Abstract
Since their discovery three decades ago, NK cells have been classified as cells of the innate immune system. NK cells were shown to respond rapidly and non-specifically to infection, and were thought to act as a functional “bridge” to sustain the early innate immune response until the later adaptive immune responses could be mounted. In light of new findings showing how NK cells possess nearly all of the features of adaptive immunity including memory, we propose the placement of NK cells as an “evolutionary bridge” between innate and adaptive immunity.
Keywords: Adaptive immunity, Innate immunity, NK cells
Introduction
In all of the immunology textbooks, NK cells are presented in sections describing the innate immune system. Ever since Eva Klein coined the phrase “Natural Killer” in 1975 to describe the ability of a subset of lymphocytes able to spontaneously kill tumor cells without pre-conditioning [1, 2], and Klas Karre proposed the “missing self” hypothesis in 1984 to explain the rules by which these lymphocytes function [3, 4], NK cells have been considered characteristically more “innate” than “adaptive”. However, the definitions of “innate” and “adaptive” have been blurred by recent findings showing adaptive immune features in NK cells.
According to the strictest definition of the word “adaptive”, what does it really mean that an individual cell or organ system can “adapt”? The Merriam-Webster dictionary defines “adaptation” as a “modification of an organism or its parts that makes it more fit for existence under the conditions of its environment”. If an immune cell can “adapt” in its behavior in response to encounter with a pathogen and by doing so becomes more “fit” to respond against repeated pathogen exposure, does this not classify it as “adaptive”? Implicit in this argument is the fact that an immune cell must survive after encountering a pathogen in order to adapt. Immunologists have given a name to the persistence and adaptation of immune cells following infection – memory.
Immunological memory has long been a hallmark of adaptive immunity. The ability of previously activated immune cells to persist for months and even years following initial stimulation through their antigen receptors represents a defining trait of immune memory [5–7]. These long-lived cells must then be able to become re-activated and undergo further clonal expansion following subsequent stimulation. In the context of an infection, the secondary or recall response should be more efficacious (i.e. demonstrate more rapid expansion and/or enhanced effector function) and protect the host against a re-encounter with the pathogen. These characteristics have previously only been demonstrated in T and B cells, and there was little evidence that these traits existed in other cell subsets of the immune system.
Recent findings
In a mouse model of contact hypersensitivity, von Andrian and colleagues first provided evidence that NK cells can retain a “memory” of previous exposure to a specific antigen [8]. Contact-hypersensitivity responses, a form of delayed-type sensitivity thought to be mediated by T and B cells containing antigen-specific receptors recognizing haptenated proteins, could be generated in RAG-deficient mice (which lack T and B cells), but not in RAG and IL-2 receptor common-γ chain double-deficient mice (which lack T, B, and NK cells)[8]. This hapten-specific “memory” could be transferred via liver NK cells from a sensitized donor such that a naïve recipient mouse acquired the ability to mediate contact hypersensitivity independent of T and B cells [8]. The ability to mount recall responses were shown to persist for one month and could only be elicited by haptens previously used to sensitize the host, but not by an unrelated hapten, demonstrating specificity [8]. Together, these findings indicate that NK cells have the ability to mount adaptive and antigen-specific secondary responses.
However, the mechanism responsible for NK cell-mediated contact hypersensitivity, as well as the precise NK cell subsets responsible for the hapten-specific “memory”, were not defined in this study. Furthermore, these observations left us to ponder: What are the NK cell receptors recognizing haptens on the surface of target cells and what is the nature of the hapten ligand? Why do NK cells from the liver, but not the spleen, contain the ability to mount recall responses? Why would natural selection evolve mechanisms in NK cells to mediate secondary responses against chemical haptens? Wouldn’t NK cells more likely have developed ligand specificity and immune memory against pathogens during the course of evolution?
