Abstract
Purpose
Recently, the majority of protein coding genes were sequenced in a collection of pancreatic cancers, providing an unprecedented opportunity to identify genetic markers of prognosis for patients with adenocarcinoma of the pancreas.
Experimental Design
We previously sequenced over 750 million base pairs of DNA from 23,219 transcripts in a series of 24 adenocarcinomas of the pancreas. In addition, 39 genes that were mutated in more than one of these 24 cancers were sequenced in a separate panel of 90 well-characterized adenocarcinomas of the pancreas. Of these 114 patients, 89 underwent pancreaticoduodenectomy, and the somatic mutations in these cancers were correlated with patient outcome.
Results
When adjusted for age, lymph node status, margin status, and tumor size, SMAD4 gene inactivation was significantly associated with shorter overall survival (Hazard ratio [95% C.I.] = 1.92 [1.20 – 3.05], p=0.006). Patients with SMAD4 gene inactivation survived a median of 11.5 months, compared to 14.2 months for patients without SMAD4 inactivation. By contrast, mutations in CDKN2A, TP53, or the presence of multiple (≥4) mutations or homozygous deletions among the 39 most frequently mutated genes were not associated with survival.
Conclusions
SMAD4 gene inactivation is associated with poorer prognosis in patients with surgically-resected adenocarcinoma of the pancreas.
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer death in the United States (1). World-wide pancreatic cancer is responsible for over 213,000 deaths each year (2). The 5-year overall survival rate for all patients diagnosed with pancreatic cancer is less than 4 percent (3). Surgical resection offers the best hope for long-term survival, with a 17% 5-year survival rate in most surgical series (4–10). A number of pathological features have been shown to correlate with outcome following surgery (4, 11–14). For example, the completeness of resection (margin status), size of the cancer, degree of differentiation, vascular invasion, lymph node status and tumor stage are all independent prognostic indicators following pancreaticoduodenectomy for pancreatic cancer(4, 11–16). These factors have been useful guides in the clinical management of patients with pancreatic cancer. Genetic markers that could be used as prognostic indicators of outcome would be useful in establishing an individualized treatment plan for a patient. For example, a more aggressive surgical approach, such as vascular resection and reconstruction, may be considered for a patient with a reduced risk of systemic recurrence.
In an attempt to establish such genetic makers, we took advantage of the recently completed mutational analysis of the pancreatic cancer coding genome(17). The results of this “pancreatic cancer genome project,” provide a unique opportunity to determine if any genes with somatic changes correlate with patient outcome following surgical resection (17). This previous study included the sequencing of the protein-coding exons from 20,661 genes in 24 advanced adenocarcinomas of the pancreas as well as copy number analyses using high density oligonucleotide arrays (17). In addition, 39 of the genes mutated in two or more of the 24 cancers were sequenced in an additional panel of 90 well-characterized infiltrating adenocarcinomas of the pancreas (17).
In the current study, we correlated these mutational data with patient survival following surgical resection. The identification of somatic genetic alterations that are associated with patient outcome may lead to useful clinical tools to guide the treatment of patients with pancreatic cancer, and may provide insight into alterations underlying the aggressive behavior of this cancer.
MATERIALS AND METHODS
This study was approved by the Johns Hopkins Institutional Review Board.
Patients
Our previous sequencing study included 114 patients (24 in the Discovery Screen and 90 from the Validation Screen) treated between February 1989 and May 2007(11, 17, 18). Ninety-one of these 114 patients underwent a pancreaticoduodenectomy (Whipple procedure), 12 a distal pancreatectomy, 3 a total pancreatectomy, while 8 were autopsied. The current study considered only the 91 patients from this previous study who underwent a pancreaticoduodenectomy(11, 18). There were two perioperative deaths in this group, defined as death during the patient’s initial hospitalization or within 30 days of surgery. The 89 remaining pancreaticoduodenectomy patients were included in the current study.
As reported previously, most of the patients included in this study were evaluated by a multidisciplinary group (surgery, medical oncology, radiation oncology, and pathology) and postoperative chemoradiation therapy was encouraged (4, 11). The majority of the patients received standard chemoradiation protocols consisting of 4000 to 5000 cGy of external beam radiation to the tumor bed given with two 3-day courses of 5-fluorouracil (5-FU; 500 mg/m2/day) followed by weekly bolus 5-FU for four additional months (4, 11). Other therapies included more intensive 5-FU plus leucovorin-based chemoradiation, gemcitabine-based chemotherapy, and chemoradiation including 5-FU, mitomycin C, leucovorin, and dipyridamole (4, 11, 19). Overall survival rates have been reported to be similar for all forms of adjuvant chemoradiation.
Somatic Mutations
The “Discovery Screen” of the previous sequencing study included the sequencing of 20,661 protein coding genes in a series of 24 adenocarcinomas of the pancreas (17). In the “Validation Screen”, 39 genes that were mutated more than once in the Discovery Screen were sequenced in an additional panel of 90 well-characterized adenocarcinomas of the pancreas (17). The current study focused on the 39 genes included in both the Discovery and Validation Screens, and on the 89 patients who underwent a pancreaticoduodenectomy and survived at least 30 days.
