Abstract
Investigations aimed at identifying regulators of nuclear architecture in Drosophila demonstrated that cells lacking H3K9 methylation and RNA interference (RNAi) pathway components displayed disorganized nucleoli, ribosomal DNA (rDNA) and satellite DNAs. The levels of H3K9 dimethylation (H3K9me2) in chromatin associated with repeated DNAs decreased dramatically in Su(var)3–9 and dcr-2 (dicer-2) mutant tissues compared with wild type. We also observed a substantial increase in extrachromosomal circular (ecc) repeated DNAs in mutant tissues. The disorganized nucleolus phenotype depends on the presence of Ligase 4 and ecc DNA formation is not induced by removal of cohesin. We conclude that the structural integrity and organization of repeated DNAs and nucleoli are regulated by the H3K9 methylation and RNAi pathways, and other regulators of heterochromatin-mediated silencing. In addition, repeated DNA stability involves suppression of non-homologous end joining (NHEJ) or other recombination pathways. These results suggest a mechanism for how local chromatin structure can regulate genome stability, and the organization of chromosomal elements and nuclear organelles.
Nuclei and chromosomes maintain specific and dynamic architectures that are required for many essential functions1. Nuclear bodies are involved in diverse biological processes and exhibit dynamic mobility, and individual chromosomes occupy distinct domains within interphase nuclei2. Chromosomes in the metazoan interphase nucleus are comprised of two types of cytologically and functionally distinct chromatin, euchromatin and heterochromatin3. Patterns of posttranslational histone modifications associated with these domains are strongly correlated with functions such as gene regulation, chromosome inheritance and replication timing4. For example, regions that display heterochromatin-mediated gene silencing are rich in histone H3K9 methylation and lack many histone acetylations, whereas histones in transcriptionally active euchromatic regions are highly acetylated and methylated at H3K4 (ref. 5).
The first indication that chromosome organization can affect gene expression stems from the discovery of position effect variegation (PEV) in Drosophila6. PEV describes the epigenetic inactivation or silencing of a euchromatic gene that has been positioned close to or within heterochromatin, or a heterochromatic gene moved to distal chromosome locations. PEV is exquisitely sensitive to the dosage of genetic modifiers, known as suppressors and enhancers of variegation (Su(var)s and E(var)s)7. PEV modifiers regulate heterochromatin formation and functions. The Su(var)3–9 family encodes a histone methyltransferase (HMTase) responsible for H3K9 methylation, and Su(var)2–5 encodes heterochromatin protein 1 (HP1). Methylated H3K9 and Su(var)3–9 bind to HP1, providing a molecular mechanism for maintaining the silenced epigenetic state5. Components of the RNAi pathway also act as Su(var)s to influence silencing of tandem repeats8. Recent studies have shown that the RNAi pathway and double-stranded RNAs are required for the initial recruitment of H3K9 methyltransferases, and establishment and maintenance of heterochromatin9.
The nucleolus, the site of ribosome assembly, is an example of an essential nuclear organelle. The nucleolus organizer region (NOR) contains tandemly repeated ribosomal DNAs (rDNAs), and is embedded in heterochromatin in most eukaryotes. Single rDNA genes inserted into euchromatin are able to form mini-nucleoli through a self-assembly process that is likely to be initiated by rRNA transcription10. Epigenetic regulation of rDNA transcription is well documented in yeast, plant and mammalian systems — for example, a mammalian nucleolar remodelling complex (NoRC) regulates heterochromatin formation and rRNA expression by controlling histone H4 deacetylation, H3K9 dimethylation and de novo DNA methylation at rDNA11. In addition, nucleolar dominance is observed when only one NOR forms a nucleolus in interspecies hybrids12. rDNA magnification occurs in yeast and flies with low rDNA content; this process is likely to be mediated by unequal sister-chromatid recombination13. Finally, mutations in a protein that regulates silencing in Saccharomyces cerevisiae (SIR2) result in ecc rDNA formation, which is thought to influence cell senescence and aging14.
It is surprising that rDNA produces the overwhelming majority of RNAs in the cell, despite its association with `silenced' heterochromatin. This paradox suggests that the evolutionarily conserved positioning of NORs in heterochromatin may regulate important, unknown features of nucleolus formation. Here, we test the hypothesis that heterochromatin and associated proteins regulate the organization of nucleoli and repeated DNAs in Drosophila. Our results demonstrate that a subset of Su(var) proteins, including the Su(var)3–9 HMTase, HP1 and the RNAi pathway, are required for the normal organization of nucleoli and satellite DNAs in the nucleus. Furthermore, these regulators of heterochromatin suppress ecc DNA formation from repeated DNAs, which is mediated by NHEJ or other recombination/repair pathways.
RESULTS
Multiple nucleoli are present in Su(var) mutant cells
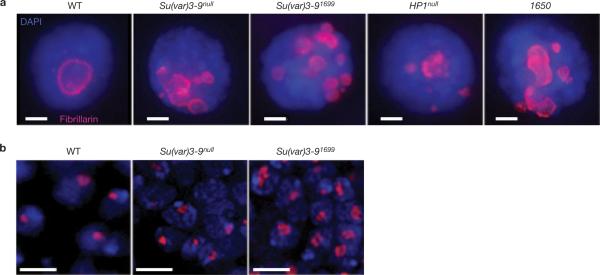
Indirect immunofluorescence microscopy was used to examine nucleolar organization in whole-mount (three dimensional) imaginal disc tissues and polytene larval salivary glands from wild-type and Su(var) mutant larvae (see Methods). Staining for fibrillarin, a component of the rRNA processing machinery15, confirmed that wild-type polytene and diploid cells contain single nucleoli (Fig. 1). In contrast, salivary gland cells from animals homozygous for mutations in the Su(var)3–9 histone H3K9 methyltransferase or HP1/Su(var)2–5 genes contained 1–12 nucleoli (Fig. 1a and see Supplementary Information, Table S1). The average numbers of nucleoli in mutant cells (Su(var)3–9null = 2.7, Su(var)3–91699 = 5.0, HP1null = 2.8) were significantly higher than in wild type (average = 1, see Supplementary Information, Table S1). Increases in nucleolar numbers were accompanied by a proportional increase in both nuclear and nucleolar volumes, even though the ratios remained constant (data not shown). Irregularly shaped, multilobed nucleoli were observed in 44% of Su(var)3–9null mutant diploid imaginal disc cells, compared with only 10% in wild-type cells (Fig. 1b).