More recently, in a mouse model of CMV infection, we have demonstrated that NK cells possess nearly all of the hallmark features of the adaptive immune system [9]. Using an adoptive transfer system, where antigen-experienced NK cells can be tracked with a congenic marker, we have observed that Ly49H-bearing NK cells can undergo robust expansion leading to long-lived memory cells persisting in the host for at least months after infection [9]. Furthermore, memory NK cells have the capacity to become re-activated, and produce more IFN-γ and degranulate more robustly than resting NK cells from naive mice [9]. Upon re-encounter with murine CMV (MCMV), memory NK cells can undergo vigorous secondary expansion with comparable kinetics to naive NK cells, but are more protective than resting NK cells following virus challenge [9]. Analogous to previously measured endogenous or TCR-transgenic T-cell responses, NK cells possess the feature of “memory’ found in CD8+ and CD4+ T cells that defines them as being adaptive immune cells. Based on transcriptional microarray analyses, T cells have been shown to change their molecular profile as they become activated, undergo expansion and contraction, and become memory cells [10]. Similarly, future studies characterizing the gene transcript expression in resting, activated, and memory NK cells will give us clues into how NK cells become long-lived and persist for many months following virus infection. Together with the von Andrian study [8], these observations prompted us to re-examine, in this mini-review, our current criteria for partitioning the immune system.
Defining “innate” and “adaptive”: a matter of semantics?
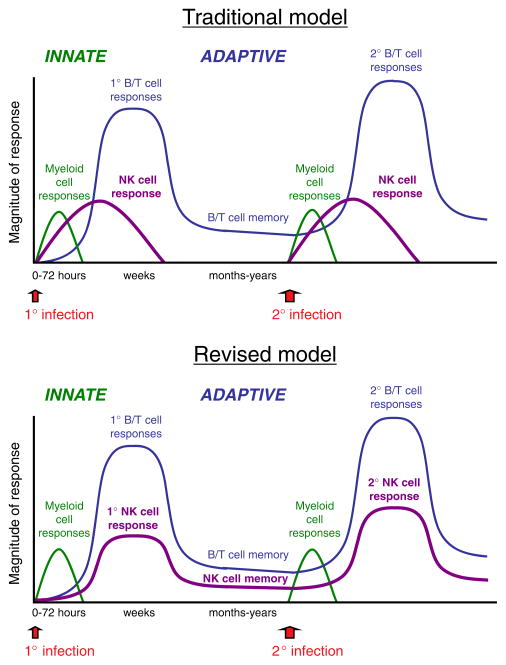
Cells of the innate immune system generally respond rapidly and “non-specifically” to “stress” or to conserved structures shared by microbial organisms (PAMP) usually manifested by inflammation following tissue damage and pathogen invasion. Many of these cells (mainly of the myeloid lineage, but including NK cells) are present at the site of injury or are immediately recruited, quickly mediate their effector functions, and are thought to disappear after a few hours or days (Fig. 1).
Figure 1.
Traditional and revised models of innate and adaptive immunity. Graphs show the immune response against primary and secondary infections. The response of myeloid cells (neutrophils, macrophages, and dendritic cells) is depicted by the green lines. The NK cell response is shown in purple, and the response of B and T cells is shown in blue. The magnitude of the response refers to the qualitative nature of the immune response (i.e. efficacy (protective capability) and effector cytokine production) rather than the absolute number of cells.
Meanwhile, cells of the adaptive immune system (from the lymphoid lineage), which have never previously encountered the antigen or pathogen, typically mount a response much later, after the antigen has been delivered by antigen-presenting cells to the draining lymph nodes or spleen where antigens (either whole or processed) are presented to B and T cells (Fig. 1). These cells (possessing somatically rearranged receptors) become activated, proliferate to increase the number of effectors, and traffic to sites of infection, mediating highly specific anti-pathogen responses. Following pathogen clearance, cells of the adaptive immune system reside in lymphoid and non-lymphoid tissues for months and years, ready to quickly respond when the pathogen is re-encountered (Fig. 1). NK cells, previously thought to respond and then disappear after one or two weeks, have now been demonstrated to reside in multiple organs as long-lived memory cells [9], ready to mount robust secondary responses against pathogen invasion (Fig. 1).