Deletions were identified in the Discovery Screen using high-density oligonucleotide arrays as previously described (17). High-density oligonucleotide array analysis was not performed on the Validation Screen samples. Therefore, for purposes of analysis in this study deletions in the samples of the Validation Screen were determined by patterns of sequencing failure. For the sequencing in the Validation Screen, eleven sets of primers were used to amplify SMAD4, nine sets for TGFβR2 and SMAD3, eight sets for TGFβR1, and one set of primers was used for CDKN2A. For SMAD4, TGFβR2, SMAD3 and TGFβR1, we defined a homozygous deletion as having failed sequencing reactions (with less than 20% of the region of interest having a phred score < 20) for three or more consecutive amplicons. For CDKN2A, we defined a homozygous deletion as having a failed sequencing reaction (with less than 20% of the region of interest having a phred < 20) for the single amplicon that was analyzed. When these criteria were applied to the 90 cancers included in the Validation Screen, 33% of the cancers were categorized as having a CDKN2A deletion, 19% a SMAD4 deletion, 1% a TGFβR2 deletion, 1% a TGFβR1 deletion, and 1% a SMAD3 deletion. These percentages are similar to those observed with the high-density oligonucleotide arrays in the Discovery Phase (17). In addition, when these criteria were used, we did not observe cancers with both a homozygous deletion and an inactivating intragenic somatic mutation.
Statistical analyses
Survival curves for the 89 patients who underwent a pancreaticoduodenectomy and survived at least 30 days after surgery were constructed using the Kaplan-Meier method. Although 39 genes were sequenced in the Validation phase, the majority of the genes were altered in only one or two patients. Consequently, several approaches were used to examine if genetic alterations affected patient outcome. First, a set of genes hypothesized to differentiate survival, SMAD4, TP53, CDKN2A and members of the transforming growth factor beta (TGFβ) signaling pathway, were each correlated with survival. Next, in a subanalysis, the members of the TGFβ pathway were combined, and patients were grouped by whether they had a mutation or homozygous deletion in any gene in the pathway. Finally, all 39 genes sequenced in the Validation phase were combined, and patients were grouped by whether they had fewer than 4, or 4 or more mutations and/or homozygous deletions among the 39 genes.
Survival curves were compared between groups of patients using a log-rank statistic, for descriptive purposes. Coefficients from Cox proportional hazards models were used to test for the effect of genetic alterations on survival while adjusting for age, tumor size, margins, lymph nodes and grade. We considered p-values < 0.01 to be statistically significant, adjusting for the multiple survival comparisons performed in this study.
Results
Patients
The mean (standard deviation) age of the 89 patients was 65.3 (10.5) years (range: 36 – 85 years) (Table 1). There were 43 males (48%) and 46 females (52%), with the racial distribution being 80 white (90%), and 9 of other races (10%). The mean (standard deviation) size of the tumors was 3.6 (1.7) cm. Seventy-one patients (80%) had lymph node metastases. Fifty-five of the carcinomas were well- or moderately-differentiated, and 34 were poorly differentiated. Fifty-eight of the 89 patients had negative margins, and 31 had positive margins.
Table 1.
Characteristics of patients who underwent a pancreaticoduodenectomy and survived at least 30 days after surgery.
| Characteristic | Patients N = 89 |
|---|---|
| Age – Mean (SD) | 65.3 (10.5) |
| Gender – No. (%) | |
| Male | 43 (48.3) |
| Female | 46 (51.7) |
| Race – No. (%) | |
| White | 80 (89.9) |
| Other Race | 9 (10.1) |
| Grade – No. (%) | |
| Poor | 34 (38.2) |
| Moderate/Well | 55 (61.8) |
| Tumor Size – No. (%) | |
| < 3 cm | 32 (36.0) |
| 3–5 cm | 44 (49.4) |
| > 5 cm | 12 (13.5) |
| Unknown | 1 (1.1) |
| Margin – No. (%) | |
| Negative | 58 (65.2) |
| Positive | 31 (34.8) |
| Positive Lymph Nodes – No. (%) | |
| Yes | 71 (79.8) |
| No | 18 (20.2) |
| No. of Positive Lymph Nodes – Mean (SD) | 3 (3.8) |
| No. of Lymph Nodes Examined– Mean (SD) | 15 (8.5) |
| Median (Range) follow-up in months | 12.9 (3.1, 56.0) |
Follow-up was available on all 89 patients, ranging from 3.1 months to 56.0 months, with a median of 12.9 months. Follow-up was extensive, as 80 (90%) of the 89 patients included in this study were followed until the time of death. The five-year survival rate for all 89 patients in this series (Figure 1) is less than the 17–20% five-year survival rate we have previously reported for all patients treated by pancreatoduodenectomy for adenocarcinoma of the pancreas at our institution (4–6, 11, 18, 20). This may reflect the reported trend for clinically more aggressive malignancies to successfully grow as xenografts and cell lines (21, 22).