Figure 1.
Su(var) mutants contain multiple nucleoli. (a) Immunofluorescence microscopy with antibodies against the nucleolus marker fibrillarin (red) in whole-mount salivary-gland nuclei from wild-type (WT), Su(var)3–9null, Su(var)3–91699, HP1null and Su(TDA-PEV)1650 homozygous mutants. Wild-type cells have one nucleolus, whereas the mutants display multiple nucleoli. DAPI, blue. The scale bar represent 10 μm. (b) Immunofluorescence microscopy of fibrillarin in whole-mount imaginal disc and brain tissues from wild-type and Su(var)3–9 mutants are shown. The single, wild-type nucleolus tended to be round, whereas nucleoli in the mutants were irregular (lobed) and larger. Quantitative analysis showed that 44% of Su(var)3–9null mutant nuclei contained lobed nucleoli (n = 55), versus 10% for wild type (n = 51; P <0.001). The scale bars represent 5 μm.
Fibrillarin staining in salivary glands of Su(TDA-PEV)1650 (Fig. 1a) and Su(var)2–10/dPIAS mutant cells displayed the multiple nucleoli phenotype, whereas mutations in seven other Su(var) loci and two Polycomb group (PcG) genes had no effect on nucleolar organization (see Supplementary Information, Table S1). We conclude that many but not all regulators of gene silencing are required for the formation of a normal, single nucleolus.
Ectopic nucleoli in Su(var) mutants contain rDNA
Multiple, ectopic nucleoli associated with Su(var) mutations could result from dispersion or fission of nucleolar material initially formed around a single rDNA cluster, or from mislocalization of rDNA. These hypotheses were distinguished by combined fibrillarin immunofluorescenc microscopy and rDNA fluorescent in situ hybridization (FISH). Wild-type polytene nuclei displayed single rDNA sites within each nucleolus, whereas nuclei from Su(var)3–9, HP1 and Su(TDA-PEV)1650 mutants contained multiple, dispersed rDNA foci associated with ectopic nucleoli (Fig. 2a). Similarly, 33% of Su(var)3–9null diploid disc cells contained multiple rDNA sites (average = 1.44 ± 0.72), compared with only 2% of wild type (average = 1 ± 0.1, Fig. 2b, c). These results demonstrate that ectopic nucleoli in Su(var) mutants are nucleated independently by mislocalized rDNA, including the multilobed nucleoli observed in mutant diploid cells.
Figure 2.
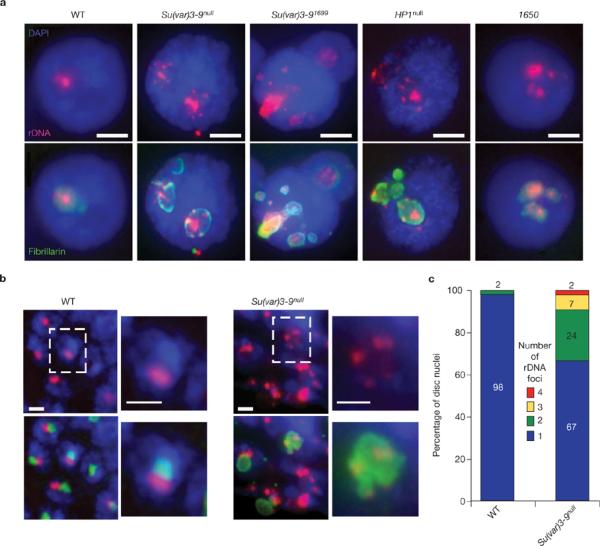
Su(var) mutants have dispersed rDNA foci, each of which forms an ectopic nucleolus. (a) FISH of rDNA (red) and immunfluorescence microscopy of fibrillarin (green) were performed on whole-mount salivary glands from wild-type, Su(var)3–9null, Su(var)3–91699, HP1null and Su(TDA-PEV)1650 homozygous mutants. DAPI, blue. There is a single site of rDNA in >98% of wild-type nuclei, whereas the Su(var) mutant nuclei contain multiple rDNA foci, which are all surrounded by fibrillarin. The scale bars represent 15 μm. (b) Combined rDNA FISH (red) and immunofluoresence microscopy of fibrillarin (green) of whole-mount imaginal disc and brain tissues from wild-type and Su(var)3–9null mutant larvae. Wild-type nucleoli contain a single, compact rDNA focus, whereas Su(var)3–9null mutants frequently display multiple rDNA foci. The scale bars represent 3 μm. Boxed nuclei are shown at higher magnification to the right of each image. (c) Quantitative analysis of the number of rDNA foci in wild-type and Su(var)3–9null diploid nuclei. 98% of wild-type cells (n = 96) contain one rDNA signal, compared with 67% of Su(var)3–9null nuclei, and the percentages with two, three and four rDNA signals were 24%, 7% and 2%, respectively (average = 1.44 ± 0.73 rDNA foci per mutant nucleus, n = 53, P <0.001).
Su(var)3–9 mutants disrupt the organization of other repeated DNAs
The severe disruption of rDNA and nucleolar organization raised the possibility that the three-dimensional spatial relationships of other heterochromatic DNAs are also affected by Su(var) mutations. FISH analysis on polytene nuclei was performed using probes against tandemly repeated satellite DNAs (Fig. 3a), which localize to the heterochromatic chromocenter16. An average of two sites were observed in wild-type nuclei for satellites 1.688 and 1.686, compared with three sites in Su(var)3–9null mutants (Fig. 3b, c; P <0.001). Similar observations were made with satellites AACAC and AATAT (data not shown). Distances between signals for each satellite increased significantly in Su(var)3–9null mutants (1.688 = ninefold, 1.686 = threefold; Fig. 3b, d; P <0.001). Notably, mislocalized satellite DNA and rDNA were not restricted to a specific nuclear compartment. We conclude that heterochromatic repeated DNAs become dispersed and disorganized in Su(var)3–9 mutants, as observed for rDNA and nucleoli.
Figure 3.
