Abstract
Sperm proteins that interact with zona pellucida 3 (ZP3) have not been clearly identified in humans. In the present study, the yeast two-hybrid (Y2H) system was used to identify human sperm proteins that interact with human ZP3. Human ZP3 cDNA was cloned into pAS2-1 yeast vector and used as bait to find reactive proteins in the human testis cDNA library. Six specific clones were obtained that were further confirmed for interaction using the mammalian two-hybrid system. These six clones showed homologies with several proteins in the GenBank database. Of these, the strongest ZP3 interacting protein, that shows 97% homology with ubiquitin associated protein-2 like (UBAP2L), was tested in the hemizona assay. UBAP2L antibodies significantly (p < 0.001) inhibited human sperm-zona binding in this assay. We conclude that the Y2H system is a useful strategy for identifying novel genes encoding proteins that interact with ZP proteins. To our knowledge, this is the first study using the Y2H system to identify sperm proteins that interact with human oocyte ZP3. Novel proteins identified using this system may find applications in elucidating the fertilization cascade, development of a new generation of non-steroidal contraceptives, and specific diagnosis and treatment of human infertility.
Keywords: Fertilization, Yeast two-hybrid system, Human ZP3, Sperm proteins, Contraception
1. Introduction
Mammalian fertilization is a complex cascade of molecular events which enables the sperm cell to undergo capacitation, recognition, attachment, and binding to oocyte zona pellucida (ZP) and to undergo acrosomal exocytosis, penetrate ZP, fuse with the oocyte plasma membrane, and fertilize the egg (Yanagimachi, 1994). The molecules and mechanisms involved in sperm-egg recognition and binding have not been clearly delineated (Dean, 2006; Tulsiani et al., 2006). Various pathways and molecules have been proposed to be involved in this process in several mammalian species (Naz and Ahmad, 1994; Naz et al., 2000; Yi et al. 2006; Tulsiani et al., 2006; Xu et al., 2006). In humans, ZP3 has been identified as one of the primary zona pellucida components that is involved in sperm binding and acrosomal exocytosis (Gupta et al., 2006; Dean, 2006). Both specific sugar residues as well as peptide moieties have been proposed as mediators of sperm-egg recognition, attachment and binding, and in acrosomal exocytosis (Chapman et al., 1998; Chakravarty et al., 2005., Gupta et al., 2006). cDNA encoding ZP3 has been cloned and sequenced from several species including humans (Harris et al., 1994; 1999; Gupta et al., 2006). Our laboratory found that, in humans, there are at least four sperm proteins that interact with cognate human ZP3 (Naz and Ahmad, 1994). Although several molecules have been proposed as potential candidates, the sperm proteins and glycoproteins involved in binding to oocyte ZP3 have not been clearly elucidated.
The yeast two-hybrid system (Y2H) is a genetic method used to identify proteins that interact with a target protein expressed in yeast as a hybrid with a DNA-binding domain (Guarente, 1993; Fields and Sternglanz, 1994; Allen et al., 1995). It has been widely used to examine protein-protein interactions. A reporter gene expression is activated via reconstitution of a functional transcription factor when two hybrid proteins associate. Typically, a gene encoding a protein of interest is fused to the DNA-binding domain of a transcription factor (such as GAL4, LEXA), while another gene is fused to a transcriptional activation domain (such as GAL4, VP16) (Allen et al., 1995). The activation-domain hybrid is introduced into a yeast strain expressing the DNA-binding domain hybrid and a productive interaction between the two proteins of interest localizes the activation domain to the DNA-binding domain. Subsequent transcription of an adjacent reporter gene, typically lacZ or a nutritional marker provides an identifiable phenotype. Besides studying the protein-protein interactions, the Y2H system has also been extensively used to identify novel genes encoding proteins that interact/bind/associate with a protein of interest (Suter et al., 2008; Bao et al., 2009).
The aim of the present study was to identify human sperm genes encoding proteins that interact with human ZP3 using Y2H (MATCHMAKER GAL4-based yeast two-hybrid system, Clontech Laboratories Inc., Mountain View, CA, USA). The long-term objective is to delineate sperm proteins that can be used as targets for the development of novel contraceptives and for the specific diagnosis and treatment of human infertility.