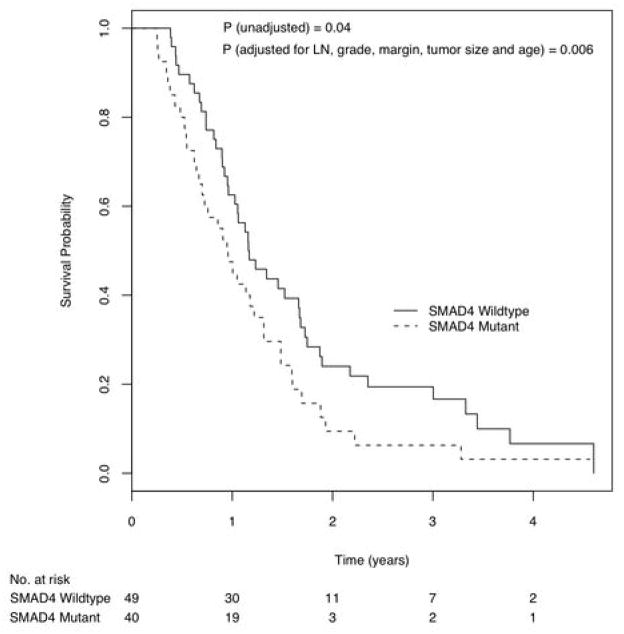
Figure 1.
Kaplan Meier curves of survival probability by patients with and without mutation or deletion in the SMAD4 gene.
SMAD4 and the TGFB pathway vs. Survival
The survival curves of patients with and without SMAD4 gene inactivation were significantly different (Log rank p = 0.04). When adjusted for lymph node status, tumor grade, margin status, tumor size and age in a Cox proportional hazards model, SMAD4 gene mutation status remained significantly associated with overall survival (Figure 1, Hazard ratio = 1.92 [95% C.I., 1.20 – 3.05], p=0.006). Patients with intact (wild-type) SMAD4 gene status had a median survival time of 14.2 [95% C.I., 12.5 – 20.5] months, while patients with genetically inactivated SMAD4 gene (either by intragenic mutation or by homozygous deletion) had a median survival time of 11.5 [95% C.I., 8.5 – 16.0] months.
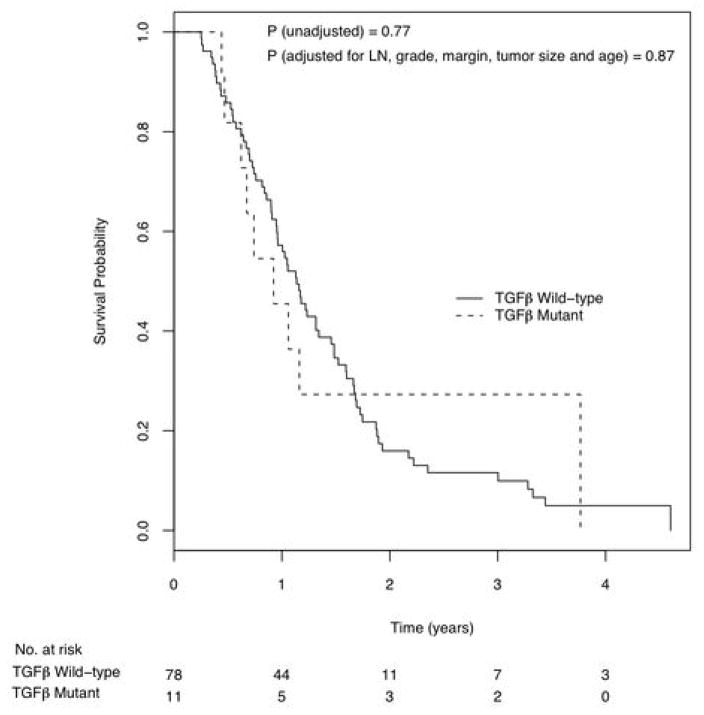
As the SMAD4 gene encodes for a protein that is an important member of the transforming growth factor beta (TGFβ) pathway, we examined all of the members of this pathway (SMAD3, SMAD4, TGFβR1 and TGFβR2) individually and together for additional alterations in these tumors. Patients were grouped by whether they had a mutation or deletion in any of these four genes. Inactivation of a member of the (TGFβ) signaling pathway did not correlate with survival when adjusted for other factors (Hazard ratio = 1.56 [95% C.I., 0.96 – 2.52], p=0.07). We then examined each gene in the TGFβ signaling pathway individually. One patient had a mutation on SMAD3 and another had a homozygous deletion on SMAD3. There were no patients with mutations or deletions on TGFβR1. However, there were 11 patients (9 with mutations, 2 with homozygous deletions) with alterations on the TGFβR2 gene by itself, and, although the numbers are relatively small, there was no difference in survival for the TGFβR2 mutant versus TGFβR2 wild-type cases (Figure 2, Hazard Ratio = 0.94 [95% C.I., 0.45 – 1.96], p=0.87). This observation highlights the conclusion that not all of the inactivating mutations in members of a pathway have the same effect on survival.