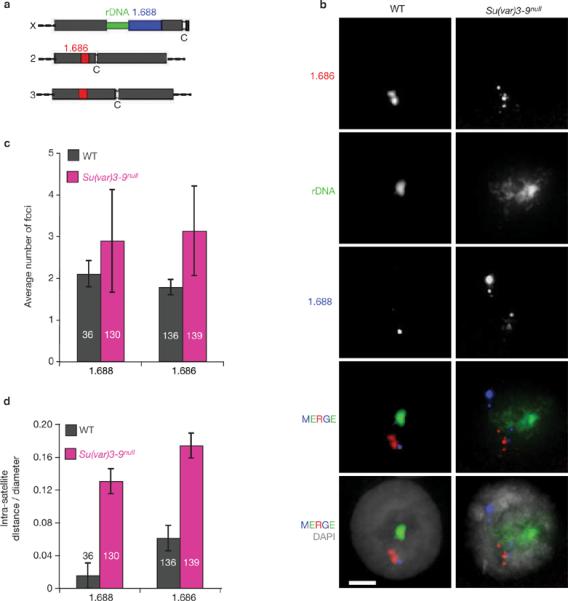
Satellite DNA organization is disrupted in Su(var)3–9null mutant nuclei. (a) Schematic representation of the locations of rDNA and satellite DNAs in the Drosophila melanogaster genome (not to scale). The rDNA is located in the heterochromatin of the X and Y sex chromosomes, the 1.688 satellite (359-bp repeats) is next to the X rDNA, and the 1.686 satellite is in the heterochromatin of chromosomes 2 and 3. (b) FISH was performed on whole-mount polytene salivary glands isolated from wild-type and Su(var)3–9null mutants. In wild-type nuclei, specific satellite DNAs are localized at single sites, and the different satellite signals are close to each other. In Su(var)3–9null mutant nuclei, individual satellite DNAs are dispersed to multiple sites and are not clustered with other satellites. DAPI, grey. FISH probe colours correspond to the diagram in a. The scale bars represent 15 μm. (c) The number of 1.688 and 1.686 foci were significantly higher in mutant nuclei compared with wild type (P <0.001). (d) Distances between satellite signals were quantified in three-dimensional reconstructions. The intra-satellite distances in Su(var)3–9null mutant nuclei were significantly higher than in wild type (P <0.001). The n values are indicated within the histograms and error bars correspond to s.d.
H3K9me2 levels at rDNA and satellite DNAs decrease significantly in Su(var)3–9 mutants
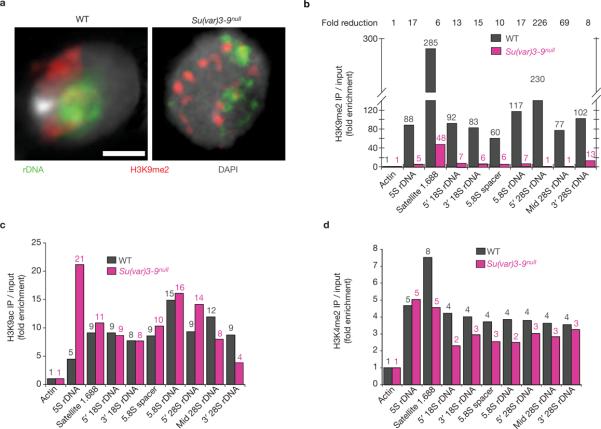
Heterochromatic nucleosomes in a variety of organisms, including Drosophila, are enriched for the H3K9me2 modification. Thus, HP1 and Su(var)3–9 could control rDNA and nucleolar organization indirectly by regulating the flanking heterochromatin, or could act directly on rDNA chromatin. To address this question, we determined whether rDNA and satellite DNAs contained methylated H3K9, and if this modification was disrupted in Su(var)3–9 mutants. Combined immunofluorescence microscopy and FISH studies indicated that H3K9me2 partially overlapped with rDNA in wild-type diploid cells, and was significantly reduced in Su(var)3–9null mutants (Fig. 4a).
Figure 4.
Analysis of histone modifications in chromatin containing repeated DNA in wild-type and Su(var)3–9null cells. (a) Immunofluorescence microscopy using antibodies that specifically bind H3K9me2 (red) combined with FISH for rDNA (green) in squashed diploid nuclei. In wild-type nuclei, rDNA chromatin partially overlaps with the H3K9me2 signals (correlation coefficient = 0.61 ± 0.08), and the amount of overlap was significantly reduced in Su(var)3–9null mutants (correlation coefficient = 0.40 ± 0.03; P <0.01). The scale bar represents 3 μm. (b) ChIP analysis of H3K9me2 levels in wild-type and Su(var)3–9null mutant imaginal disc tissues. The graph shows H3K9me2 levels for the repeated DNAs examined by PCR, standardized to actin and HDAC single-copy controls (see Methods); values are averages of five ChIP experiments and are shown above each bar. In wild-type cells, the 1.688 satellite (359-bp repeats), 5S rDNA (in chromosome 2 euchromatin) and the rDNA on the sex chromosomes contain significant enrichment for H3K9me2, compared with input chromatin and controls. H3K9me2 levels in chromatin derived from Su(var)3–9null mutant tissues were significantly reduced (6–226-fold) compared with wild type. (c, d) ChIP analysis of two modifications associated with active or open chromatin (H3K9ac and H3K4me2). Small enrichment for these modifications was observed on repeated DNAs in wild-type chromatin, compared with input and single-copy controls. For most of the repeated DNAs, levels were not significantly altered in Su(var)3–9null mutant chromatin (P >0.5 for all regions). H3K9ac levels were significantly increased in 5S rDNA in the mutants (P <0.05), and H3K4me2 was significantly decreased for the 1.688 satellite (P <0.05). Values are averages of two experiments and are shown above each bar.
Quantitative chromatin immunoprecipitation (ChIP) showed that rDNA, 5S rDNA and satellite DNAs were enriched for H3K9me2 in wild-type diploid cells, and that this modification was not well represented at single-copy gene controls (actin and HDAC3; Fig. 4b). Moreover, the levels of H3K9me2 on all repeated DNAs decreased substantially in chromatin isolated from Su(var)3–9null mutant discs. The reductions in H3K9me2 levels varied between the different repeats and within the rDNA (6–226-fold:Fig. 4b), most likely reflecting known redundancy in the HMTases responsible for this modification in flies17. Taken together, the ChIP and combined immunofluorescence microscopy and FISH (IF–FISH) results suggest that the effects of Su(var)3–9 on rDNA and nucleolar organization are mediated through the chromatin structure of the rDNA itself, rather than solely through the flanking heterochromatin.