2. Materials and methods
2.1. Construction of bait plasmid (pAS21-ZP3)
The ZP3 cDNA was obtained from the National Institute of Technology and Evaluation (NITE), National Biological Resource Center (NBRC), Japan (http://www.nbrc.nite.go.jp/e/hflcdna-e.html). This has human zona pellucida glycoprotein 3A precursor cDNA cloned into pME18SFL3 vector at EcoRI and XbaI sites (Accession number: AK056788; FLJ number: FLJ32226; Clone ID: PLACE6004380). It has Homo sapiens zona pellucida glycoprotein 3A precursor cDNA of 1845 bp. Analysis in the GenBank database using BLAST (http://www.ncbi.nlm.gov.blast) revealed that in the ~1000 bp region, at the 3' end, it has a significant homology with previously published ZP3 sequences including human. This region has a 332 aa long peptide sequence that shows a 96% homology with ZP3. This sequence was PCR-amplified using sense (5'CCGGAATTCATGGCCGGCAGCATTAACT 3') and antisense (5'CGCGGATCCCTTCTTTTATTCGGAAGCAG 3') primers, incorporating EcoRI and BamHI sites, respectively, and cloned into pAS2-1 GAL4 binding domain vector following the manufacturer’s protocol (Clontech Laboratories Inc., Mountain View, CA, USA). PCR amplification cycles involved: initial denaturation at 94° C for 5 min, 30 cycles at 94° C for 1 min, 55° C for 1 min, 72° C for 1 min, and the final extension at 72° C for 10 min. The authenticity and right orientation of the cloned sequence was confirmed by restriction digestion, and by sequencing (Northwoods DNA Inc., Solway, MN, USA).
2.2. Expression of ZP3 protein in yeast
To examine the ZP3 protein expression, the yeast cells were transformed with bait ZP3 plasmid (pAS2-1-ZP3) by BD Yeastmaker™ Yeast transformation system 2 (Clontech Laboratories) following the manufacturer’s protocol. The single isolated colony of bait strain AH109, having ZP3 plasmid (pAS2-1-ZP3), was isolated, inoculated into SD/-Trp medium, and incubated at 30°C in a shaker at 220 rpm overnight. The culture was then transferred into YPDA medium and incubated at 30°C until the A600nm reached 0.4–0.6. The culture was centrifuged and the cell pellet was used for protein extraction by urea-SDS method (Clontech Laboratories). The protein extract was resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the separated proteins were transferred onto a nitrocellulose membrane for the Western blot procedure (Arthur and Naz, 2008). The blot was blocked with 5% non-fat dried milk for 2 hr at 37° C, incubated with anti-human ZP3 rabbit polyclonal antibody (GenWay Biotech Inc., San Diego, CA, USA; catalog # 18-003-42708), and then with alkaline phosphatase-conjugated goat anti-rabbit antibodies. The immunoreactive bands were visualized using 5-bromo, 4-chloro, 3-indolyl phosphate (BCIP) and nitro blue tetrazolium (NBT) as the substrate. Anti-human ZP3 rabbit polyclonal antibody used in this assay was raised against a human ZP3 50 aa synthetic peptide epitope: NKGDCGTPSHSRRQPHVMSQWSRSASRNRRHVTEEADVTVGATDLPGQEW. This sequence showed an 88% homology with our ZP3 sequence using BLAST P. HepG-2 lysate was used as a positive control. The human ZP3 antibody recognizes a 42 kD protein band in this lysate.
2.3. Screening of human testis library and selection of ZP3 interactive clones
The commercially available human testis cDNA library, cloned into pACT-2, a GAL-4 activation domain vector, and transformed into Y-187 yeast strain, was purchased from Clontech Laboratories and used for screening in the Y2H system. The library was tested for the expression of proteins using the murine monoclonal antibody to the activation domain (aa 768–881) (Clontech Laboratories) by the Western blot procedure. It was found to express numerous proteins that were recognized by the antibody.
A fresh colony of bait strain AH109 (pAS2-1-ZP3), confirmed by sequencing as described above, was inoculated into SD/-Trp medium and incubated at 30°C in a shaker at 220 rpm overnight. It was centrifuged (3000 rpm, 5 min) and re-suspended in the medium. One ml of the testis cDNA library was mixed with 3 ml of AH109 (pAS2-1-ZP3), 45 ml of 2X YPDA/kanamycin (50 µg/ml) medium was added, and the mixture was incubated at 30°C in a shaker at 40 rpm. After 22 hr, zygote formation in the yeast mating culture was examined under a phase contrast microscope (400X). After 2 hr of further incubation at 30°C, the yeast mating culture was centrifuged (3000 rpm, 10 min) and the pellet was re-suspended in 10 ml of 0.5X YPDA/kanamycin medium. The mating mixture was plated on various selective media, namely SD/-Trp, SD/-Leu, SD/-Leu/-Trp, SD/-His/-Leu/-Trp (TDO) medium and SD/-Ade/-His/-Leu/-Trp (QDO) medium and then incubated at 30°C until the colonies appeared (3–21 days). The colonies grown on TDO and QDO media were further selected for their growth on X-α Gal selection medium.
The colonies from X-α Gal selection were inoculated into SD/-Leu liquid medium and used for plasmid extraction by the Yeastmaker™ plasmid isolation kit following the manufacturer’s protocols (Clontech Laboratories, USA). The testis cDNA inserts were amplified from the plasmid DNA using activation domain primers provided by the manufacturer. The plasmid DNA and amplified testis cDNA inserts were sequenced and the sequences were analyzed by BLAST (http://www.ncbi.nlm.gov.blast). The open reading frames of the cDNA inserts were delineated.