Figure 2.
Kaplan Meier curves of survival probability by patients with and without mutation or deletion in the TGFβR2 gene.
CDKN2A, TP53, and KRAS vs. Survival
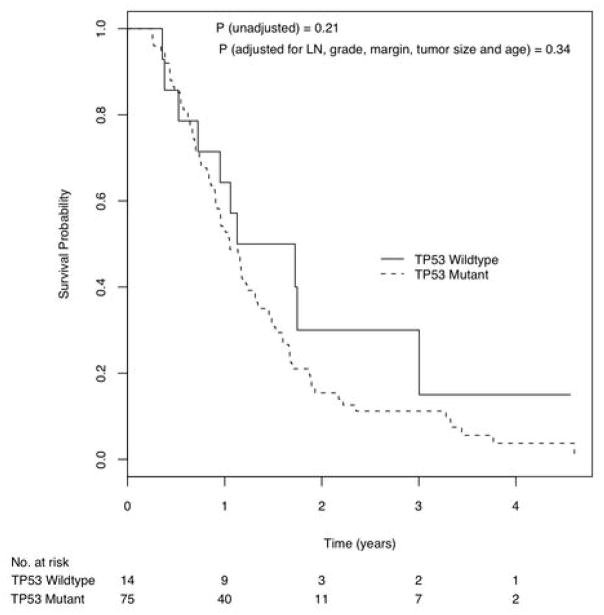
There were no significant differences in survival between patients with and without CDKN2A gene inactivation (Log rank p = 0.63). When adjusted for lymph node status, tumor grade, margin status, tumor size and age, the difference remained non-significant (Hazard ratio = 1.21 [95% C.I., 0.76 – 1.93] p=0.42). Similarly, TP53 gene mutation status did not correlate with survival (Log rank p = 0.21), even when adjusted for lymph node status, tumor grade, margin status, tumor size and age (Figure 3, Hazard ratio = 1.41 [95% C.I., 0.69 – 2.85], p=0.34). Virtually all of the cancers (98.9%) harbored a KRAS gene mutation, excluding the possibility of a relationship between KRAS gene status and prognosis.
Figure 3.
Kaplan Meier curves of survival probability by patients with and without mutation or deletion in the TP53 gene.
Multiple Mutations and Survival
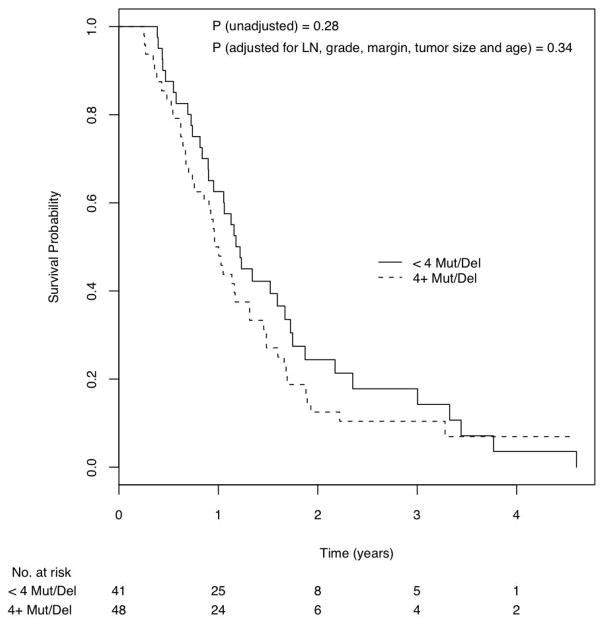
Finally, we examined the relationship between multiple gene mutations (an intragenic mutation or homozygous deletion in <4 vs. 4 or more of the 39 genes) and survival. The survival curves for these two groups of patients were not significantly different (Log rank p = 0.28), and the difference remained non-significant even when adjusted for lymph node status, tumor grade, margin status, tumor size and age (Figure 4, Hazard ratio = 1.28 [95% C.I., 0.78 – 2.12], p=0.34).
Figure 4.
Kaplan Meier curves of survival probability by patients with < 4 mutations or homozygous deletions and with ≥4 mutations or homozygous deletions.
The hazard ratios for all variables included in the multivariate models are presented in Table 2. For all models, increasing age was the only factor that was significantly associated with shorter survival, apart from SMAD4 gene status (see Table 2).
Table 2.
