 3′ end of the alternatively spliced transcript (Supplemental Fig. 1). There was no apparent compensatory increase in obscurin B mRNA expression (Supplemental Fig. 1). The ratio of the expression of the targeted RhoGEF exon to β-Actin in the morphant embryos compared to control injected embryos was 0.47 +/− 0.14 amplification units at 0.2 mM morpholino dose and 0.39 +/− 0.17 units at 0.4 mM morpholino dose, indicating average of 50–60% reduction in the expression of deleted exon. The combination of the two morpholinos was much more efficient at inducing exon skipping than either morpholino alone. By band densitometry of RT-PCR products, injection of either morpholino alone resulted in approximately equivalent amounts of exon skipped and correctly spliced products (Supplemental Fig. 2). Injection of both morpholinos together resulted in an approximately 2:1 ratio of exon-skipped to correctly spliced products. RT-PCR analysis did not detect significant amounts of splice products with retained introns (data not shown). If these splicing events occurred, they may have been degraded by non-sense mediated decay.
Fig. 1.
Targeting of the obscurin A RhoGEF domain. (I) Diagram of obscurin A demonstrating the location of the tandem immunoglobulin-like (Ig) domains, the RhoGEF domain (Rho) and the ankyrin binding (Ank) domains. (II) Diagram of the exons encoding the double homology (DH) domain of the RhoGEF module. A 117-bp exon encoding for part of the GTPase activation site of the RhoGEF domain was targeted for induced exon skipping by the injection of antisense morpholinos (black bars) directed at the splice donor and acceptor sites for that exon. (III) Splice product 1 represents the normal splicing. The morpholinos are frequently able to block normal splicing of the targeted exon resulting in the splice product 2. PCR primers (arrows) from the adjacent exons were used to detect the splice products. Both products were cloned and sequenced and the nucleotide and predicted amino acid sequences displayed. (IV) RNA from 48 hpf embryos was isolated from obscurin A RhoGEF (ObscRhoMO) morphant, uninjected (Uninj) control morpholino-injected (CMO) embryos. Note that the band (<) corresponding to splice product 2 is only present in the RNA isolated from the morphant embryos.
Rescue of the observed defects using an obscurin miniconstruct
When both morpholinos were injected together, the percentage of 72 hpf embryos with skeletal muscle, cardiac and neurologic abnormalities was 81, 42, and 87% at 0.2 mM of ObscRhoMOs (N=492) and 98, 86, and 97% at 0.4 mM (N=224) (Fig. 2A). Morphant embryos were shorter and thinner with small heads and eyes and abnormalities of cardiac structure and function.
To verify that the observed developmental defects were due to the loss of obscurin RhoGEF activity, a miniconstruct encoding for the RhoGEF and carboxy terminal protein interaction domains (RhoGEF/Ank) was injected into control and ObscRhoMOs (0.2 mM) embryos. Expression of the intact miniconstruct was confirmed by RT-PCR and western analysis (Fig. 2E,F). Injection of RhoGEF/Ank resulted a significant reduction in the number of affected embryos to 19% (Ni = 5 injections; Ne = 371). The amount of expression construct injected was based on a dose response curve and was the dose that best rescued the morphant phenotype without causing observable defects when injected alone.
To confirm that the rescue was due to RhoGEF activity, a single base pair insertion to shift the reading frame of the rescue construct was created. Co-injection of this construct (Nonsense) did not result in significant rescue with 70% of the embryos demonstrating phenotypic abnormalities (Ni = 2 injections; Ne = 144). Injection of a rescue construct with a translation termination codon inserted after the RhoGEF domain (RhoGEF) resulted in partial rescue with abnormalities noted in 41% of the embryos (Ni = 2 injections; Ne = 145). The inability of this construct to more fully rescue the phenotype may be due to differences in mRNA or protein stability or reduced specificity of cellular localization due to the absence of the carboxy terminal protein interaction domains.
Failure of rescue by a construct with mutations introduced into the GTPase activation site provided the most direct evidence of the requirement of RhoGEF activity for the observed rescue. As noted above, the targeted exon encodes for a significant portion of the obscurin RhoGEF GTPase activation site. Within this targeted domain, a conserved lysine-tyrosine (KY) motif was changed to threonine-methionine (TM) by site-directed mutagenesis (Fig. 2H). This KY motif appears to have important structural and catalytic activities within the GTP binding and activation domain (Liu et al., 1998). The lysine corresponds to K-1732 of Trio GEF1 and has been demonstrated to be required for Rac1 activation. The tyrosine (Y-1733) is a conserved hydrophobic domain within the core of the activation domain that appears to be an important determinant of the architecture of the binding fold and affects binding fold access and activation of all small GTPases (Liu et al., 1998). The conversion of the KY motif to TM prevented phenotypic rescue with 74% of embryos affected (Ni = 2 injections; Ne = 136). Injection of 2250 pg of this construct (RhoGEF/Ank-m) alone did not result in developmental defects.
Skeletal muscle defects
We have previously characterized the skeletal muscle phenotype of embryos depleted of obscurin A (Raeker et al., 2006). In this study, we compared the muscle phenotype of those embryos (ObscMO) to the phenotype of embryos treated with morpholinos targeting the active site of the obscurin A RhoGEF domain (ObscRhoMO) to determine the specific functions of that domain. As in ObscMO embryos, ObscRhoMO embryos demonstrated abnormalities of skeletal muscle development (Fig. 3,4) but with a distinct morphology. One of the most consistent findings of in vivo downregulation or absence of obscurin (Kontrogianni-Konstantopoulos et al., 2006; Lange et al., 2009; Raeker et al., 2006) is the mispatterning of the sarcoplasmic reticulum (SR) which may be due to interaction of the globular carboxy terminal domain of obscurin with small ankyrins in the sarcoplasmic reticulum (Bagnato et al., 2003; Kontrogianni-Konstantopoulos et al., 2003). Defects in the patterning of the SR were a prominent feature of the ObscMO skeletal muscle phenotype (Raeker et al., 2006). The normal appearance of the SR in the skeletal muscle of the ObscRhoMO embryos likely indicates that the carboxy terminus of the targeted obscurin is functionally intact and suggests that the RhoGEF signaling domain does not have a major role in SR organization.