2.4. Confirmation of interacting clones using the mammalian two-hybrid system
The authenticity of the ZP3-interacting testis clones, obtained using the Y2H system, was further confirmed by the mammalian two-hybrid system. The ZP3 insert was cloned into pM, a mammalian GAL-4 binding domain vector, and the interacting testis cDNAs were cloned into pVP-16, a mammalian GAL-4 activation domain vector (Clontech Laboratories). The primers to amplify the interacting testis clones were designed based on nucleotide sequences at the open reading frames. The list of primers used for this purpose is given in Table 1. Chinese hamster ovaries (CHO)-k1 cells were used to study the interaction. The cells were incubated in F-12/glutamax medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen Corp., Carlsbad, CA, USA) at 5% CO2 incubator. Before transfection, the cells were suspended to obtain 50–80% confluence in a fresh medium. The mixture of pM-ZP3 and different clones of pVP-16 were co-transfected with pG5SEAP reporter vector by the calcium phosphate-mediated method. After 12 hr, the transfection mixture was replaced with fresh medium and incubated at 5% CO2 incubator. The cell supernatants were collected after 48–72 hr to assay for the secreted alkaline phosphatase (SEAP) activity using the chemiluminescent SEAP detection kit following the manufacturer’s protocol (Roche Diagnostics, Indianapolis, IN, USA). Absorbance460nm reading was converted to standard deviation (SD) units by the following formula: SD unit = absorbance value of test sample - mean absorbance value of control group/SD of control group (Naz and Chauhan, 2002). SD values of ≥ + 2 were considered positive.
Table 1.
Sequence of primers used for PCR-amplification of the interacting clones.
| Clone | Sequence of primers | Restriction sites | Size of amplicon |
|---|---|---|---|
| GS-2 | Sense: 5’AAAACTGCAGTGGCAGTAGCCTGGGC 3’ Anti-sense: 5’GCTCTAGATTACTGGAGGTGACTGAA 3’ |
PstI XbaI |
652 bp |
| GS-6 | Sense: 5’AAAACTGCAGCGGGAGCTACAAGCATG 3’ Anti-sense: 5’CCCAAGCTTCTAAAAATTCAAAAAAA 3’ |
PstI HindIII |
228 bp |
| GS-7 | Sense: 5’ ACGCGTCGACTGTTGTTGGCCCTGGT 3’ Anti-sense: 5’CCCAAGCTTCTAGAAGTCACAGCCATC 3’ |
SalI HindIII |
440 bp |
| GS-12 | Sense: 5’ ACGCGTCGACCACACACAGTCTTTGC 3’ Anti-sense: 5’ CCCAAGCTTTTATTCTTTCAGGAAA 3’ |
SalI HindIII |
470 bp |
| GS-18 | Sense: 5’ACGCGTCGACGCTCCCTGGATGGCT 3’ Anti-sense: 5’ GCTCTAGATCACTTTCTCACAGCC 3’ |
SalI XbaI |
326 bp |
| Col99 | Sense: 5’ ACGCGTCGACACGAGAAGCACCTACT 3’ Anti-sense: 5’CCCAAGCTTTTAAGGAACCGAGAT 3’ |
SalI HindIII |
479 bp |
2.5. Hemizona assay
The hemizona assay was performed as described earlier (Kadam et al., 1995; Naz et al., 2000). Human oocytes were obtained from the patients participating in the in vitro fertilization and embryo transfer (IVF-ET) program. The study was approved by the Institutional Review Board (IRB). Human spermatozoa were collected from healthy fertile donors. The population of motile sperm (0.5×106 sperm/ml) was used in the hemizona assay. The effect of antibody to the strongest reacting protein (Ubiquitin associated protein-2 like (UBAP2L) on human sperm-ZP binding was examined in the hemizona assay. Affinity-purified rabbit polyclonal antibody to human UBAP2L were purchased from Bethyl Laboratories, Montgomery, TX, USA (catalog number: A310-153A). Immunoglobulins from normal rabbits were used as controls. Sperm (0.5×106 motile sperm/100-µl drop) were incubated for 4 hr with 7–15 µg of antibody, control immunoglobulins, or BSA. Sperm were then centrifuged and incubated (37°C, 1 hr) with one half of the hemizona pair. The experimental hemizona was incubated with sperm treated with anti-UBAP2L antibody, and the other half of the hemizona pair from the same oocyte served as a control and was incubated with untreated sperm. The hemizonae were then washed (x3) to remove the loosely attached sperm. The number of sperm tightly bound to the outer surface of the hemizona was determined and expressed as the hemizona index: HZI = [number of sperm tightly bound on experimental hemizona / number of sperm bound on control hemizona]×100.