Hazard ratios from multivariate Cox proportional hazards models. Each shaded block represents a set of variables that were included in a single model.
| Hazard Ratio | 95% C.I. | P value | |
|---|---|---|---|
| Model 1 | |||
| SMAD4 mutant vs. wild-type | 1.92 | [1.20, 3.05] | 0.006 |
| Metastases vs. no metastases | 1.00 | [0.57, 1.75] | > 0.99 |
| Positive vs. negative margins | 1.29 | [0.78, 2.10] | 0.31 |
| Poor vs. moderate/well grade | 1.08 | [0.66, 1.77] | 0.76 |
| Age | 1.03 | [1.01, 1.05] | 0.002 |
| Tumor size ≥ 3.5 vs. < 3.5 cm | 1.42 | [0.89, 2.27] | 0.14 |
| Model 2 | |||
| TGFβ mutant vs. wild-type | 0.94 | [0.45, 1.96] | 0.87 |
| Metastases vs. no metastases | 1.05 | [0.59, 1.86] | 0.86 |
| Positive vs. negative margins | 1.25 | [0.78, 2.04] | 0.36 |
| Poor vs. moderate/well grade | 1.17 | [0.73, 1.89] | 0.51 |
| Age | 1.03 | [1.01, 1.05] | 0.007 |
| Tumor size ≥ 3.5 vs. < 3.5 cm | 1.38 | [0.87, 2.22] | 0.17 |
| Model 3 | |||
| TP53 mutant vs. wild-type | 1.41 | [0.69, 2.85] | 0.34 |
| Metastases vs. no metastases | 1.03 | [0.59, 1.80] | 0.91 |
| Positive vs. negative margins | 1.25 | [0.77, 2.01] | 0.37 |
| Poor vs. moderate/well grade | 1.16 | [0.73, 1.86] | 0.53 |
| Age | 1.03 | [1.01, 1.05] | 0.01 |
| Tumor size ≥ 3.5 vs. < 3.5 cm | 1.40 | [0.88, 2.21] | 0.16 |
| Model 4 | |||
| CDKN2A mutant vs. wild-type | 1.21 | [0.76, 1.93] | 0.42 |
| Metastases vs. no metastases | 1.01 | [0.58, 1.77] | 0.96 |
| Positive vs. negative margins | 1.26 | [0.78, 2.03] | 0.34 |
| Poor vs. moderate/well grade | 1.19 | [0.74, 1.90] | 0.48 |
| Age | 1.03 | [1.01, 1.05] | 0.006 |
| Tumor size ≥ 3.5 vs. < 3.5 cm | 1.37 | [0.87, 2.18] | 0.18 |
| Model 5 | |||
| ≥4 vs. < 4 mutations or deletions | 1.28 | [0.78, 2.12] | 0.34 |
| Metastases vs. no metastases | 0.95 | [0.53, 1.72] | 0.87 |
| Positive vs. negative margins | 1.22 | [0.75, 1.98] | 0.41 |
| Poor vs. moderate/well grade | 1.15 | [0.72, 1.84] | 0.57 |
| Age | 1.03 | [1.01, 1.05] | 0.006 |
| Tumor size ≥ 3.5 vs. < 3.5 cm | 1.31 | [0.81, 2.11] | 0.26 |
Discussion
Pancreatic cancer is often thought of as a homogeneous disease in which all patients develop early metastases and rapidly progress to death (23). Recent comprehensive studies have shown that molecularly defined sub-groups of pancreatic cancer can identify patients with distinct clinical features, including prognosis and response to therapy (24–29). For example, Franko et al. found that high levels of allelic loss in ductal adenocarcinomas correlate with poorer prognosis following resection, and Sato et al. have reported that aberrant methylation of Reprimo, a gene involved in p53-induced G2 cell cycle arrest, is associated with significantly worse prognosis(26, 27). While efforts to identify genetic alterations of prognostic significance have examined one gene at a time, the recent sequencing of the coding regions of the pancreatic cancer genome provided the opportunity to correlate the mutational status of a large number of genes with patient survival after surgery (17).
We correlated the mutational status of 6 the 39 genes included in both the Discovery and Validation Screen phases of the pancreatic cancer genome initiative with the survival of 89 patients in this previous study who underwent a pancreaticoduodenectomy (17). We found that patients whose cancers had SMAD4 gene inactivation, by either intragenic mutation or homozygous deletion, had significantly worse survival than patients with intact SMAD4 (Figure 1). This result builds on previous observations that the loss of expression of the smad4 protein by immunolabeling is associated with poor prognosis (30, 31). Tascilar et al. immunolabeled 249 surgically resected ductal adenocarcinomas of the pancreas for the smad4 protein and found that patients with cancers with intact smad4 protein expression survived significantly longer than did patients with cancers lacking smad4 (median survival 19.2 months vs. 14.7 months, p=0.03) (31). We also noted that while SMAD4 gene inactivation was strongly correlated with worse survival, inactivation of the TGFβR2 gene, another member of the TGFβ signaling pathway, did not show an association with survival. TGFβ signaling is mediated by the canonical pathway for which smad4 is a central mediator, and non-canonical pathways such as the Erk, JNK and p38 MAPK kinase pathways, among others (32). While further work is needed to investigate the role of TGF downstream pathways on patient outcome, our observation of a relationship between shorter survival and SMAD4 gene inactivation suggests a role for canonical TGFβ signaling in pancreatic cancer.