H3K4me2 and H3K9 acetylation (H3K9ac) have been characterized as modifications associated with active or open chromatin. ChIP analysis showed that chromatin associated with repeated DNAs contained low levels of H3K9ac (Fig. 4c) and H3K4me2 (Fig. 4d), which remained mostly unchanged in Su(var)3-9null mutants. The exceptions were significant increases in 5S rDNA H3K9ac levels in the mutants (P <0.03) and H3K4me2 decreases for the 1.688 satellite (P <0.04).
Su(var)3–9 mutations cause significant increases in the amount of ecc repeated DNA
Loss of H3K9me2 and defective heterochromatin structure could alter the organization of repeated DNAs and nucleoli by formation of ecc DNAs, or chromatin decondensation could cause dispersal of repeated DNAs in the cytoplasm. To examine these hypotheses, ecc DNA was quantified in mutant and wild-type cells using `Hirt' supernatants, which separates ecc DNA from genomic DNAs18. Wild-type polytene tissues contained ecc 5S rDNA and satellite 1.688 DNA, as observed previously19, but very low levels of ecc 18S–5.8S–28S rDNA (Fig. 5a). The amounts of ecc DNA increased dramatically in Su(var)3–9null mutant tissues versus wild type for rDNA (46–78-fold) and satellite 1.688 (20-fold;Fig. 5b; P <0.05 for all regions), which was not observed for the single-copy genes, actin and HDAC. For diploid cells, ecc repeated DNAs were approximately twofold higher in Su(var)3–9 mutant tissues than in wild type (Fig. 5c; P <0.05 for all regions). Lower levels of ecc DNA in mutant diploid cells are likely to result from the absence of endoreplication and loss during mitosis (see Discussion). We conclude that loss of H3K9me2 from chromatin containing repeated DNAs results in ecc DNA formation, and that the increased ecc rDNA leads to the formation of ectopic nucleoli.
Figure 5.
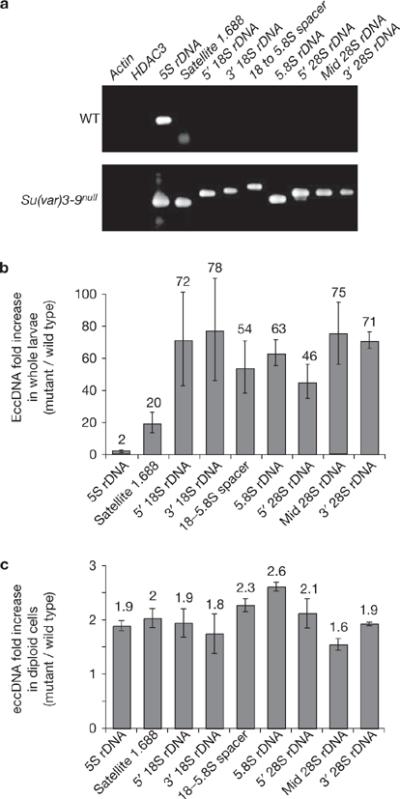
Levels of extrachromosomal repeated DNAs are significantly increased in Su(var)3–9null mutant tissues compared with wild type. (a) Extrachromosomal DNA was isolated from wild-type and Su(var)3–9null mutant larvae using the Hirt supernatant method, and PCR reactions, terminated at logarithmic phase of amplification, were performed to evaluate the amounts of eccDNA corresponding to specific sequences (see Methods). The gel shows an example of the PCR reactions for the specific regions examined. EccDNAs from the single-copy genes (actin and HDAC3) were not detected in either wild-type or mutant larvae. The asterisk indicates that the band in the 1.688 satellite lane corresponds to the primers, not the PCR products. (b) Quantification demonstrates that the amount of eccDNA for the 1.688 satellite and different regions of the rDNA are significantly higher in Su(var)3–9null mutants compared with wild-type (20–78-fold enrichment); the increase for 5S rDNA was only twofold because wild-type larvae contain high levels of ecc 5S rDNA. The values, as shown above each bar, are averages of three sample extractions. (c) Quantification of PCR products indicates that the amount of eccDNA in Su(var)3–9null mutant diploid cells is about twofold higher than in wild type. The values, shown above each bar, were averages of three sample extractions, and P values were <0.05 for the regions examined. The error bars correspond to s.d.
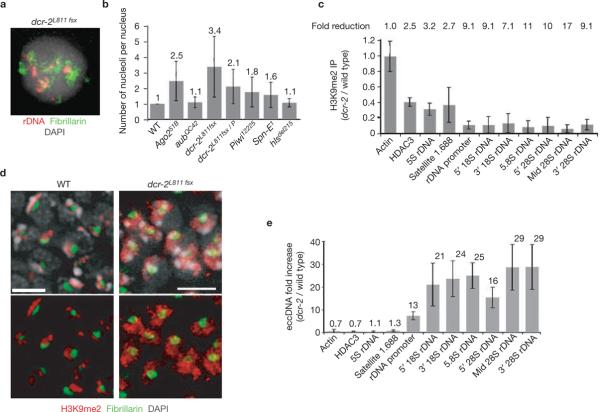
RNAi mutants display multiple nucleoli, loss of H3K9me2 and increases in ecc rDNA
The targeting of H3K9me2 by the RNAi pathway9 and the presence of small RNAs homologous to the 1.688 satellite and other repeats20 led us to examine RNAi mutants for disorganized nucleoli. All RNAi loci examined displayed significantly increased nucleolus numbers when mutated, in comparison with wild type (P <0.001 for all except for hlsΔ215, P <0.08; Fig. 6a, b and see Supplementary Information, Table S1). Dicer-2 is required for siRNA-directed mRNA cleavage and ChIP analysis of dcr-2L811fsx mutants revealed significant H3K9me2 reduction in the rDNA (P <0.05, Fig. 6c); unlike Su(var)3–9 mutants, reductions were not observed for 5S rDNA and satellite 1.688. Interestingly, H3K9me2 was concentrated in heterochromatin in wild-type cells, but was more broadly distributed in dcr-2L811fsx homozygous mutants (Fig. 6d). Finally, ecc rDNA increased significantly in dcr-2L811fsx mutants (13–29-fold, Fig. 6e), consistent with ectopic nucleolus formation.