Each sample was tested with 7–9 hemizonae obtained from 3–5 women in three different experiments performed on separate days using sperm from 2–3 fertile men.
2.6. Statistical analysis
Significance of the difference between the experimental (UBAPL2 antibody/control immunoglobulins) and BSA control hemizona indices was analyzed by using unpaired and paired Student’s t-test. A p value of < 0.05 was considered significant.
3. Results
3.1. Construction of bait plasmid (pAS2-1-ZP3)
The 1000 bp sequence at the 3' end of 1845 bp human zona pellucida glycoprotein 3A precursor, which showed up to 96% homology with the published ZP3 sequence, was amplified from pME18SFL3 vector and cloned into pAS2-1 yeast vector at EcoRI and BamHI sites (Fig. 1 and Fig. 2). Authenticity of the insert in the clone was confirmed by restriction digestion (Fig. 2) and sequencing (Fig. 1). The ZP3 cDNA insert had an open reading frame (ORF) translating into 332 amino acids.
Figure 1.
The sequence of ZP3 that was cloned from pME18SFL3 vector into pAS2-1 vector. The ~1000 bp at the 3' end of 1845 bp that showed homology with the published ZP3 sequence was cloned into pAS2-1 vector. The nucleotide and amino acid sequences of the cloned ZP3 are shown in bold. It translates into 332 amino acids with a stop codon before and after the coding sequence.
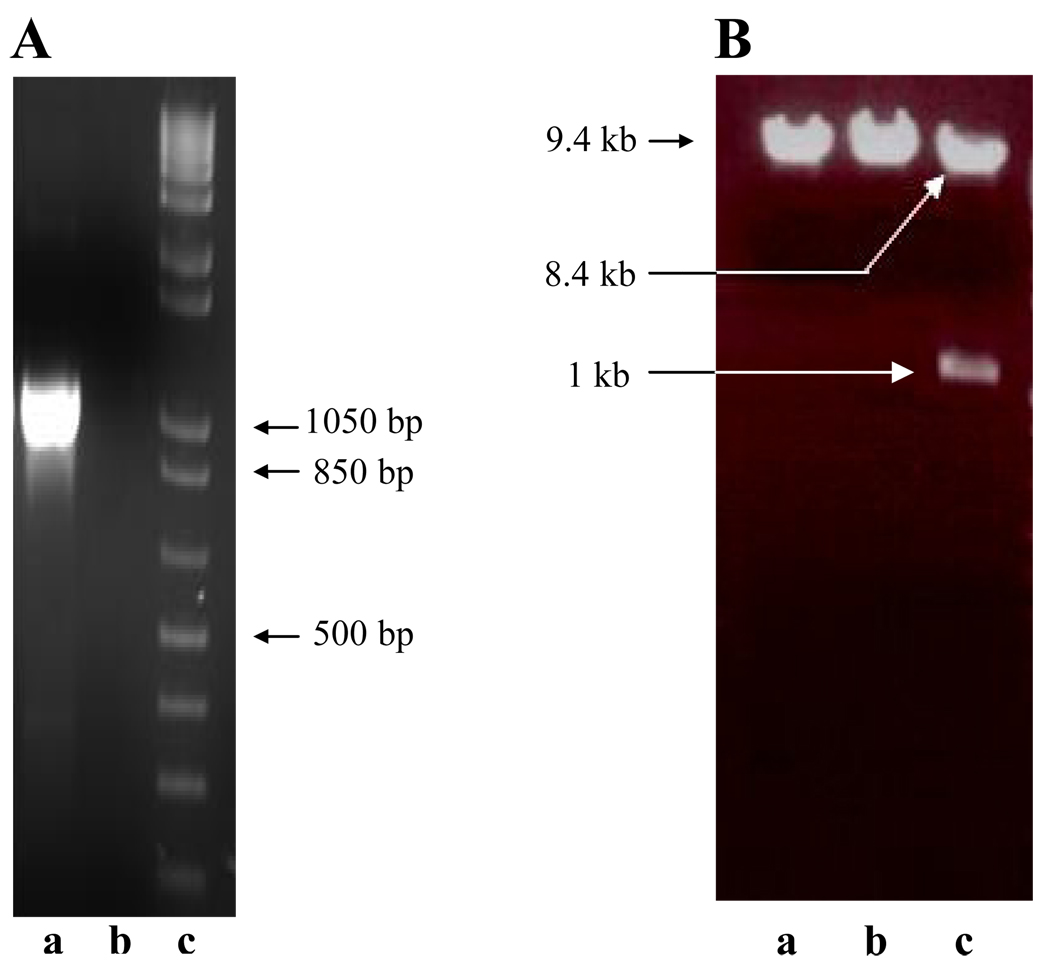
Figure 2.