The association of SMAD4 gene inactivation with poorer prognosis may relate to an increased propensity of pancreatic cancers with inactivated SMAD4 to metastasize widely. Iacobuzio-Donahue et al. recently studied the patterns of metastases in a series of 76 patients with pancreatic cancer who underwent a rapid autopsy (33). Remarkably, 12% of these patients had no evidence of metastatic disease, but instead died of complications of locally advanced pancreatic cancer (33). Among patients with metastases, there was a significant range of metastatic disease, with some patients having only a few metastases (1 to 10), while others had hundreds to thousands (33). The patterns of failure found at autopsy (local versus metastatic) correlated with the smad4 immunolabeling pattern of the patient’s primary carcinoma, with loss of smad4 expression correlating with widespread metastases (33). Similarly, Maitra et al. showed that loss of smad4 expression correlates with progression to metastasis in colorectal carcinoma (34). In this latter study, none of the adenomas or stage I colorectal adenocarcinomas showed loss of smad4 expression, whereas five of 23 (22%) of stage IV cancers showed loss of smad4 expression (34).
The association of SMAD4 gene inactivation with poorer prognosis and an increased propensity to metastasize has direct clinical implications. Some patients with pancreatic cancer have “borderline” resectable tumors- they have resectable pancreatic head cancers that are at high risk for a margin-positive resection (35). While further work is needed, our results, combined with those previously reported in the literature, suggest that patients with borderline resectable pancreatic cancers and SMAD4 gene inactivation might be spared the risk of surgery since their cancer is more likely to metastasize, while patients with borderline resectable pancreatic cancers and intact SMAD4 may benefit from the local control provided by neoadjuvant therapy and surgical resection.
In addition to the SMAD4 gene, we correlated the mutational status of 4 other genes individually and a summary of all 39 genes with survival (17). The mutational status of these 4 other genes did not correlate with survival (Figures 2–4). While previous reports have suggested that the mutational status of KRAS, TP53 and of CDKN2A each correlate with patient survival, we found no such correlations (36–48). These results are perhaps not surprising since the KRAS gene is almost universally activated, and the CDKN2A gene, if one includes methylation, is also nearly universally inactivated in pancreatic cancer (17, 49).
In summary, we correlated the mutational status of the most frequently mutated genes in pancreatic carcinoma with survival and found that only SMAD4 gene mutational status significantly correlated with outcome.
Acknowledgments
Supported by the NIH SPORE (Specialized Programs of Research Excellence) in Gastrointestinal Cancer Grant CA62924, The Sol Goldman Pancreatic Cancer Research Center, The Lustgarten Foundation for Pancreatic Cancer Research, the Monastra Foundation, the Michael Rolfe Pancreatic Cancer Foundation and NIH grant CA121113.
The authors thank Scott E. Kern for his insightful suggestions.
Footnotes
Statement of Translational Relevance
The identification of somatic genetic alterations that are associated with patient outcome may lead to useful clinical tools to guide the treatment of patients with pancreatic cancer, and may provide insight into alterations underlying the aggressive behavior of this cancer.
Reference List
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–6. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 3.Hruban RH, Pitman MB, Klimstra DS. Fourth Series, Fascicle. 6. Washington, DC: American Registry of Pathology and Armed Forces Institute of Pathology; 2007. Tumors of the pancreas. Atlas of tumor pathology. [Google Scholar]
- 4.Sohn TA, Yeo CJ, Lillemoe KD, et al. Resected adenocarcinoma of the pancreas - 616 patients: results, outcome, and prognostic indications. J Gastroenterol Surg. 2000;4:567–79. doi: 10.1016/s1091-255x(00)80105-5. [DOI] [PubMed] [Google Scholar]