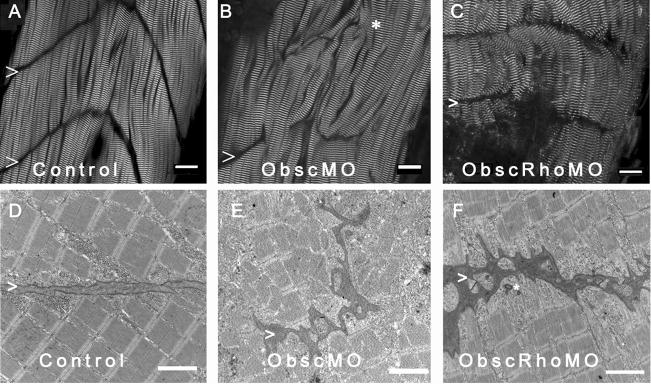
Fig. 3.
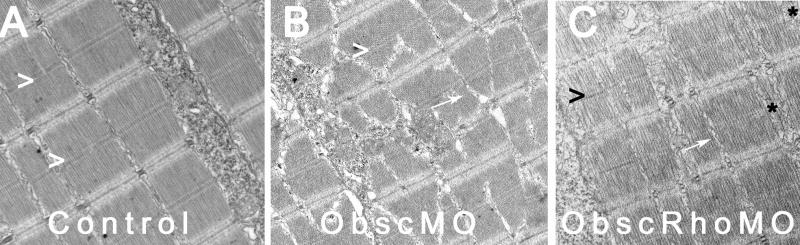
Architecture of the myotendinous junction (MTJ). Confocal analysis using α-actinin (a Z disk component) immunolabeling (A–C) to assess myofibril morphology and ultrastructural analysis (D–F) to define the architecture of the MTJ was performed in control (A,D), obscurin A morphant (B,E) and obscurin A RhoGEF morphant (C,F) embryos at 72 hpf. In control embryos, the skeletal myocytes are aligned in parallel and the myotendinous junctions are well defined (A,D: >). The skeletal myocytes in obscurin A (ObscMO) morphant embryos vary in length and in their relationship to each other (*) with elongated myocytes extending past very rudimentary somite boundaries (B,E:>). When compared to the ObscMO embryos, the obscurin A RhoGEF (ObscRhoMO) morphant embryos demonstrate a more consistent relationship between adjacent myocyte (more uniform parallel arrangement) and normal positioning of a complete or nearly complete somite boundary (>). However, the boundary is not sharply defined as in control embryos and often appears electron dense and irregular, not unlike the rudimentary boundaries/MTJs noted in ObscMO embryos. Scale bars are 20 μm (A–C) and 2 μm (D–F).
Fig. 4.
Skeletal muscle phenotype in obscurin A RhoGEF morphant embryos at 72 hpf. Ultrastructural analysis was performed at 72 hpf on control, obscurin A (ObscMO) and obscurin A RhoGEF (ObscRhoMO) morphant embryos. In control embryos, the myofibrils are aligned in register across the sarcoplasm (A) with well defined M bands and finely “stitched” Z disks (D). In the obscurin morphant embryos (B) there is marked disturbance of myofibril organization with areas of misalignment of the M bands of adjacent myofibrils (E:^). By comparison, the obscurin A RhoGEF morphants display myofibril organization (C) similar to control embryos but reduced myofibrillar content with more sarcoplasm devoid of myofibrils and more rounded nuclei (C,G: ^). There were also mild sarcomeric abnormalities including, infrequently, an indistinct appearance of the Z disk and absence of the M band (F:*) which could occur even within myofibrils demonstrating mature Z disks and M bands (F:^). Note that in skeletal myocytes stained with DAPI, the nuclei in the obscurin A RhoGEF morphants (RhoMO) are closer together and more rounded (G) than in controls (H) consistent with reduced myofibril volume. Scale bars 2 μm (A–C), 0.5 μm (D–F) and 10 μm (G,H).
Another notable feature of ObscMO embryos was the elongated skeletal myocytes that extended beyond primitive and highly irregular vertical myoseptae, the zebrafish equivalent of the myotendinous junction (Raeker et al., 2006). By comparison, the vertical myoseptae in the ObscRhoMO embryos were correctly positioned and were usually complete, in that they extended almost the full width of the myotome. Yet they were late to achieve a mature “chevron” shape and were occasionally deficient between the most superficial myocytes. On electron microscopic analysis, the vertical myoseptae in the more severely affected ObscRhoMO embryos were similar in character to those in the ObscMO embryos in that they were much more electron-dense and less organized than those in controls (Fig. 3). Given the abnormalities of the myotendinous junctions in both morphant phenotypes, the structural and RhoGEF signaling properties of obscurin A appear to cooperatively influence the reciprocal and coordinated remodeling of the intracellular and extracellular architecture that occurs during striated muscle development.
In addition, there were notable reductions in myofibril density in the ObscRhoMO embryos. Skeletal myocytes in ObscRhoMO embryos appeared thinner than those in the corresponding somites of control embryos at 72 hpf with reduced myofibrillar volume and increased sarcomere-free areas of the myoplasm (Fig. 4C,F,G). The structure of the myofibrils and relationship of the myocytes to each other was very similar to controls. In contrast to the skeletal myofibrils of ObscMO embryos, the myofibrils in the ObscRhoMO embryos were aligned in transverse register across the sarcoplasm with rates of misalignment that were comparable to controls (<5%). The higher order structure of the skeletal muscle was also similar to controls with ObscRhoMO myofibrils and myocytes demonstrating a more consistent parallel arrangement compared to the “twisting” of myofibrils and myocytes noted in ObscMO skeletal muscle (Fig. 4B,E).
Compared to the dramatic alteration of skeletal muscle architecture in the ObscMO embryos, the abnormalities of sarcomere structure in the ObscRhoMO embryos was much more subtle and could only be detected by electron microscopic analysis. For instance, Z disks in control and ObscMO embryos rapidly achieved a “stitched” or interdigitated appearance shortly after their assembly. In contrast, Z disks of newly formed sarcomeres added at the terminal ends of the myofibril and formed by lateral addition to existing myofibrils often displayed a “fuzzy” appearance in the ObscRhoMO morphants (Fig. 4F). This appeared to be the result of a delay in maturation as opposed to a permanent structural abnormality as the Z disks of mature myofibrils were indistinguishable from those in controls.