Confirmation of ZP3 insert cloned into yeast pAS2-1 vector (8.4 kb) at EcoRI and BamHI sites. (A) The ZP3 insert of 1 kb was PCR-amplified from pME18SFL3 using specific primers (lane a). The negative control (lane b) and the DNA molecular weight ladder (lane c) are included for comparison. (B) The pAS2-1-ZP3 yeast clone (9.4 kb) digested with EcoRI (lane a) or BamHI (lane b) alone did not show, but the clone digested with both the EcoRI and BamHI restriction enzymes (lane c), showed the expected 1 kb ZP3 band in the agarose gel electrophoresis. The authenticity of the clone was further confirmed by sequencing.
The expression of ZP3 protein in AH109 (pAS2-1-ZP3) clone was confirmed using commercially available anti-ZP3 antibody. Anti-ZP3 antibody specifically recognized a band of ~53 kD in the AH109 culture extract (Fig. 3, Panel A, lane a) and no band in the pAS2-1 vector alone culture extract (Fig. 3, Panel A, lane b). This band was not recognized by the control immunoglobulins (Fig. 3, Panel B, lane a).
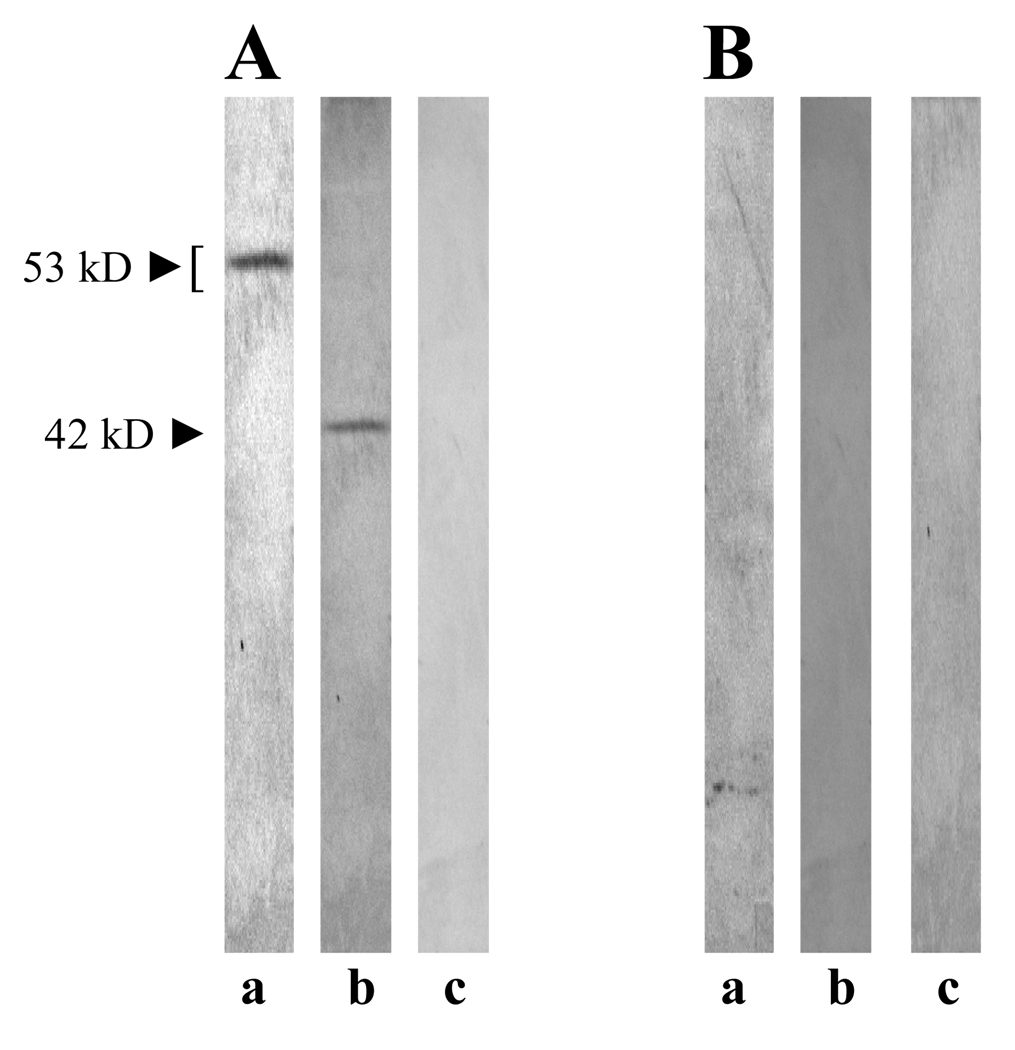
Figure 3.