- 5.Yeo CJ, Abrams RA, Grochow LB, et al. Pancreaticoduodenectomy for pancreatic adenocarcinoma: postoperative adjuvant chemoradiation improves survival. A prospective, single-institution experience. Ann Surg. 1997;225:621–33. doi: 10.1097/00000658-199705000-00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeo CJ, Sohn TA, Cameron JL, Hruban RH, Lillemoe KD, Pitt HA. Periampullary adenocarcinoma: analysis of 5-year survivors. Ann Surg. 1998;227:821–31. doi: 10.1097/00000658-199806000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlon KC, Klimstra DS, Brennan MF. Long-term survival after curative resection for pancreatic ductal adenocarcinoma. Clinicopathologic analysis of 5-year survivors. Ann Surg. 1996;223:273–9. doi: 10.1097/00000658-199603000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lüttges J, Schemm S, Vogel I, Hedderich J, Kremer B, Klöppel G. The grade of pancreatic ductal carcinoma is an independent prognostic factor and is superior to the immunohistochemical assessment of proliferation. J Pathol. 2000;191:154–61. doi: 10.1002/(SICI)1096-9896(200006)191:2<154::AID-PATH603>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Matsuno S, Egawa S, Fukuyama S, et al. Pancreatic Cancer Registry in Japan: 20 years of experience. Pancreas. 2004;28:219–30. doi: 10.1097/00006676-200404000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Sener SF, Fremgen A, Menck HR, Winchester DP. Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985–1995, using the National Cancer Database. J Am Coll Surg. 1999;189:1–7. doi: 10.1016/s1072-7515(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 11.Yeo CJ, Cameron JL, Sohn TA, et al. Six hundred fifty consecutive pancreaticoduodenectomies in the 1990s: pathology, complications, and outcomes. Ann Surg. 1997;226:248–57. doi: 10.1097/00000658-199709000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benassai G, Mastrorilli M, Quarto G, et al. Factors influencing survival after resection for ductal adenocarcinoma of the head of the pancreas. J Surg Oncol. 2000;73:212–8. doi: 10.1002/(sici)1096-9098(200004)73:4<212::aid-jso5>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa O, Ohigashi H, Sasaki Y, et al. Practical grouping of positive lymph nodes in pancreatic head cancer treated by an extended pancreatectomy. Surgery. 1997;121:244–9. doi: 10.1016/s0039-6060(97)90352-4. [DOI] [PubMed] [Google Scholar]
- 14.Magistrelli P, Antinori A, Crucitti A, et al. Prognostic factors after surgical resection for pancreatic carcinoma. J Surg Oncol. 2000;74:36–40. doi: 10.1002/1096-9098(200005)74:1<36::aid-jso9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Pongprasobchai S, Pannala R, Smyrk TC, et al. Long-term survival and prognostic indicators in small (<or=2 cm) pancreatic cancer. Pancreatology. 2008;8:587–92. doi: 10.1159/000161009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakao A, Harada A, Nonami T, et al. Lymph node metastases in carcinoma of the head of the pancreas region. Br J Surg. 1995;82:399–402. doi: 10.1002/bjs.1800820340. [DOI] [PubMed] [Google Scholar]
- 17.Jones S, Zhang X, Parsons DW, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman JM, Swartz MJ, Hsu CC, et al. Analysis of fluorouracil-based adjuvant chemotherapy and radiation after pancreaticoduodenectomy for ductal adenocarcinoma of the pancreas: results of a large, prospectively collected database at the Johns Hopkins Hospital. J Clin Oncol. 2008;26:3503–10. doi: 10.1200/JCO.2007.15.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrams RA, Grochow LB, Chakravarthy A, et al. Intensified adjuvant therapy for pancreatic and periampullary adenocarcinoma: survival results and observations regarding patterns of failure, radiotherapy dose and CA19-9 levels. Int J Radiat Oncol Biol Phys. 1999;44:1039–46. doi: 10.1016/s0360-3016(99)00107-8. [DOI] [PubMed] [Google Scholar]
- 20.Winter JM, Cameron JL, Campbell KA, et al. 1423 pancreaticoduodenectomies for pancreatic cancer: a single institution experience. J Gastrointest Surg. 2006;10:1199–211. doi: 10.1016/j.gassur.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Wistuba II, Behrens C, Milchgrub S, et al. Comparison of features of human breast cancer cell lines and their corresponding tumors. Clin Cancer Res. 1998;4:2931–8. [PubMed] [Google Scholar]
- 22.Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–74. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 23.Niederhuber JE, Brennan MF, Menck HR. The National Cancer Data Base report on pancreatic cancer. Cancer. 1995;76:1671–7. doi: 10.1002/1097-0142(19951101)76:9<1671::aid-cncr2820760926>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 24.Liang JJ, Wang H, Rashid A, et al. Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res. 2008;14:7043–9. doi: 10.1158/1078-0432.CCR-08-0381. [DOI] [PubMed] [Google Scholar]
- 25.Fujii S, Mitsunaga S, Yamazaki M, et al. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008;99:1813–9. doi: 10.1111/j.1349-7006.2008.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franko J, Krasinskas AM, Nikiforova MN, et al. Loss of heterozygosity predicts poor survival after resection of pancreatic adenocarcinoma. J Gastrointest Surg. 2008;12:1664–72. doi: 10.1007/s11605-008-0577-9. [DOI] [PubMed] [Google Scholar]
- 27.Sato N, Fukushima N, Matsubayashi H, Iacobuzio-Donahue CA, Yeo CJ, Goggins M. Aberrant methylation of Reprimo correlates with genetic instability and predicts poor prognosis in pancreatic ductal adenocarcinoma. Cancer. 2006;107:251–7. doi: 10.1002/cncr.21977. [DOI] [PubMed] [Google Scholar]