The other subtle sarcomeric abnormality noted was an inconsistent appearance of the M bands. In the vast majority of skeletal myofibrils in the ObscRhoMO embryos, well-formed M bands could be identified. However, unlike control and ObscMO embryos, ObscRhoMO embryos displayed a small number of skeletal myofibrils that lacked well-defined M bands on ultrastructural analysis at 72 hpf (Fig. 5). Absent or poorly defined M bands most commonly occurred in newly formed skeletal myofibrils (Fig. 4F), but were also noted in more mature myofibrils (Fig. 5C), indicating that the obscurin A RhoGEF domain may participate in the recruitment and stabilization of protein complexes within the M band. The absence of the M band was reminiscent of C. elegans Unc-89 mutants that lack a well formed M band (Benian et al., 1996).
Fig. 5.
Absence of consistent M bands in obscurin A RhoGEF skeletal muscle at 72 hpf. Control embryos (A) consistently demonstrate well-formed M bands (>) in skeletal myofibrils by 72 hpf. Obscurin A morphant embryos (B), although they display marked abnormalities in myofibrillar architecture, also consistently demonstrate mature M bands (>). In contrast, skeletal myofibrils in obscurin A RhoGEF morphant embryos (C) demonstrated occasional absence of M bands (*) in otherwise mature-appearing myofibrils. Note that the extrajunctional SR in the ObscMO embryo is markedly disorganized (B: arrow), but is much more normal appearing in the ObscRhoMO embryos (C: arrow).
Cardiac defects
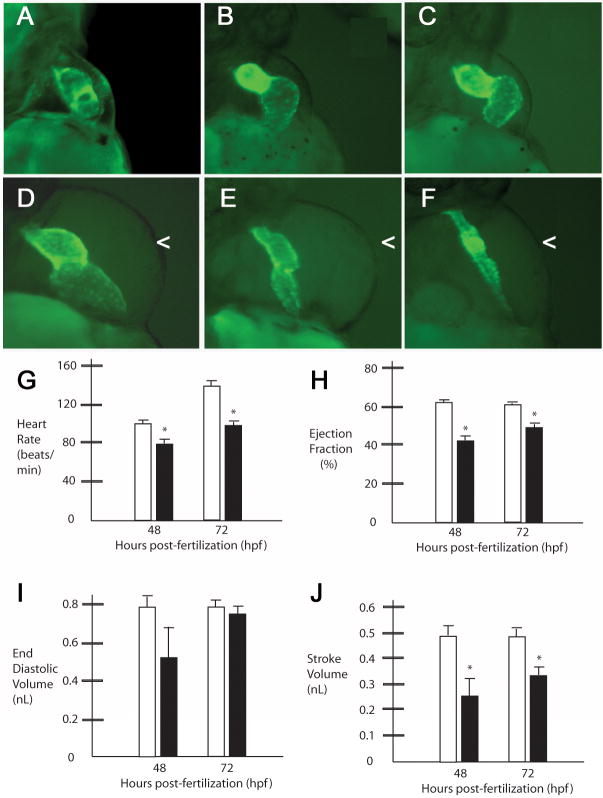
The cardiac defects noted in the RhoGEF morphant embryos were grossly similar to those previously noted in the obscurin A morphant embryos (Raeker et al., 2006). Cardiac abnormalities ranged from mild cardiac dilatation to severe cardiac hypoplasia (Fig. 6). Cardiac hypoplasia often resulted in a stretched appearance to the heart as the rest of the embryo continued to grow and develop. In the more severely affected embryos (Fig. 6E,F), there was markedly reduced cardiac output. Since the diminished flow through the heart can affect its function, the functional properties of the morphant hearts were assessed in the more mildly affected embryos (Fig. 6B–D). Even in embryos with end-diastolic volumes similar to controls (Fig. 6H), there were noted to be lower heart rates and reduced ejection fractions at 48 and 72 hours (Fig. 6G, I). The reduced calculated stroke volume (Fig. 6J) and lower heart rates would predict a decrease in cardiac output that may have contributed to the observed pericardial edema (Fig. 6D–F).
Fig. 6.
Obscurin A RhoGEF signaling is required for normal cardiac development. Cmlc2-EGFP transgenic zebrafish embryos injected with obscurin A RhoGEF morpholinos (B–F) were compared to control-injected transgenic embryos (A) at 72 hpf. Morphant embryos demonstrated a spectrum of cardiac abnormalities ranging from reduced cardiac function with cardiac dilatation and mild pericardial edema (B–D) to mild (E) or severe (F) ventricular hypoplasia and pericardial edema (<). (G–J) Cardiac functional assessments were performed in morphant (black bars) and control embryos (white bars) at 48 and 72 hpf. In the more mildly affected embryos with mild ventricular hypoplasia as determined by comparable end diastolic volume measurements relative to controls (I), there were significant reductions in heart rate (G), ejection fraction (H) and stroke volume (J) at 48 and 72 hpf compared to controls (*t-test; P<.05).