Expression of ZP3 protein in AH109 (pAS2-1-ZP3). The expression of ZP3 protein in AH109 was confirmed using anti-ZP3 antibody. (A) Anti-ZP3 antibody specifically recognized a protein band of ~53 kD in AH109 culture extract (lane a). The antibody recognized the expected protein band of 42 kD in HepG-2 lysate that was used as a positive control, supplied by the manufacturer (lane b). The antibody did not react with any protein band in vector (pAS2-1) alone culture lysate without ZP3 (lane c). (B) Control rabbit immunoglobulins did not react with any band in AH109 culture extract (lane a), HepG-2 lysate (lane b), or pAS2-1 vector alone culture extract (lane c).
3.2. Screening of human testis library
Expression of different proteins in the human testis library used for screening was examined using antibody to activation domain, supplied by the manufacturer. The testis library was found to express numerous proteins in yeast culture extracts by the Western blot procedure (data not shown).
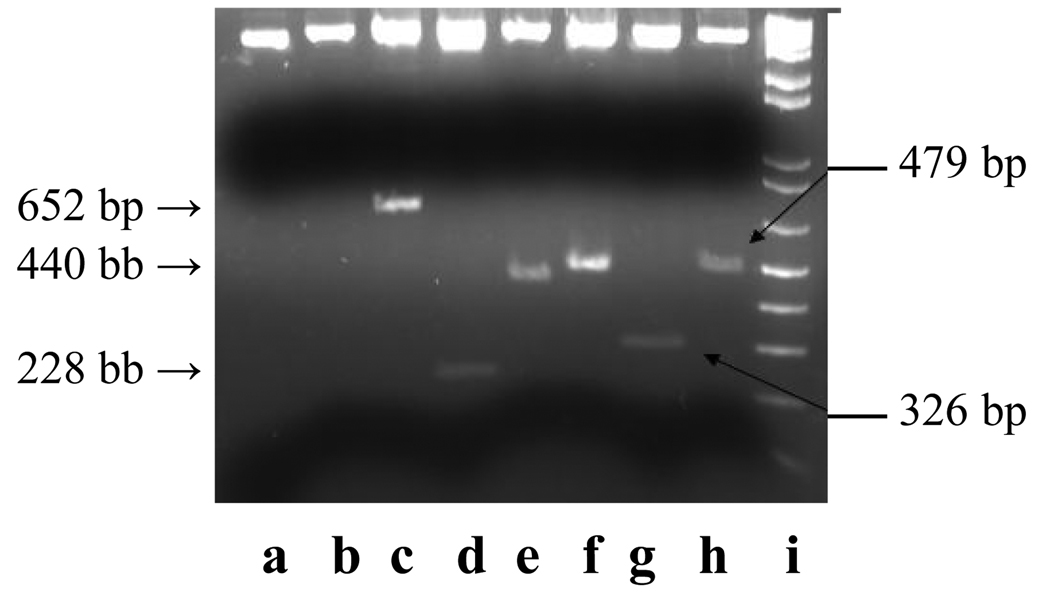
Several clones were found to react with the AH109 (pAS2-1-ZP3) bait, as identified by the presence of blue colonies. Plasmids and direct cultures of the blue colonies from X-α gal selection on SD/-Ade/-His/-Leu/-Trp were screened using activation domain primers. The amplicons of various sizes were identified in the interacting blue colonies (Fig. 4). In the primary screening, 33 blue colonies were obtained after X-α gal selection and inserts from only 23 colonies could be amplified by PCR. Further multiple screening using high stringency, yielded six colonies, designated as GS-2, GS-6, GS-7, GS-12, GS-18, and Col99, respectively. These colonies were PCR-amplified and the inserts were sequenced using primers described in Table 1. The amino acid sequences of the interacting clones were decided based on the longest ORF. The sequences (nucleotide and amino acid) of these six interacting clones were analyzed by BLAST for their similarity with Homo sapiens sequences in the GenBank. The sequences of these six clones were found to have a significant similarity with already available sequences in the GenBank (Table 2). GS-2 showed a significant homology with ubiquitin associated protein-2 like isoforms/ NICE-4 protein (97%), hCG 1811260, KIAA1491 protein from human unidentified gene-encoded (HUGE) database, and unnamed protein product from the human genome sequencing (HGS) project (72%). GS-6 showed a significant homology (60%–89%) with mitochondrial ribosomal protein, unnamed protein product from HGS project, hCG 1811730, unnamed protein product from HGS project, hCG 2038839 protein, and putative calcium sensing receptor like 1. GS-7 showed a significant homology (100% identity) with sperm acrosome associated 3/lysozyme. GS-12 showed a significant homology with olfactory receptor, family 2, members 3, 8, and 2 and olfactory receptor ORI-53 (75%–96%). GS-18 showed a significant homology (97%–100%) with protocadherin 17. Col99 showed a significant homology with KIAA1668 protein from HUGE database, MICAL like 1 protein interacting with Rab13, and hypothetical testis protein.