What are the attributes that immunologists have formulated to distinguish innate from adaptive immunity (Table 1)? Does the rate of the responding cell dictate whether that particular cell type is considered “innate” or “adaptive”? Or does the specificity with which the cell recognizes pathogens and the location at which these cells are primed determine the nomenclature and categorization? Does the longevity of the cell once it becomes activated as a result of infection or immunization and the ability to become re-activated following subsequent infection define it as a cell of adaptive immunity? Or must a cell contain genetically rearranged receptors for it to be categorized as a part of the adaptive immune system? In the end, the question really becomes how stringent our parameters are by which we define cells of the immune system (Table 1).
Table 1.
The properties of the NK-cell response compared with innate and adaptive immune responses
| Property | Innate immunity | Adaptive immunity | NK-cell response |
|---|---|---|---|
| Specificity | No? | Yes | Yes |
| Clonal expansion | No? | Yes | Yes |
| Long-lived progeny | No? | Yes | Yes |
| Recall response | No? | Yes | Yes |
| Receptor rearrangement | No | Yes | No |
Do NK cells belong to innate or adaptive immunity?
NK cells are known to respond rapidly against tumor cells and provide a front-line defense against pathogens. However, the speed at which NK cells respond to infection should not dictate that they are cells of innate immunity. True, innate immune cells do generally respond quickly against pathogen encounter; however, the definition of innate immunity should not be confined to this characteristic alone. Indeed, while NK cells can rapidly secrete effector cytokines such as IFN-γ during inflammation due to high levels of type I-IFN or IL-12 and IL-18 in the environment [11, 12], memory CD8+ T cells possess the same ability to secrete cytokines in the absence of cognate antigen [13–15], and no one would argue that this latter cell type should be classified as “innate”. Furthermore, functional parallels exist between NK cells and CD8+ T cells. When an NK cell encounters its cognate ligand on a target cell, it mediates cytotoxicity (typically by directional release of perforin and granzyme) and secretes effector cytokines (such as IFN-γ) in a manner nearly identical to that of effector and memory CD8+ T cells [16–18].
What about “specificity” (or lack there of) as a definition of adaptive versus innate immunity? If the cells of adaptive immunity are strictly identified as those containing antigen-specific receptors that have productively rearranged their genes during development or are able to undergo somatic hypermutation, then our definition is clear and simple. T and B cells contain such receptors – every other cell in the immune system does not. By this definition, T and B cells are the only cells that are highly specialized and allow the host to be completely adaptable in its response to potentially every possible infection and antigen, and thus are unambiguously the only cells belonging to the adaptive immune system. However, if “specificity” is defined by the cognate recognition of an immune receptor for a unique ligand encoded by a specific pathogen, which is not shared by other pathogens, then NK cells demonstrate immune specificity as well. The Ly49H receptor expressed on a subset of NK cells appears to be exquisitely specific for the MCMV-encoded m157 glycoprotein [19, 20]. Ly49H has not been found to recognize viral ligands encoded by any virus other than MCMV, and an m157-related protein has not been identified in any other virus, although it is impossible to completely exclude that Ly49H will be found to recognize ligands encoded by pathogens other than MCMV. Crystal structures of the m157 glycoprotein have revealed that it bears many striking similarities to MHC class I proteins, although it does not bind peptides or associate with β-2-microglobulin [21]. The specificity of the Ly49H receptor for m157 has been demonstrated using a mutant strain of MCMV lacking the m157 gene; Ly49H+ NK-cell responses were abrogated in mice infected with an m157-deleted virus [9, 22]. Recently, another activating receptor on mouse NK cells, Ly49P, has been demonstrated to recognize MCMV-infected cells, and this specific interaction is dependent on the viral component m04 [23]. In humans, specific NK cell subsets that preferentially expand during CMV infection have been documented as well [24–27]. Furthermore, analogous to the TCR, which requires ITAM-bearing CD3 subunits to transmit a complete signal in T cells following TCR ligation of peptide-MHC, activating Ly49 receptors require the ITAM-bearing signaling molecule DAP12 to initiate a full activation signal in NK cells [28]. Thus, NK cells, at least with respect to Ly49H+ NK cells, share the adaptive immune criteria of “specificity” with B and T cells (Table 1).