Figure 6.
The RNAi pathway is also required to maintain the structural integrity of repeated DNAs and the nucleolus. (a) Combined rDNA FISH (red) and immunofluorescence microscopy of fibrillarin (green) shows that dcr-2L811fsx polytene nuclei contain multiple rDNA foci and ectopic nucleoli. (b) The histogram shows the average number of nucleoli in different RNAi mutants examined (absolute values are indicated above each bar). Mutations at all loci contained significantly more nucleoli than wild type (P <0.001, except aubQC42, P <0.004). The hlsdel215 allele of SpnE1 had a mild phenotype (P = 0.083). (c) ChIP analysis reveals reduced H3K9me2 levels in dcr-2L811fsx chromatin compared with wild type (P <0.05), more so for rDNA than the 5S rDNA and satellite 1.688. Values are averages of four PCR reactions from two ChIP experiments and the overall fold reductions are shown above. (d) Immunofluorescence microscopy for H3K9me2 (red) and fibrillarin (green) on whole-mount brains and imaginal discs from wild-type and dcr-2L811fsx mutants. H3K9me2 localizes predominantly in DAPI-bright heterochromatin regions in wild-type nuclei, but is more broadly distributed in dcr-2L811fsx nuclei. The scale bar represents 5 μm. (e) ecc rDNA levels in dcr-2L811fsx mutant larvae are significantly higher than in wild type (13–29-fold increases), but eccDNA levels for 5S rDNA and satellite 1.688 did not increase. The values indicated above each bar are averages of two PCR experiments from two sample extractions (P <0.05). The error bars correspond to s.d.
Ligase 4 mutations partially suppress the Su(var)3–9 multiple nucleolus phenotype
Extrachromosomal DNA formation is likely to arise from somatic recombination, as suggested by the observation that sir2-dependent ecc rDNA formation in S. cerevisiae requires the RAD52 complex14,21 (sir2 encodes an NAD-dependent histone deacetylase that is required for epigenetic silencing, and the RAD52 complex is required for repair of double-strand breaks). Efforts to identify recombination proteins required for ecc DNA formation in Drosophila have been unsuccessful22 and Drosophila RAD52 homologues have not been identified. A recent study identified Ligase IV, an essential regulator of NHEJ, as necessary for ecc DNA formation in mammals23. Fibrillarin immunofluorescence microscopy showed that polytene nuclei from Lig4 mutants (transheterozygous for null alleles 57 and 29) contained a single nucleolus (Fig. 7a). However, cells from Lig4–Su(var)3–9 double mutants displayed an average of 1.7 nucleoli (± 0.8; n = 83), which is significantly lower than the 2.7 nucleoli observed in Su(var)3–9null single mutants (P <0.001). Thus, loss of Lig4 partially suppresses the formation of multiple nucleoli in Su(var)3–9 mutants. We conclude that Lig4 and the NHEJ pathway, and perhaps other repair machinery, participate in ecc DNA formation in Drosophila (see Discussion).
Figure 7.
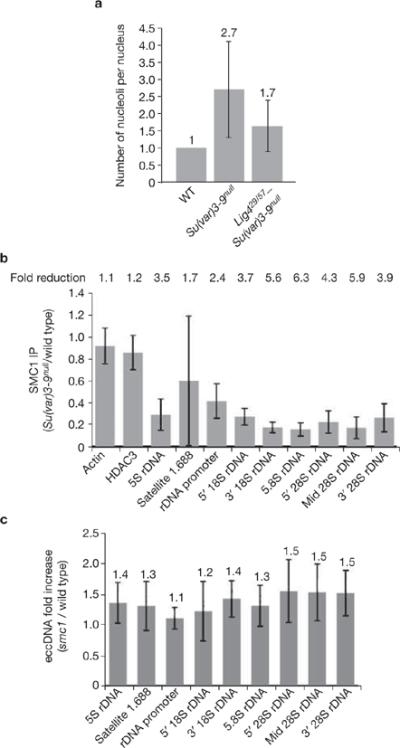
Effects of Ligase 4 and cohesin mutants on ectopic nucleolus and eccDNA formation. (a) Ligase 4 mutations partially suppress ectopic nucleolus formation in Su(var)3–9 mutants. Average nucleolus number of Lig4null–Su(var)3–9null polytene nuclei is 1.7 (n = 83), which is significantly lower (P <0.001) than the average 2.7 (n = 54) nucleoli observed in Su(var)3–9null mutant nuclei. Absolute values are indicated above each bar. (b) ChIP analysis shows reduced SMC1 levels at repeated DNAs in Su(var)3–9null chromatin, relative to wild type; fold reductions are shown (P <0.05 for all repeated DNA except 5S rDNA). Values are averages of four PCR reactions from two ChIP experiments. (c) The amount of eccDNA from satellite 1.688 and rDNA in smc1exc46l mutant tissues do not differ significantly from wild type. Absolute values are indicated above each bar. Error bars indicate s.d.
Su(var)3–9 mutations reduce cohesin levels, but a cohesin mutation does not increase ecc DNAs
Sister-chromatid cohesion inhibits ecc rDNA formation in S. cerevisiae24, and H3K9me2 and the HP1 homologue SWI6 are required for cohesin recruitment in S. pombe25. Thus, recruitment of cohesin to pericentric heterochromatin by the H3K9me pathway could also regulate repeated DNA structural integrity in Drosophila. ChIP analysis showed that levels of the SMC1 (structural maintenance of chromosme 1) cohesin subunit26 were significantly reduced in Su(var)3–9null chromatin (16–29% of wild-type levels, Fig. 7b). However, the amount of ecc DNA isolated from animals homozygous for the smc1exc461 mutation did not differ significantly from wild type (Fig. 7c). Therefore, we conclude that Su(var)3–9 and H3K9 methylation are required for cohesin recruitment at repeated DNAs in Drosophila, but cohesin is not essential for repressing ecc DNA formation.