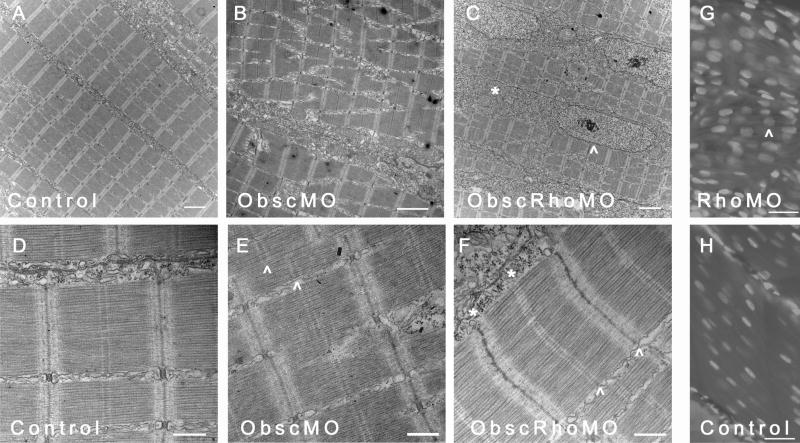
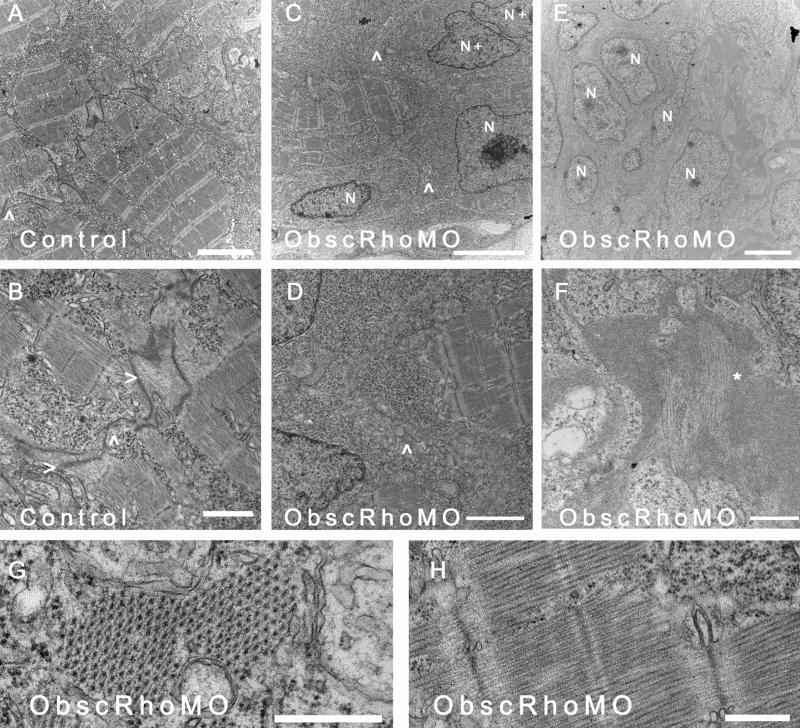
Ultrastructural analysis was performed at 72 hpf using transmission electron microscopy (Fig. 7). As with the skeletal myocytes, there was a decreased ratio of myofibrillar area to nuclear area within each myocyte suggestive of a reduction in the number of mature contractile units/cell (Fig. 7C,D). There also appeared to be significant abnormalities in the formation of cell-cell contacts. Unlike cardiac myocytes in control embryos that had formed mature intercalated disks with identifiable gap and adherens junctions by 72 hpf (Fig. 7A,B), cardiac myocytes in ObscRhoGEF embryos had very indistinct cell-cell contacts (Fig. 7C,D) with no adherens junctions. This is again consistent with close structural and signaling interactions between obscurin A and transmembrane cell adhesion complexes.
Fig. 7.
Ultrastructural analysis of cardiac myocytes at 72 hpf. (A) Cardiac myocytes in control embryos have formed intercalated disks (^) and filled the sarcoplasm with mature myofibrils. (B) Higher magnification of an intercalated disk (ID) from a control embryo demonstrates adherens (>) and gap (^) junctions. (C,D) In contrast, cardiac myocytes from mildly affected obscurin A RhoGEF morphant embryos have reduced myofibrillar content [compare the nuclear (N) area to the myofibrillar area within each myocyte] and very poorly defined regions of cell-cell (^) contact with no identifiable IDs. Examples of recent nuclear division (N+) were more frequent in the obscurin RhoGEF morphant embryos. (E,F) In more severely affected embryos, cardiac myocytes appear markedly abnormal with loosely arranged contractile filaments (F:*). (G,H) The mature cardiac myofibrils that did form in the obscurin A RhoGEF morphant embryos were not significantly different than control with a normal appearing lattice of thick filaments on cross-section (G) and laterally aligned Z disks and M bands on longitudinal section (H). Scale bars are 4 μm (A–C), 2 μm (D,E), 1 μm (F) and 0.5 μm (G,H).
Although reduced in number, the morphology of the mature cardiac myofibrils that did form was not appreciably different in ObscRhoMO embryos than in controls (Fig. 7E,F). The lattice of myosin thick filaments was well organized and evenly spaced in contrast to what has previously been noted in skeletal myotubes depleted of the full obscurin A (Kontrogianni-Konstantopoulos et al., 2006; Raeker et al., 2006). This suggests that the arrangement of the thick filaments is more dependent upon the structural properties of obscurin A than the signaling properties of its RhoGEF domain.
The constellation of abnormalities noted in the obscurin A RhoGEF-depleted cardiomyocytes would suggest that they were less differentiated than cardiomyocytes from control embryos of the same developmental stage (as determined by hpf and somite number). The absence of mature cell-cell contacts and small cells with sparse myofibrils is consistent with a less differentiated state. Whether or not cardiac myocytes depleted of obscurin RhoGEF activity might eventually fully differentiate is more difficult to determine as the marked decrease or absence of blood flow due to the ventricular hypoplasia often resulted in ventricular chamber collapse as has been noted in response to mechanical interruption of flow during cardiac development (Hove et al., 2003). The capacity for other signaling mechanisms to eventually compensate for the loss of obscurin RhoGEF signaling may account for the difference in phenotype between the ObscRhoMO embryos and the recently described obscurin null mouse (Lange et al., 2009).
Neural development
In addition to the above-noted delays in striated muscle development, obscurin A RhoGEF morphant embryos were noted to have abnormalities of neural development. In control embryos, obscurin A mRNA (Raeker et al., 2006) and protein expression (Fig. 9A,E) was noted in the developing brain and retina in a pattern that mirrored RhoA expression (Fig. 9B). Due to its well-characterized pattern of differentiation in zebrafish embryos, the developing retina was examined as model of the role of obscurin A RhoGEF signaling in the brain.
Figure 9.