In addition to specificity and immune memory, NK cells undergo developmental and peripheral tolerance mechanisms similar to B and T cells to safeguard against autoimmunity. During development, adaptive immunity relies on the “selection” of immune cells that can distinguish between the host’s own cells and unwanted invaders. Whereas T cells undergo selective pressures during development in the thymus (where checkpoints ensure self-tolerance and prevent autoimmunity) [29], NK cells have also been shown to undergo similar “selection” processes during their development. As with T cells, education of NK cells in the bone marrow also ensures that these cells will be tolerant of “self” in the periphery [30, 31]. Immature NK cells have been shown to require interactions between their inhibitory receptors with MHC class I molecules to become fully functional in the periphery; without these interactions, NK cells are rendered hypo-responsive [30, 31]. Furthermore, akin to negative selection of thymocytes and peripheral T-cell anergy, when in experimental models in which developing Ly49H+ NK cells encountered their cognate viral ligand m157 in the bone marrow, tolerance mechanisms were put in place to delete these immature cells or keep them unresponsive in the periphery [32, 33]. Further regulatory mechanisms of self-tolerance controlling peripheral NK cells (as has been demonstrated in regulatory T cells) remain to be defined. Additionally, studies examining the environmental cues for NK-cell activation, proliferation, and homeostasis will give much insight into how these cells are triggered and subsequently regulated as they differentiate into effector and memory cells. Understanding the molecular mechanisms governing the survival and function of NK cells will allow us to harness the protective capacity of this cell subset in vaccination against infectious pathogens. Together, the many developmental, phenotypic, and functional similarities that exist between NK cells and T cells suggest that NK cells are a good candidate to occupy the missing evolutionary link between innate and adaptive immunity.
A paradigm shift?
The innate immune system is widely believed to be evolutionarily more ancient than the adaptive immune system. Whereas innate immunity is found in nearly all organisms (including jawless fish, worms, insects, and even starfish), adaptive immunity is thought to have first arisen in the jawed vertebrates [34–37]. Because reproduction is required for the perpetuation of a complex species and more time is required for vertebrates to reproduce (compared with less complex creatures), the primitive immune system might have evolved the ability to recognize and “remember” specific pathogens in order to mount stronger attacks each time a pathogen is encountered. The ability of adaptive immunity to adapt its defenses against future viral, parasitic, or bacterial challenges ensures that the higher organism will survive long enough to allow for the propagation of its progeny.
Although we may never fully understand how the cells of adaptive immunity evolved from their predecessors, there is evidence of these mechanisms when comparing higher animals to lower organisms – at some point during the evolution of species, T and B cells appeared, having acquired recombinase proteins necessary to rearrange V(D)J genetic elements and diversify antigen-sensing receptors [34, 36, 38, 39]. Even within the study of our own immune system, we can find clues in potential “predecessor cells” such as NK cells, which possess traits of both innate and adaptive immunity. If there is a subset of cells that constitutes an evolutionary “bridge” between innate and adaptive immune cells, this population would presumably contain characteristics of both systems. Indeed, NK cells can mediate both “specific” adaptive-like responses, as well as “non-specific” innate-like responses. In addition to the cognate recognition of m157 and subsequent adaptive immune responses mounted by Ly49H+ NK cells, Ly49H− NK cells that do not encounter cognate antigen can still participate as “bystanders” [40, 41], producing IFN-γ in response to inflammatory cytokines such as IL-12 and IL-18 secreted by activated myeloid cells [11, 12, 42], but not clonally expanding. A recent study suggests that such “bystander” activation of NK cells can result in “memory-like” properties [42]; however, how these in vitro cytokine-stimulated NK cells compare with NK cells following viral infection in vivo needs to be further evaluated. Even macrophages have recently been proposed to possess “adaptive” features in their ability to regulate inflammation [43]. Reciprocally, unique T and B cell subsets (γδ, CD8αα, and NKT cells; and B1 and marginal zone B cells) have been discovered that are limited in their antigen-specific receptor repertoire, function early in the immune response, and thus could be considered “innate”-like [44, 45]. Altogether, these cells form both an evolutionary as well as a functional “bridge” between innate and adaptive immunity.