DISCUSSION
Posttranslational histone modifications have been correlated with regulation of gene expression4. However, recent discoveries of distinct centromeric histone modifications, and the requirement of H3K9me2 in transcriptional elongation, indicate that some patterns of modifications defy simple global interpretations27,28. In addition, we have limited knowledge of the impact of chromatin structures on other nuclear functions, such as genome stability, and the three-dimensional organization of sequences, chromosomes and nuclear organelles. Here, we show that the Su(var)3–9 H3K9 methyltransferase, its binding partner HP1, and five components of the RNAi pathway are required for the normal organization of rDNA, satellite DNAs and nucleoli in Drosophila. When animals lack these components, repeated DNAs and nucleoli become dispersed to multiple nuclear locations. ChIP and IF–FISH showed that H3K9me2 levels in chromatin associated with repeated DNAs are strongly reduced in Su(var)3–9 and dcr-2 mutant animals. We also observed significantly increased amounts of extrachromosomal repeated DNAs in these mutants. We conclude that the H3K9 methylation and RNAi pathways directly regulate nuclear architecture, by affecting chromatin structure and repressing formation of rDNA and satellite ecc DNAs.
We observed that other genes involved in heterochromatin structure and function are also required to maintain the structural integrity of repeated DNA and nucleoli, specifically the dPIAS SUMO E3 ligase29,30 and Su(TDA-PEV)1650 (function unknown). Further studies are required to determine whether these proteins effect the integrity of repeated DNA and nucleoli through H3K9 methylation or other pathways. Mutations in nine out of 18 loci associated with gene silencing had no effect on nucleolar organization, including dsir2 and two Polycomb-group genes (see Supplementary Information, Table S1). Interestingly, loss of the SUV4-20 H4K20 methyltransferase did not produce multiple nucleoli; this result demonstrates that H3K9 methylation is the primary histone modification responsible for maintaining repeated DNA integrity, and not H4K20 trimethylation by SUV4-20, which requires H3K9me2 (ref. 31). In addition, Drosophila sir2 mutants did not contain multiple nucleoli, despite SIR2 repression of ecc rDNA formation in S. cerevisiae14. We conclude that rDNA organization and nucleolar architecture are regulated by some but not all proteins involved in heterochromatin structure and function.
We observed that Dicer-2 regulates H3K9me2 levels and ecc DNA formation at some but not all repeated DNAs, in contrast with the broad impact of Su(var)3–9. For example, Su(var)3–9 mutants displayed increased levels of ecc rDNA, 5S DNA and 1.688 satellite, whereas only ecc rDNA levels increased significantly in dcr-2 mutants The dcr-2 locus has been shown to affect production of small interfering RNAs (siRNAs) more than micro RNAs (miRNAs)32; thus, Dcr-2 and other RNAi components may preferentially direct H3K9 methylation at rDNA through the siRNA pathway in Drosophila. In contrast, the stability of other repeats such as satellites and 5S DNA seems to be regulated by Su(var)3–9 and H3K9 methylation through other pathways or components, independent of the RNAi pathway.
The overall distributions of H3K9me2 observed with immunofluorescence microscopy analysis were also strikingly different in Su(var)3–9 and dcr-2 mutants. H3K9me2 levels were significantly reduced in Su(var)3–9 mutant nuclei, although visible amounts were retained in the heterochromatin (Fig. 4a). In contrast, H3K9me2 levels were not reduced in dcr-2 mutant diploid cells, and instead were mislocalized to a larger portion of the nucleus (Fig. 6d). This observation is surprising, as H3K9me is not detectable in RNAi mutants in S. pombe33. We conclude that the absence of Dicer-2 alters the specificity of siRNA-mediated targeting of H3K9me2 in Drosophila.
Mice deleted for both Suv3–9 genes exhibit genome instability and early embryonic lethality34. In contrast, Drosophila Su(var)3–9null flies are viable and fertile, despite very low levels of H3K9me2, presumably due to the presence of redundant H3K9 methyltransferases17. Our studies show that the absence of Su(var)3–9 has dramatic effects on nuclear organization. The presence of fibrillarin around ectopic rDNAs suggests that transcription and processing of ribosomal RNA occurs in ectopic nucleoli. Similarly, ectopically integrated rDNA forms functional `mini-nucleoli'10 and rescues defects in X–Y pairing in male meiosis caused by endogenous rDNA deletion35. These observations suggest that increased nucleolar volumes and ecc rDNA do not cause significant growth abnormalities. However, more developmental or physiological phenotypes may yet be discovered. For example, we have observed that non-recombinant chromosomes display significantly increased levels of meiotic non-disjunction in Su(var)3–9null females (G.H.K., unpublished observations), consistent with previous studies demonstrating heterochromatin's participation in achiasmate segregation36,37.
Another explanation for the absence of dramatic phenotypic abnormalities in Su(var)3–9null animals arises from our observation that diploid tissues display lower levels of ectopic nucleoli and ecc DNA compared with polytene cells. ecc DNAs lack functional centromeres and should be poorly transmitted in rapidly dividing diploid cells, but would be retained in the non-mitotic polytene cells. We propose that the levels of ecc DNA and ectopic nucleoli in mitotic larval cells that give rise to most adult tissues are not high enough to affect viability and fecundity.
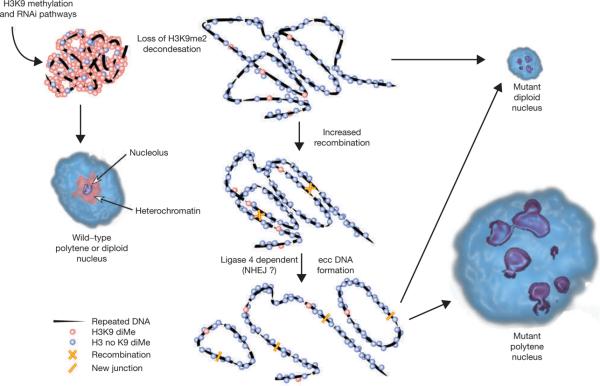
Our findings demonstrate that chromatin structures regulated by Su(var)3–9, HP1, the RNAi pathway and H3K9 methylation are required to maintain the structural integrity of tandemly repeated, heterochromatic sequences (Fig. 8). HP1-mutant cells display increased restriction enzyme accessibility in heterochromatin, consistent with chromatin decondensation and loss of gene silencing38. We propose that H3K9 methylation, and perhaps other heterochromatic properties and components, generate a chromatin structure that normally restricts access of DNA repair proteins to repeated DNA substrates, or locally inhibits their activity (Fig. 8). The observation that mutations in Lig4, an essential player in the NHEJ pathway, partially suppress ectopic nucleolus formation in Su(var)3–9null mutants supports a role for DNA repair. It is somewhat surprising that the NHEJ pathway, rather than homologous recombination, would participate in ecc DNA formation from repeats. However, Lig4 is associated with proteins required for double-strand-break repair and V(D)J site-specific recombination39, and may affect other repair pathways in Drosophila. Alternatively, repeated ecc DNA formation in Drosophila may not involve homologous recombination. Finally, we also showed that cohesins are significantly reduced at repeated DNA chromatin in Su(var)3–9null nuclei. However, complete loss of the SMC1 cohesin component did not lead to increases in ecc DNA. This suggests that cohesins do not suppress ecc DNA formation in Drosophila, contrary to observations in S. cerevisiae24.