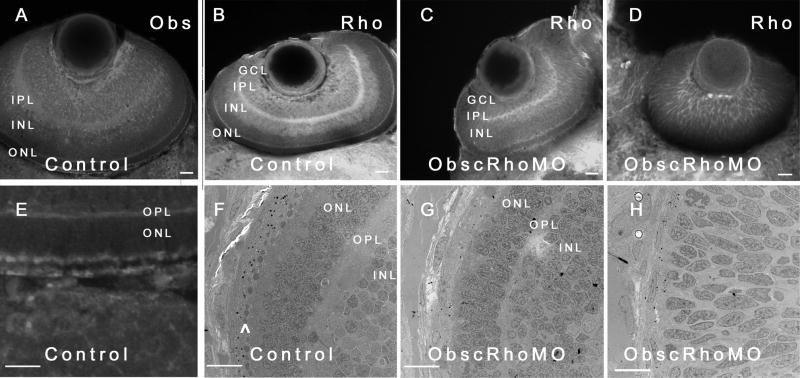
Eye phenotype of obscurin A RhoGEF embryos. Control (A,B,E) and obscurin RhoGEF (C,D) morphant embryos were immunolabeled for Obscurin (A,E) or RhoA (B–D) at 72 hpf. In the control embryo, distinct retinal layers are noted [ganglion cell layer (GCL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), and outer nuclear layer (ONL)]. Obscurin is expressed in each of the layers, usually at the cell periphery with the greatest abundance in the axon-rich IPL (A) and at the apical and basal aspects of the ONL (E) in a distribution similar to RhoA (A,B). In the mildly affected obscurin A RhoGEF morphant embryos (C), rudimentary GCL, IPL, INL, OPL, and ONL are noted while the more severely affected embryos do not develop identifiable layers (D). On ultrastructural analysis, there is significant pigment deposition and a well developed photoreceptor layer (^) in the retina of a control embryo (F) while even a mildly affected morphant embryo (G) has very little pigmentation and no identifiable photoreceptors. A more severely affected embryo (H) lacks even rudimentary layers and the cells remain elongated and poorly differentiated. Scale bars are 20 μm (A–D) and 10 μm (E–H).
Obscurin was expressed in all layers of the developing retina but was most abundant in the inner plexiform layer and at the apical and basal surfaces of the outer nuclear layer. The inner plexiform layer is filled with axons and it is likely that obscurin and RhoA signaling have important roles in extending axons. Depletion of the obscurin A RhoGEF signaling domain resulted in morphant embryos with small, poorly developed eyes (Fig. 8) with persistent cellularity of the lens, hypoplasia of the optic nerve, and poorly defined layers within the retina. In the more severely affected embryos, the cells were elongated and relatively uniform in appearance, lacking any of the identifiable layers normally present at this stage of development (Fig. 9D,H). Of note, this was not associated with a global delay in development as the embryos had the same number of somites as the control embryos and only moderate abnormalities of skeletal muscle development as noted above.
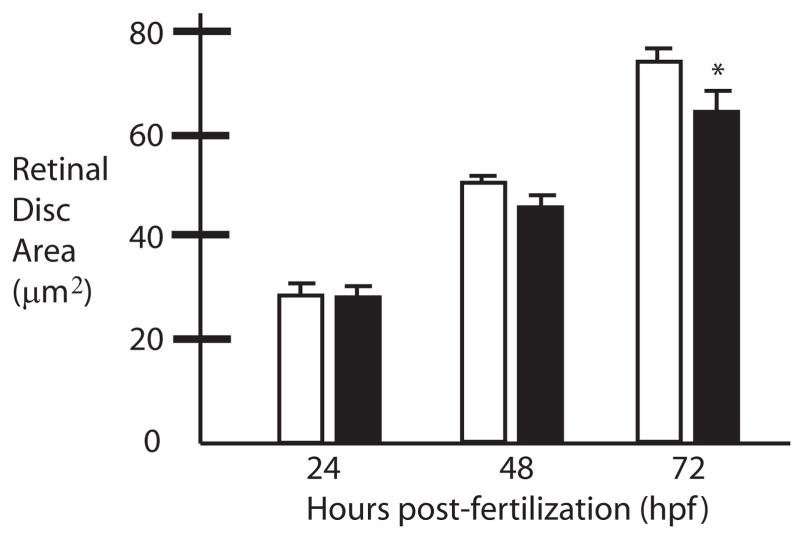
Figure 8.
Small eye phenotype of obscurin A RhoGEF morphant embryos. Compared to control (white bars) embryos the retinal disks of morphant embryos (black bars) exhibit slowed growth. The growth discrepancy reaches statistical significance by 72 hpf (*t-test: p<0.05).
Retinal development is dependant upon the interaction of secreted and cell-contact signals directing complex morphogenetic changes in retinal neuroblasts that lead to their lamination (layering) and differentiation (Galli-Resta et al., 2008; Hitchcock and Raymond, 2004; Neumann and Nuesslein-Volhard, 2000). In even the most severely affected ObscRhoMO embryos, retinal development had advanced beyond the invagination of the optic cup and the development of two identifiable cell layers, the retinal pigment epithelium of the outer retina and the inner retinal neuroepithelium. However, in the more severely affected morphant embryos (4 out of 12 examined by EM or immunohistochemical analysis), there was no significant differentiation beyond that point by 72 hpf with cells in the retinal neuroepithelium remaining elongated and poorly differentiated with no discernable lamination (Fig. 9D,H). In contrast, control embryos at that stage of development had formed easily identifiable ganglion cell (GCL), inner and outer plexiform (IPL and OPL), and inner and outer nuclear (INL and ONL) layers (Fig. 9B,F).
Previous studies have demonstrated that retinal neuroblasts are anchored apically via cell-extracellular matrix interactions to the inner limiting membrane and at the basal surface that apposes the pigmented epithelium are linked to each other by a network of adherens junctions (cell-cell contacts) (Galli-Resta et al., 2008). This polarity is essential for the correct orientation of future cell divisions that will lead to retinal layering. In the more mildly affected embryos, layering does proceed in a normal but delayed fashion, with the appearance of a ganglion cell layer, inner and outer plexiform layers and inner and outer nuclear layers (Fig. 9C,G). The inner nuclear layer further divides into two sublayers, potentially representing amacrine and bipolar/horizontal cell precursors. In the ObscRhoMO embryos, none of the layers are highly differentiated but they do appear to all be present and to be correctly positioned, suggesting that obscurin A RhoGEF signaling primarily determines the rate of retinal differentiation rather than affecting cell fate determination or the organization of the layers. Since obscurin A is expressed in the developing zebrafish eye (Raeker et al., 2006) and retinal development is not entirely dependent on brain development after the development of an optic cup (with formation of the retinal pigment epithelium and the retinal neuroepithelium), these abnormalities appear to be consistent with a primary delay in the progressive differentiation of the primitive retinal layers due to impaired obscurin RhoGEF-mediated small GTPase signaling.