A fundamental distinction between NK cells versus B cells and T cells is the ability of B cells and T cells to generate an almost infinite repertoire of antigen-specific receptors as a consequence of somatic gene rearrangement. In contrast, NK cells are limited to the receptors that are encoded by genes pre-existing in the genome. Therefore, the receptors expressed by NK cells must be selected for at a population level, rather than the individual level. However, the murine Ly49 genes and the primate KIR genes are polygenic and exhibit extensive allelic polymorphism, thereby providing for considerable diversity within the species. Of note, the activating receptors within the Ly49 and KIR gene families are evolving more rapidly than the inhibitory receptors from which they arose, likely due to selective pressure exerted by pathogens [46]. If NK cells are restricted to receptors encoded by germline genes, this obviously limits their total repertoire potential, and implies that selection of these genes will be driven at the population level. This selective pressure might have resulted in the recognition of host-encoded rather than pathogen-encoded ligands, for example the recognition of the host’s stress-induced ligands of NKG2D. The evolution of NK receptors for pathogen-encoded ligands could be either for broadly cross-reactive molecules, such as PAMP (which has not yet been documented for NK-cell receptors), or possibly for selected pathogens that persist in the host and must be controlled throughout the lifetime of the host, such as herpesviruses.
As immunologists, we sometimes too strictly partition cells and cell subsets, often in a binary fashion (e.g. Th1 versus Th2; B1 versus B2, etc.), pigeon-holing or artificially imposing forced-categorization that oversimplify the intricacies of the immune system, and thus hazard overlooking new biological concepts. When placing stringent boundaries on how specific cell subsets can be categorized and named, we become susceptible to continuously arguing semantics. Rather than fretting over how to classify, we should instead celebrate the unexpected observation that furthers our understanding of the natural world, and discover how these new concepts in biology can be translated towards advancing human health and fighting infectious disease.
Acknowledgments
L.L.L is an American Cancer Society Research Professor and the Irvington Institute Fellowship Program of the Cancer Research Institute supports J.C.S. Supported by NIH grants AI066897, AI068129, CA095137, and AI64520.
Abbreviation
- MCMV
murine CMV
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5:112–117. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- 2.Kiessling R, Klein E, Pross H, Wigzell H. Natural killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975;5:117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 3.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 4.Ljunggren HG, Karre K. In search of the ’missing self ’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 5.Harty JT, Badovinac VP. Shaping and reshaping CD8+T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 6.Sprent J, Surh CD. T cell memory. Annu Rev Immunol. 2002;20:551–579. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- 7.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 8.O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell-and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 9.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 11.Cousens LP, Orange JS, Su HC, Biron CA. Interferon-alpha/beta inhibition of interleukin 12 and interferon-gamma production in vitro and endogenously during viral infection. Proc Natl Acad Sci USA. 1997;94:634–639. doi: 10.1073/pnas.94.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orange JS, Biron CA. Characterization of early IL-12, IFN-alphabeta, and TNF effects on antiviral state and NK cell responses during murine cytomegalovirus infection. J Immunol. 1996;156:4746–4756. [PubMed] [Google Scholar]
- 13.Berg RE, Crossley E, Murray S, Forman J. Memory CD8+T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J Exp Med. 2003;198:1583–1593. doi: 10.1084/jem.20031051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kambayashi T, Assarsson E, Lukacher AE, Ljunggren HG, Jensen PE. Memory CD8+T cells provide an early source of IFN-gamma. J Immunol. 2003;170:2399–2408. doi: 10.4049/jimmunol.170.5.2399. [DOI] [PubMed] [Google Scholar]