Figure 8.
A schematic representation of a model for regulation of nuclear architecture by the H3K9 methylation and RNAi pathways. In wild-type diploid and polytene nuclei, the majority of the heterochromatin contains H3K9me2 and a single nucleolus forms around the rDNA. Loss of H3K9me2 from repeated DNA, due to Su(var)3–9, HP1 or RNAi mutations, causes chromatin decondensation and elevated recombination between repeated DNA copies. The recombination process results in formation of eccDNAs that localize throughout the nucleoplasm, causing dispersal of satellite DNAs (not shown) and, in the case of rDNA, the formation of ectopic nucleoli. Decondensation is proposed to be primarily responsible for the lobed structure of rDNA and nucleoli in diploid cells, with a minor contribution from low levels of ecc rDNA formation (dotted line). In polytene cells, decondensation is likely to be a prerequisite for increased recombination, but the much higher levels of ecc rDNA is proposed to generate the majority of the ectopic nucleoli.
Chromatin decondensation (for example, `looping') and increased recombination are likely to occur in both diploid and polytene cells in response to loss of H3K9 methylation (Fig. 8). The much larger increases in ecc DNAs and ectopic nucleoli in polytene tissues probably reflect a stronger requirement for repressing DNA recombination or NHEJ. In highly endoreplicated nuclei, euchromatic sequences are present in thousands of copies, satellite sequences are replicated at most twice and rDNA is replicated to intermediate levels (~250 copies)16. This differential endoreplication results in stalled forks at euchromatin-heterochromatin junctions40, and presumably between the rDNA and adjacent sequences. Stalled forks and associated single-stranded DNA in S. cerevisiae have been shown to provide substrates for repeated ecc DNA formation41, and could have similar roles in polytene cells that lack Su(var)3–9 and H3K9 methylation. Diploid cells would not be expected to generate as much ecc DNA and ectopic nucleoli as endoreplicating cells, because DNA copy numbers are much lower, and they would not contain as many stalled replication forks. Furthermore, diploid nuclei would not retain ecc DNAs, as they are likely to be lost during cell division; maintenance of ecc DNAs in polytene nuclei would be higher, as they are non-mitotic.
Other examples of heterochromatic silencing mechanisms affecting recombination have been reported. Some combinations of Su(var) mutations increase meiotic recombination in Drosophila heterochromatin42, and loss of gene-silencing components in budding and fission yeasts increases both meiotic and somatic recombination in the rDNA33,43. Similarly, the G9a H3K9 methyltransferase regulates accessibility of the V(D)J recombination machinery during mouse lymphocyte development44. Here, we expand on previous studies by demonstrating the impact of these mechanisms on nucleolar organization and the spatial arrangement of repeated sequences in the nucleus of a developing animal. In addition, these findings may have broader significance to genome stability; the extensive sequence homology inherent to repeated DNAs would presumably generate translocations and other chromosome aberrations in somatic and/or germ cells if exchange was not repressed by heterochromatic structures.
METHODS
Fly stocks
All fly stocks were raised at 22 °C. Information regarding the fly strains are described in the Supplementary Information, Table S1. The suv4–20, Pc, Ph, Lig4, aub, Spn-E1 and piwi flies were from the Bloomington stock center (Bloomington, IN) and the dcr-2 P element insertion was from the Harvard fly center (Boston, MA).
Antibodies
The human anti-fibrillarin antibody (dilution 1:500 for immuofluorescence microscopy) was a gift from K. M. Pollard (TSRI, San Diego, CA), and the rabbit anti-H3K9me2 antibody (dilution 1:100 for immuofluorescence microscopy and 1:1000 for ChIP) was provided by T. Jenuwein (IMP, Vienna, Austria)45. Rabbit antibodies against H3K9ac and H3K4me2 were purchased from Upstate (Charlottesville, VA). Rabbit antibodies against SMC1 (1:1000 for ChIP) were a gift from D. Dorsett (St Louis University School of Medicine, St Louis, MI)46.
FISH and IF-FISH
FISH was performed as previously described37 using 100 ng of each probe. In combined IF–FISH experiments, immunofluorescence was performed before the FISH treatment. FISH probes were made by nick translation and terminal labelling, using previously described materials10.
Volumetric, distance and colocalization analysis
All images were captured using an Applied Precision Deltavision Workstation and deconvolved using the conservative algorithm with eight iterations. SoftWorx software was used to measure colocalization and distances, which were normalized to nuclear diameter. The deconvolved, stacked images were converted to TIFF files, three-dimensionally reconstructed, and volumes of nuclei (DAPI signals) and nucleoli (fibrillarin signals) were measured using Metamorph software. All statistical comparisons and P values were calculated using the two-sample t test, assuming unequal variance.
Chromatin immunoprecipitation
Modifications of a previously described protocol were used47. Brain and disc tissues were dissected from fifty third-instar larvae, then fixed in 1.8% paraformaldehyde in PBST for 10 min at room temperature. The tissues were washed twice in cold PBST, then once in cold TE and RIPA lysis buffer. Sonication in a 1 ml volume was performed with a Branson Sonifier 450 (six times with a 90% duty cycle and a 5.5 power output; each cycle included a 2-min rest interval) or the Bioruptor (Diagenode, Philadephia, PA); 30 s on–off cycles for 12 min at high intensity). Sheared chromatin (300 μl) was used for immunoprecipitaion. The input and immunoprecipitated DNAs were resuspended in 100 μl, 1 μl of which was used in 25-μl PCR reactions that were terminated at the logarithmic phase of amplification. Signals from the PCR products were captured using a BioRad Gel Doc workstation and analyzed with Quantity One software. Values were calculated as a percentage of input. Real-time PCR was also performed to confirm our results. Primer sequences are available on request.