- 28.Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer. 2005;41:2213–36. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 29.Yatsuoka T, Sunamura M, Furukawa T, et al. Association of poor prognosis with loss of 12q, 17p, and 18q, and concordant loss of 6q/17p and 12q/18q in human pancreatic ductal adenocarcinoma. Am J Gastroenterol. 2000;95:2080–5. doi: 10.1111/j.1572-0241.2000.02171.x. [DOI] [PubMed] [Google Scholar]
- 30.Biankin AV, Morey AL, Lee CS, et al. DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J Clin Oncol. 2002;20:4531–42. doi: 10.1200/JCO.2002.12.063. [DOI] [PubMed] [Google Scholar]
- 31.Tascilar M, Skinner HG, Rosty C, et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2001;7:4115–21. [PubMed] [Google Scholar]
- 32.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 33.Iacobuzio-Donahue CA, Fu B, Yachida S, et al. DPC4 Gene Status of the Primary Carcinoma Correlates With Patterns of Failure in Patients With Pancreatic Cancer. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.17.7188. (Iin press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maitra A, Molberg K, Albores-Saavedra J, Lindberg G. Loss of Dpc4 expression in colonic adenocarcinomas correlates with the presence of metastatic disease. Am J Pathol. 2000;157:1105–11. doi: 10.1016/S0002-9440(10)64625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varadhachary GR, Tamm EP, Abbruzzese JL, et al. Borderline resectable pancreatic cancer: definitions, management, and role of preoperative therapy. Ann Surg Oncol. 2006;13:1035–46. doi: 10.1245/ASO.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Gerdes B, Ramaswamy A, Ziegler A, et al. p16INK4a is a prognostic marker in resected ductal pancreatic cancer: an analysis of p16INK4a, p53, MDM2, an Rb. Ann Surg. 2002;235:51–9. doi: 10.1097/00000658-200201000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu YX, Watanabe H, Ohtsubo K, et al. Frequent loss of p16 expression and its correlation with clinicopathological parameters in pancreatic carcinoma. Clin Cancer Res. 1997;3:1473–7. [PubMed] [Google Scholar]
- 38.Naka T, Kobayashi M, Ashida K, Toyota N, Kaneko T, Kaibara N. Aberrant p16INK4 expression related to clinical stage and prognosis in patients with pancreatic cancer. Int J Oncol. 1998;12:1111–6. doi: 10.3892/ijo.12.5.1111. [DOI] [PubMed] [Google Scholar]
- 39.Bold RJ, Hess KR, Pearson AS, et al. Prognostic factors in resectable pancreatic cancer: p53 and bcl–2. J Gastrointest Surg. 1999;3:263–77. doi: 10.1016/s1091-255x(99)80068-7. [DOI] [PubMed] [Google Scholar]
- 40.Campani D, Boggi U, Cecchetti D, et al. p53 overexpression in lymph node metastases predicts clinical outcome in ductal pancreatic cancer. Pancreas. 1999;19:26–32. doi: 10.1097/00006676-199907000-00004. [DOI] [PubMed] [Google Scholar]
- 41.Gansauge F, Gansauge S, Link KH, Beger HG. p53 in relation to therapeutic outcome of locoregional chemotherapy in pancreatic cancer. Ann N Y Acad Sci. 1999;880:281–7. doi: 10.1111/j.1749-6632.1999.tb09532.x. [DOI] [PubMed] [Google Scholar]
- 42.Sato Y, Nio Y, Song MM, et al. p53 protein expression as prognostic factor in human pancreatic cancer. Anticancer Res. 1997;17:2779–88. [PubMed] [Google Scholar]
- 43.Linder S, Parrado C, Falkmer UG, Blasjo M, Sundelin P, von Rosen A. Prognostic significance of Ki-67 antigen and p53 protein expression in pancreatic duct carcinoma: a study of the monoclonal antibodies MIB-1 and DO-7 in formalin-fixed paraffin-embedded tumour material. Br J Cancer. 1997;76:54–9. doi: 10.1038/bjc.1997.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinicrope FA, Evans DB, Leach SD, et al. bcl-2 and p53 expression in resectable pancreatic adenocarcinomas: association with clinical outcome. Clin Cancer Res. 1996;2:2015–22. [PubMed] [Google Scholar]
- 45.Lundin J, Nordling S, von Boguslawsky K, Roberts PJ, Haglund C. Prognostic value of immunohistochemical expression of p53 in patients with pancreatic cancer. Oncology. 1996;53:104–11. doi: 10.1159/000227545. [DOI] [PubMed] [Google Scholar]
- 46.Nakamori S, Yashima K, Murakami Y, et al. Association of p53 gene mutations with short survival in pancreatic adenocarcinoma. Jpn J Cancer Res. 1998;86:174–81. doi: 10.1111/j.1349-7006.1995.tb03036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiGiuseppe JA, Hruban RH, Goodman SN, et al. Overexpression of p53 protein in adenocarcinoma of the pancreas. Am J Clin Pathol. 1994;101:684–8. doi: 10.1093/ajcp/101.6.684. [DOI] [PubMed] [Google Scholar]
- 48.Jeong J, Park YN, Park JS, Yoon DS, Chi HS, Kim BR. Clinical significance of p16 protein expression loss and aberrant p53 protein expression in pancreatic cancer. Yonsei Med J. 2005;46:519–25. doi: 10.3349/ymj.2005.46.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–30. [PubMed] [Google Scholar]