- 15.Lertmemongkolchai G, Cai G, Hunter CA, Bancroft GJ. Bystander activation of CD8+T cells contributes to the rapid production of IFN-gamma in response to bacterial pathogens. J Immunol. 2001;166:1097–1105. doi: 10.4049/jimmunol.166.2.1097. [DOI] [PubMed] [Google Scholar]
- 16.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 17.Lanier LL. Back to the future – defining NK cells and T cells. Eur J Immunol. 2007;37:1424–1426. doi: 10.1002/eji.200737418. [DOI] [PubMed] [Google Scholar]
- 18.Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol. 2004;5:996–1002. doi: 10.1038/ni1114. [DOI] [PubMed] [Google Scholar]
- 19.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 20.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams EJ, Juo ZS, Venook RT, Boulanger MJ, Arase H, Lanier LL, Garcia KC. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc Natl Acad Sci USA. 2007;104:10128–10133. doi: 10.1073/pnas.0703735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bubic I, Wagner M, Krmpotic A, Saulig T, Kim S, Yokoyama WM, Jonjic S, Koszinowski UH. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol. 2004;78:7536–7544. doi: 10.1128/JVI.78.14.7536-7544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielczewska A, Pyzik M, Sun T, Krmpotic A, Lodoen MB, Munks MW, Babic M, et al. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J Exp Med. 2009;206:515–523. doi: 10.1084/jem.20080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 25.Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, Lopez-Botet M. Expansion of CD94/NKG2C+NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006;107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- 26.Guma M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, Lopez-Botet M. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J Infect Dis. 2006;194:38–41. doi: 10.1086/504719. [DOI] [PubMed] [Google Scholar]
- 27.Kuijpers TW, Baars PA, Dantin C, van den Burg M, van Lier RA, Roosnek E. Human NK cells can control CMV infection in the absence of T cells. Blood. 2008;112:914–915. doi: 10.1182/blood-2008-05-157354. [DOI] [PubMed] [Google Scholar]
- 28.Smith KM, Wu J, Bakker AB, Phillips JH, Lanier LL. Ly-49D and Ly-49H associate with mouse DAP12 and form activating receptors. J Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- 29.Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 32.Sun JC, Lanier LL. Tolerance of NK cells encountering their viral ligand during development. J Exp Med. 2008;205:1819–1828. doi: 10.1084/jem.20072448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripathy SK, Keyel PA, Yang L, Pingel JT, Cheng TP, Schneeberger A, Yokoyama WM. Continuous engagement of a self-specific activation receptor induces NK cell tolerance. J Exp Med. 2008;205:1829–1841. doi: 10.1084/jem.20072446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein RM, Schluter SF, Bernstein H, Marchalonis JJ. Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc Natl Acad Sci USA. 1996;93:9454–9459. doi: 10.1073/pnas.93.18.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Kasahara M, Suzuki T, Pasquier LD. On the origins of the adaptive immune system: novel insights from invertebrates and cold-blooded vertebrates. Trends Immunol. 2004;25:105–111. doi: 10.1016/j.it.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 37.van den Berg TK, Yoder JA, Litman GW. On the origins of adaptive immunity: innate immune receptors join the tale. Trends Immunol. 2004;25:11–16. doi: 10.1016/j.it.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Cannon JP, Haire RN, Rast JP, Litman GW. The phylogenetic origins of the antigen-binding receptors and somatic diversification mechanisms. Immunol Rev. 2004;200:12–22. doi: 10.1111/j.0105-2896.2004.00166.x. [DOI] [PubMed] [Google Scholar]
- 39.Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 40.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 41.Sun JC, Lanier LL. Cutting edge: viral infection breaks NK cell tolerance to missing self. J Immunol. 2008;181:7453–7457. doi: 10.4049/jimmunol.181.11.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 44.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 45.Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
- 46.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]