DISCUSSION
This is the first study to demonstrate a role for the obscurin A RhoGEF domain in the signaling processes that direct organogenesis in the brain, skeletal muscle and heart. Much of the work on obscurin to date has focused on its role in the assembly, organization and structural support of striated myofibrils (Bagnato et al., 2003; Bang et al., 2001; Borisov et al., 2004; Borisov et al., 2003; Borisov et al., 2006; Kontrogianni-Konstantopoulos and Bloch, 2005; Kontrogianni-Konstantopoulos et al., 2004; Kontrogianni-Konstantopoulos et al., 2006; Kontrogianni-Konstantopoulos et al., 2003; Raeker et al., 2006). In these studies it was determined that obscurin promotes the stable integration of myosin filaments into the myofibril (Borisov et al., 2006; Kontrogianni-Konstantopoulos et al., 2006) and that it may act as a primary scaffold for the thick filament array (Borisov et al., 2004; Borisov et al., 2006) in part through interactions with M band titin and myomesin (Fukuzawa et al., 2008). Therefore, it is not surprising that inhibition of obscurin A-mediated RhoGTPase activation reduced the assembly of new myofibrils. Cardiac myocytes in the most severely affected embryos were deficient in mature sarcomeric structures with contractile filaments forming disorganized tangles within the sarcoplasm. Even the more mildly affected embryos demonstrated reduced myofibrillar content in both the heart and skeletal muscle. In the myofibrils that did form, the architecture of the sarcomeres was well preserved. The most notable effects on sarcomere structure were the occasional absence of a well-formed M band and the indistinct appearance to the Z disk in newly-formed myofibrils. The absence of a well-formed M band is a common feature of C. elegans embryos with mutations of Unc-89 (Benian et al., 1996) and our study suggests that the obscurin RhoGEF domain may participate in the scaffolding and organization of the M band. The targeting of obscurin RhoGEF activity also had subtle effects on Z disk morphology in skeletal muscle. This may be due to the effect of obscurin RhoGEF signaling on the integration of the Z band region of titin. In a recent study, overexpression of the obscurin RhoGEF domain interfered with Z band titin incorporation (Bowman et al., 2008). Certainly the delay in maturation of the Z disk or the decreased number of myofibrils per cell may reflect irregularities in Z band titin incorporation. That those abnormalities weren’t more severe or more persistent may be due to partial phenotypic rescue by endogenous obscurin B RhoGEF activity, which is not affected by our obscurin A RhoGEF targeting, residual RhoGEF activity of obscurin A due to the subset of transcripts not targeted by the morpholinos or compensation by other RhoGEF proteins. It is also possible that the effects of obscurin RhoGEF on titin incorporation are not related to the signaling properties of the GEF domain but physical properties that may not have been dramatically affected by the 39 amino acid deletion.
Furthermore, as with our previous in vivo studies (Raeker et al., 2006), the current studies suggest that obscurin A also has important extrasarcomeric functions within striated muscle. In our prior work, we demonstrated that obscurin A participates in the alignment of myofibrils in register across the sarcoplasm and in the positioning of other cellular domains with respect to the contractile apparatus. Depletion of obscurin A during development resulted in the alteration of the internal cellular structure of the myocyte and had secondary effects on the interactions between adjacent myocytes and between myocytes and the extracellular matrix (see Fig. 3B,E and Fig 4B,E). The proposed relationship of obscurin with the submembrane cytoskeleton and transmembrane complexes was supported by the recent finding that obscurin localizes to the subsarcolemmal region of the neuromuscular junction (NMJ) in skeletal muscle fibers (Carlsson, 2008). The post-synaptic endplate of the NMJ is a region rich in nuclei, mitochondria, adhesive contacts and cytoskeletal elements. It overlies but is not closely associated with the myofibril. Therefore, obscurin’s abundant localization to the motor end plate supports its characterization as a cytoskeletal filament that interacts with the submembrane cytoskeleton to regulate cellular architecture and stabilize interactions between the cell and the extracellular environment, including surrounding cells and the ECM. A similarly close relationship between the invertebrate orthologue of obscurin, Unc-89, and transmembrane adhesive complexes has been proposed. Unc-89 has been noted to be recruited to integrin-based adherens junction complexes by actopaxin/pat-6 (Lin et al., 2003) and integrin-linked kinase (ILK)/pat-4 (Mackinnon et al., 2002) and the interaction of Unc-89 with these complexes likely promotes the assembly of the thick filaments into organized arrays (Benian et al., 1996).
While obscurin A, like Unc-89, has been primarily considered a striated muscle protein, it is clear from the current study that it also has important functions in the developing central nervous system where it may serve a similar role in regulating reciprocal interactions between the intracellular and extracellular environments. We have previously noted that, prior to its expression in striated muscle, obscurin is expressed in the developing brain and neural tube of zebrafish and mouse (Raeker et al., 2006). As development proceeds, its expression in the central nervous system is downregulated, eventually persisting at significant levels only in specialized regions including the caudate nucleus, cerebellum and hippocampus (Nagase et al., 2000), sites of suspected ongoing neurogenesis in the adult brain (Kuhn et al., 1996; Lois and Alvarez-Buylla, 1993; Luskin, 1993; Ponti et al., 2008).
Given the high level of expression and the important roles of small GTPase signaling in neural development [reviewed in (Govek et al., 2005)], it is not entirely unexpected that targeting the obscurin A RhoGEF domain would have an effect on the developing brain. Other RhoGEFs that have been demonstrated to have important roles in neural development include Kalirin and Trio, the vertebrate orthologues of Unc-73/Drosophila trio (Steven et al., 1998). Both trio and kalirin have two RhoGEF domains and a single serine-threonine kinase domain at their carboxy termini that are highly homologous to that of obscurin (Russell et al., 2002) suggesting a close evolutionary relationship between obscurin and trio/kalirin (Russell et al., 2002). Both trio and kalirin have been proposed to have important roles in neural and skeletal muscle development (Hansel et al., 2001; O’Brien et al., 2000), and to participate in the cytoskeletal adaptations that promote neuroblast migration and axon extension (Bateman and Van Vactor, 2001; Penzes et al., 2001). Cellular depletion of trio in developing myotubes commonly results in myotube collapse presumably due to impaired cell-cell and/or cell matrix interactions (Dalkilic et al., 2006; O’Brien et al., 2000). In support of this assertion, trio has been demonstrated to regulate focal adhesion dynamics, potentially through interactions with focal adhesion kinase (FAK) (Medley et al., 2003).