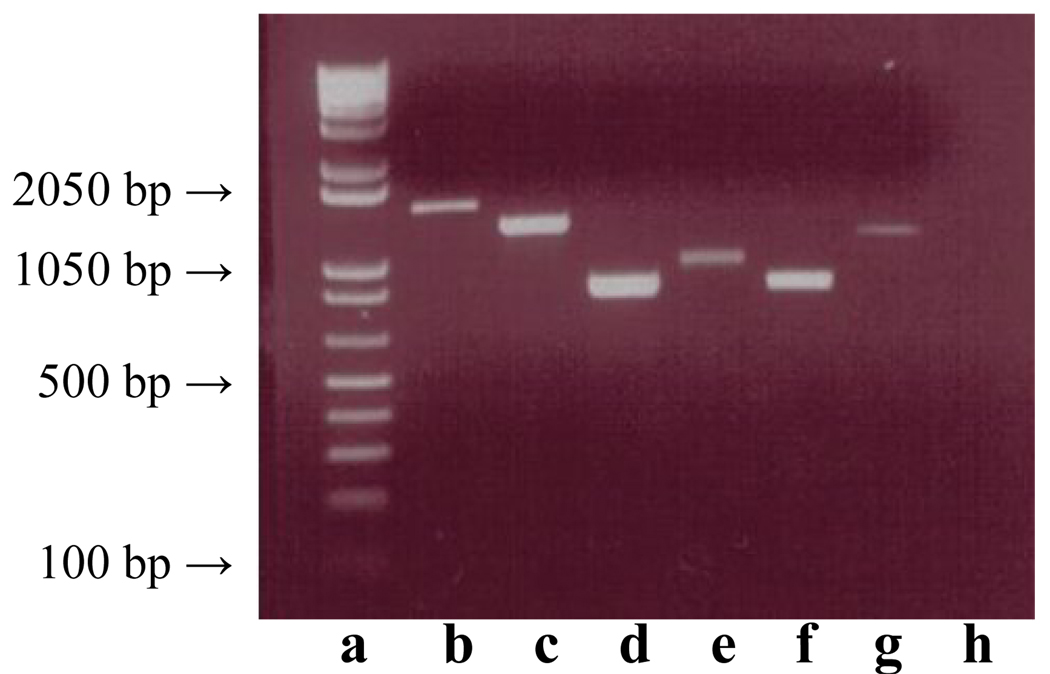
Figure 4.
The inserts of the interacting clones in the yeast two-hybrid system PCR-amplified using activation domain primers. The interacting clones showed cDNA inserts of various sizes (lanes b–g). The negative clone without an insert did not amplify any band (lane h). The DNA molecular weight ladder has been included for comparison (lane a).
Table 2.
Nucleotide and amino acid sequence homologies of six interacting clones with Homo sapiens genes/proteins in the GenBank.
| Interacting clones | Nucleotide sequence homology (BLAST N) |
Homology (%) |
Protein sequence homology (BLAST P) | Homology (%) |
|---|---|---|---|---|
| 1. GS-2 | Ubiquitin associated protein-2 like | 97 | Ubiquitin associated protein-2 like isoforms/ NICE-4 protein |
97 |
| hCG 1811260 | 72 | |||
| KIAA1491 protein from human unidentified gene-encoded (HUGE) database |
72 | |||
| Unnamed protein product from human genome sequencing (HGS) project |
72 | |||
| 2. GS-6 | Mitochondrial ribosomal protein S16 (mRPS16) |
89 | Unnamed protein product from HGS project | 60 |
| hCG 1811730 | 64 | |||
| Unnamed protein product from HGS project | 68 | |||
| hCG 2038839 | 62 | |||
| Putative calcium sensing receptor like 1 | 60 | |||
| 3. GS-7 | Sperm acrosome associated 3 | 100 | Sperm acrosome associated 3 | 100 |
| Lysozyme | 100 | |||
| 4. GS-12 | Olfactory receptor, family 2, subfamily L, member 13 |
96 | Olfactory receptor, family 2, members 3, 8, and 2 | 77–98 |
| Olfactory receptor, ORI-53 | 75 | |||
| 5. GS-18 | Protocadherin 17 (PCDH17) | 98 | Protocadherin 17 | 100 |
| 6. Col99 | MICAL like 1 (family of conserved flavoprotein oxido reductases) |
97 | KIAA1668 protein from HUGE database | 100 |
| MICAL like 1 protein interacting with Rab13 | 100 | |||
| Hypothetical protein (source: testis) | 100 |
3.3. Construction of bait plasmid (pM-ZP3) and confirmation of the yeast interacting clones in the mammalian system
The ZP3 insert from the yeast pAS2-1-ZP3 was cloned into the pM mammalian vector which was used as bait. The authenticity of the insert in the vector was confirmed by restriction digestion (Fig. 5) and sequencing.