All the individual H3K9me2 ChIP experiments showed the same trends of H3K9me2 level reduction in Su(var)3–9null chromatin (statistical analysis always showed significant reduction), although the absolute values differed. Both the cytology and ChIP results agreed that H3K9me2 levels are significantly lower in Su(var)3–9null chromatin.
Hirt ecc DNA isolation and detection
Approximately 200 larvae were frozen in liquid nitrogen, ground with a mortar and pestle, resuspended in 500 μl Hirt lysis buffer (0.6% SDS; 10 mM EDTA at pH 8)18, then incubated at room temperature for 10–20 min. 125 μl of 5M NaCl was added to the extract, which was incubated at 4 °C overnight (8–20 h). The larval extract was centrifuged at 14,000g at 4 °C for 40 min. The supernatant was phenol–chloroform extracted three times and the Hirt DNA was ethanol precipitated. To check for any genomic DNA contamination before phenol–chloroform extraction, the Hirt supernatant was subjected to methanol–acetic acid fixation on slides and examined by DAPI staining48. The precipitated Hirt DNA was also examined by standard electrophoresis agarose gel and ethidium bromide staining. 200 ng of Hirt DNA was used for each PCR reaction to probe for specific DNAs. Signals from the PCR products were captured and analyzed using a BioRad Gel Doc workstation and Quantity One software, as described above. For diploid tissues, 50 sets of brains and discs were dissected and lysed with 100 μl Hirt lysis buffer. The Hirt DNA isolated from diploid cells contains some genomic DNA, so the relative amount of ecc DNAs in mutant and wild-type tissues were quantified by comparing the results from separate PCR reactions for Hirt DNA and genomic DNA preparations.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank: J. Birchler (for hlsΔ215), R. Carthew (for DCR2L811fsx), D. Dorsett (for SMC1 antibody), S. Hawley (for smc1exc461), F. Gao (for Ago251B), T. Jenuwein (for H3K9me2 antibody), K. M. Pollard (for fibrillarin antibody), G. Reuter (for Su(var)3–9 alleles 6 and 17) and J. Rine (for dSir217); S. Weiss for experimental contributions; A. Dernburg for help with FISH experiments; and A. Blanco (formerly Metamorph) for assistance with volumetric analysis. We thank N. Bhalla, A. Dernburg, S. Erhart, P. Heun, B. Mellone, A. Minoda and W. Zhang for helpful discussions and critical comments on the manuscript. This work was supported by a National Institutes of Health (NIH) grant, R01GM061169.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS The authors declare that they have no competing financial interests.
References
- 1.Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nature Rev. Mol. Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 3.John B. The biology of heterochromatin. In: Verma RS, editor. Heterochromatin: Molecular and Structural Aspects. Cambridge University Press; Cambridge: 1988. pp. 1–147. [Google Scholar]
- 4.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Muller HJ. Types of visible variations induced by X-rays in Drosophila. J. Genet. 1930;22:299–334. [Google Scholar]
- 7.Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 2003;14:67–75. doi: 10.1016/s1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 8.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 9.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 10.Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- 11.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nature Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 12.Pikaard CS. The epigenetics of nucleolar dominance. Trends Genet. 2000;16:495–500. doi: 10.1016/s0168-9525(00)02113-2. [DOI] [PubMed] [Google Scholar]
- 13.Hawley RS, Marcus CH. Recombinational controls of rDNA redundancy in Drosophila. Annu. Rev. Genet. 1989;23:87–120. doi: 10.1146/annurev.ge.23.120189.000511. [DOI] [PubMed] [Google Scholar]
- 14.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 15.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 16.Spradling A, Orr-Weaver T. Regulation of DNA replication during Drosophila development. Annu. Rev. Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- 17.Schotta G, et al. Central role of Drosophila SU(VAR)3–9 in histone H3K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 19.Pont G, Degroote F, Picard G. Some extrachromosomal circular DNAs from Drosophila embryos are homologous to tandemly repeated genes. J. Mol. Biol. 1987;195:447–451. doi: 10.1016/0022-2836(87)90665-6. [DOI] [PubMed] [Google Scholar]
- 20.Aravin AA, et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 21.Lin YH, Keil RL. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics. 1991;127:31–38. doi: 10.1093/genetics/127.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen S, Yacobi K, Segal D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 2003;13:1133–1145. doi: 10.1101/gr.907603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen Z, Bacharach E, Lavi S. Mouse major satellite DNA is prone to eccDNA formation via DNA Ligase IV-dependent pathway. Oncogene. 2006;25:4515–4524. doi: 10.1038/sj.onc.1209485. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 25.Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 26.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid†cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nature Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson PK. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- 30.Hari KL, Cook KR, Karpen GH. The Drosophila Su(var)2–10 locus encodes a member of the PIAS protein family and regulates chromosome structure and function. Genes Dev. 2001;15:1334–1348. doi: 10.1101/gad.877901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schotta G, et al. A silencing pathway to induce H3K9 and H4K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 33.Cam HP, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nature Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 34.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 35.McKee BD, Karpen GH. Drosophila ribosomal RNA genes function as an X–Y pairing site during male meiosis. Cell. 1990;61:61–72. doi: 10.1016/0092-8674(90)90215-z. [DOI] [PubMed] [Google Scholar]
- 36.Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- 37.Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 38.Cartwright IL, et al. Analysis of Drosophila chromatin structure in vivo. Methods Enzymol. 1999;304:462–496. doi: 10.1016/s0076-6879(99)04028-8. [DOI] [PubMed] [Google Scholar]
- 39.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 40.Glaser RL, Karpen GH, Spradling AC. Replication forks are not found in a Drosophila minichromosome demonstrating a gradient of polytenization. Chromosoma. 1992;102:15–19. doi: 10.1007/BF00352285. [DOI] [PubMed] [Google Scholar]
- 41.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 42.Westphal T, Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osipovich O, et al. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nature Immunol. 2004;5:309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 45.Peters AHFM, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 46.Dorsett D, et al. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuschak TI, Kuschak BC, Smith GM, Wright JA, Mai S. Isolation of extrachromosomal elements by histone immunoprecipitation. Biotechniques. 2001;30:1064–1068. 1070–1072. doi: 10.2144/01305rr05. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.