The small GTPases activated by obscurin A were not directly examined in this study. Recent studies have identified RhoA (Ford et al., 2008) and TC10, also known as RhoQ (Coisy-Quivy et al., 2009), as targets for activation by the obscurin RhoGEF domain. Demonstration that the RhoGEF domain of Unc-89, the invertebrate orthologue of obscurin, is capable of activating Rho1, the invertebrate orthologue of RhoA (Qadota et al., 2008) further supports RhoA as a target of obscurin-mediated signaling. We did note co-localization of RhoA with obscurin in the retina and muscle (Supplemental Fig. 3) and certainly many of the developmental defects that were observed in this study would be consistent with the loss of RhoA activation. In addition to its role in neural development, RhoA has been noted to promote skeletal muscle differentiation and muscle hypertrophy through serum response factor (SRF)-mediated activation of a myogenic program (Hill et al., 1995; Wei et al., 1998). Reduced activation of this myogenic program could account for the relative atrophy of the skeletal muscle and the immature appearance of the cardiac myocytes in the developing heart (Li et al., 2005; Parmacek, 2007).
It is important to compare the findings in our zebrafish models to those of the obscurin null mouse (Lange et al., 2009). Somewhat surprisingly, the obscurin null mouse demonstrated only a mild myopathy as noted by an increased number of regenerating myofibrils. Based on the findings in the obscurin A morphant embryos (Raeker et al., 2006) and the current study, we would predict that the myopathy may be due to abnormalities of calcium handling related to disturbance of SR architecture or due to myotube detachment related to impaired cell-matrix interactions. The lack of a more severe phenotype in the null mouse may be due to compensatory processes that may be species-dependent. OBSL1 (Fukuzawa et al., 2008; Geisler et al., 2007) may be able to compensate for some of the structural properties of obscurin while trio/kalirin may be able to compensate for loss of obscurin RhoGEF signaling. The ability of compensatory mechanisms to mask protein functions in a species-specific manner is evident from muscular dystrophy models in which loss of dystrophin in the zebrafish (Bassett and Currie, 2004; Bassett et al., 2003) more closely approximates the human condition than does the null mouse model (Bulfield et al., 1984; Durbeej and Campbell, 2002). For obscurin, it is likely that both models will be important for understanding the unique and shared functions of obscurin and closely related structural and signaling molecules such as OBSL1 and trio.
Model of obscurin A function
Based on the above studies and previous characterization of obscurin A, it appears that, in addition to its role in thick filament assembly and organization, obscurin A has important functions in the structural integration of the sarcomere with the extrasarcomeric cytoskeleton, and with specific domains of the sarcoplasmic reticulum. As such, it is well situated to participate in pathways that translate mechanical signals into intracellular adaptive responses through the activation of small GTPases. Based on this study, obscurin A RhoGEF-mediated small GTPase activation does appear to be an important tissue-specific link between the transmembrane signaling pathways that initiate cellular differentiation and the morphologic changes that accompany differentiation. In the setting of reduced obscurin A RhoGEF signaling, the cells appear poised for differentiation but slow to proceed due to the alteration in small GTPase activation.
Summary
This is the first demonstration of the important role of obscurin A RhoGEF activity in heart, skeletal muscle and neural development. The immature phenotype of the cardiac myocytes and the retinal neuroblasts observed in response to molecular targeting of this domain suggests that obscurin A-mediated small GTPase signaling functions as a tissue-specific molecular switch that promotes advanced cellular differentiation. In heart and skeletal muscle, it may be capable of activating a myogenic program that promotes the maturation of cell-cell and cell-matrix contacts and the assembly of new myofibrils.
Supplementary Material
Acknowledgments
The authors acknowledge Sasha Meshinchi, Sarah Geisler and Chris Cooke for their assistance and Dr. Robert Bloch and Dr. Daniel Goldman for helpful discussions and reagents. This work was supported by grants to M.W.R. from the Muscular Dystrophy Association and the NIH (HL075093).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. Binding of an ankyrin-1 isoform to obscurin suggests a molecular link between the sarcoplasmic reticulum and myofibrils in striated muscles. J Cell Biol. 2003;160:245–53. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–72. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Bassett D, Currie PD. Identification of a zebrafish model of muscular dystrophy. Clin Exp Pharmacol Physiol. 2004;31:537–40. doi: 10.1111/j.1440-1681.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- Bassett DI, Bryson-Richardson RJ, Daggett DF, Gautier P, Keenan DG, Currie PD. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development. 2003;130:5851–60. doi: 10.1242/dev.00799. [DOI] [PubMed] [Google Scholar]
- Bateman J, Van Vactor D. The Trio family of guanine-nucleotide-exchange factors: regulators of axon guidance. J Cell Sci. 2001;114:1973–80. doi: 10.1242/jcs.114.11.1973. [DOI] [PubMed] [Google Scholar]
- Benian GM, Tinley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required fpr muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132:835–48. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Dynamics of obscurin localization during differentiation and remodeling of cardiac myocytes: obscurin as an integrator of myofibrillar structure. J Histochem Cytochem. 2004;52:1117–27. doi: 10.1369/jhc.3A6183.2004. [DOI] [PubMed] [Google Scholar]
- Borisov AB, Raeker MO, Kontrogianni-Konstantopoulos A, Yang K, Kurnit DM, Bloch RJ, Russell MW. Rapid response of cardiac obscurin gene cluster to aortic stenosis: differential activation of Rho-GEF and MLCK and involvement in hypertrophic growth. Biochem Biophys Res Commun. 2003;310:910–8. doi: 10.1016/j.bbrc.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Borisov AB, Sutter SB, Kontrogianni-Konstantopoulos A, Bloch RJ, Westfall MV, Russell MW. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: evidence from small interfering RNA-mediated gene silencing. Histochem Cell Biol. 2006;125:227–38. doi: 10.1007/s00418-005-0069-x. [DOI] [PubMed] [Google Scholar]
- Bowman AL, Catino DH, Strong JC, Randall WR, Kontrogianni-Konstantopoulos A, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin regulates assembly of titin at the Z-disk through interactions with Ran binding protein 9. Mol Biol Cell. 2008;19:3782–92. doi: 10.1091/mbc.E08-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81:1189–92. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L, Yu JG, Thornell LE. New aspects of obscurin in human striated muscles. Histochem Cell Biol. 2008;130:91–103. doi: 10.1007/s00418-008-0413-z. [DOI] [PubMed] [Google Scholar]
- Coisy-Quivy M, Touzet O, Bourret A, Hipskind RA, Mercier J, Fort P, Philips A. TC10 controls human myofibril organization and is activated by the sarcomeric RhoGEF obscurin. J Cell Sci. 2009;122:947–56. doi: 10.1242/jcs.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006;26:6522–34. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbeej M, Campbell KP. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr Opin Genet Dev. 2002;12:349–61. doi: 10.1016/s0959-437x(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Ford DL, Roche JP, Bloch RJ. The giant sarcomeric protein obscurin as a potential regulator of RhoA signaling in skeletal muscle. Biophys Soc Abst #3235 2008 [Google Scholar]
- Fukuzawa A, Lange S, Holt M, Vihola A, Carmignac V, Ferreiro A, Udd B, Gautel M. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band-implications for hereditary myopathies. J Cell Sci. 2008;121:1841–1851. doi: 10.1242/jcs.028019. [DOI] [PubMed] [Google Scholar]
- Galli-Resta L, Leone P, Bottari D, Ensini M, Rigosi E, Novelli M. The genesis of retinal architecture: an emerging role for mechanical interactions. Prog Retin Eye Res. 2008;2727:260–283. doi: 10.1016/j.preteyeres.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Geisler SB, Robinson D, Hauringa M, Raeker MO, Borisov AB, Westfall MV, Russell MW. Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin. Genomics. 2007;89:521–31. doi: 10.1016/j.ygeno.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–5. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Hansel DE, Quinones ME, Ronnett GV, Eipper BA. Kalirin, a GDP/GTP exchange factor of the Dbl family, is localized to nerve, muscle, and endocrine tissue during embryonic rat development. J Histochem Cytochem. 2001;49:833–44. doi: 10.1177/002215540104900704. [DOI] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–70. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Raymond PA. The teleost retinaas a model for developmental and regeneration biology. zebrafish. 2004;1:257–271. doi: 10.1089/zeb.2004.1.257. [DOI] [PubMed] [Google Scholar]
- Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421:172–7. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Ann Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Koh CG. Rho GTPases and their regulators in neuronal functions and development. Neuro-Signals. 2007;15:10. doi: 10.1159/000101527. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Bloch RJ. Obscurin: a multitasking muscle giant. J Muscle Res Cell Motil. 2005;26:419–26. doi: 10.1007/s10974-005-9024-7. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Randall WR, Bloch RJ. Obscurin regulates the organization of myosin into A bands. Am J Physiol Cell Physiol. 2004;287:C209–17. doi: 10.1152/ajpcell.00497.2003. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, Russell MW, Bloch RJ. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006;20:2102–11. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–48. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–33. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, Chen J. Obscurin determines the architectureof the longitudinal sarcoplasmic reticulum. J Cell Sci. 2009;122:2640–50. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A, Olson EN. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc Natl Acad Sci USA. 2005;102:1082–7. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Qadota H, Moerman DG, Williams BD. C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr Biol. 2003;13:922–32. doi: 10.1016/s0960-9822(03)00372-5. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak ET, Meadows RP, Schkeryantz JM, Janowick DA, Harlan JE, Harris EA, Staunton DE, Fesik SW. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–77. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc Natl Acad Sci USA. 1993;90:2074–7. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:8. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–89. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. [see comment] Curr Biol. 2002;12:787–97. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Medley QG, Buchbinder EG, Tachibana K, Ngo H, Serra-Pages C, Streuli M. Signaling between focal adhesion kinase and trio. J Biol Chem. 2003;278:13265–70. doi: 10.1074/jbc.M300277200. [DOI] [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Nakayama M, Hirosawa M, Ohara O. Prediction of the coding sequences of unidentified human genes. XVIII The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:273–81. doi: 10.1093/dnares/7.4.271. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–9. doi: 10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- O’Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Natl Acad Sci USA. 2000;97:12074–8. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmacek MS. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Cir Res. 2007;100:633–44. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Kambampati V, Mains RE, Eipper BA. Distinct roles for the two Rho GDP/GTP exchange factor domains of kalirin in regulation of neurite growth and neuronal morphology. J Neurosci. 2001;21:8426–34. doi: 10.1523/JNEUROSCI.21-21-08426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti G, Peretto P, Bonfanti L. Genesis of neuronal and glial progenitors in the cerebellar cortex of peripuberal and adult rabbits. PLoS ONE. 2008:3. doi: 10.1371/journal.pone.0002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Blangy A, Xiong G, Benian GM. The DH-PH region of the giant protein UNC-89 activates RHO-1 GTPase in Caenorhabditis elegans body wall muscle. J Mol Biol. 2008;383:747–52. doi: 10.1016/j.jmb.2008.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeker MO, Su F, Geisler SB, Borisov AB, Kontrogianni-Konstantopoulos A, Lyons SE, Russell MW. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev Dyn. 2006;235:2018–29. doi: 10.1002/dvdy.20812. [DOI] [PubMed] [Google Scholar]
- Russell MW, Raeker MO, Korytkowski KA, Sonneman KJ. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–46. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue CW, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–95. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Wei L, Zhou W, Croissant JD, Johansen FE, Prywes R, Balasubramanyam A, Schwartz RJ. RhoA signaling via serum response factor plays an obligatory role in myogenic differentiation. J Biol Chem. 1998;273:30287–94. doi: 10.1074/jbc.273.46.30287. [DOI] [PubMed] [Google Scholar]
- Wennerberg K, Der CJ. Rho-family GTPases: it’s not only Rac and Rho (and I like it) J Cell Sci. 2004;117:1301–12. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- Young P, Ehler E, Gautel M. Obscurin, a giant sarcomeric Rho guanine nucleotide exchange factor protein involved in sarcomere assembly. [see comment] J Cell Biol. 2001;154:123–36. doi: 10.1083/jcb.200102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.