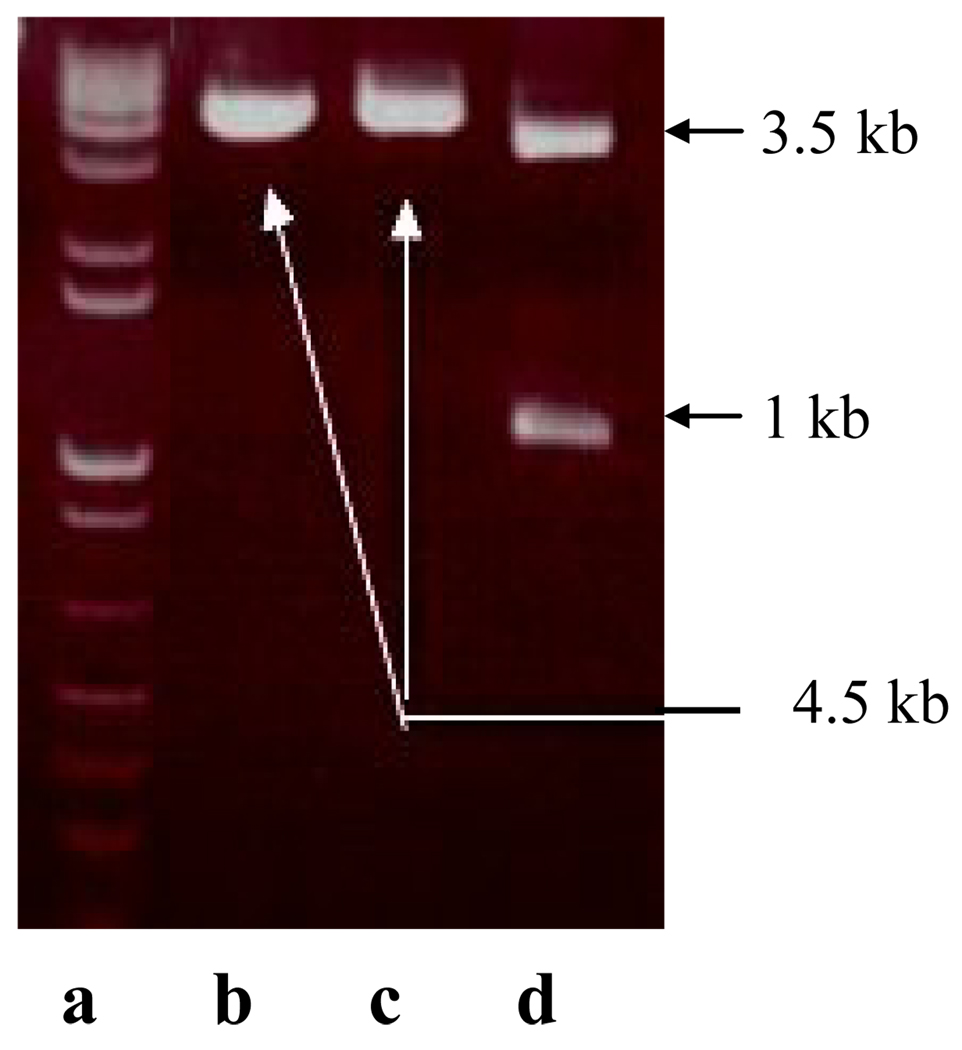
Figure 5.
Confirmation of ZP3 insert cloned into mammalian pM vector (3.5 kb) at EcoRI and BamHI sites. The pM-ZP3 mammalian clone digested with EcoRI (lane b) or BamHI (lane c) alone did not show, but the clone digested with both the EcoRI and BamHI restriction enzymes (lane d), showed the expected 1 kb ZP3 band in the agarose gel electrophoresis. The DNA molecular weight ladder has been included for comparison (lane a). The authenticity of the clone was further confirmed by sequencing.
The inserts of the six interacting clones from the Y2H system were amplified using their respective primers. The primers were incorporated with different restriction enzymes to clone them with the correct reading frame into the pVP-16 mammalian vector. The authenticity of the inserts in the recombinant vectors was confirmed by digesting them with restriction enzymes incorporated in the primers (Fig. 6) as well as sequencing. The six clones (GS-2, GS-6, GS-7, GS-12, GS-18, and Col99) were found to interact with the ZP3 cloned in the mammalian vector, as seen by the SEAP reporter gene assay (ELISA SD units, 4.1–16.8), with the GS-2 clone reacting the strongest and the GS-6 reacting the weakest in the SEAP reporter gene assay.
Figure 6.
Confirmation of the six interacting clones (GS-2, GS-6, GS-7, GS-12, GS-18, and Col99) in the pVP-16 mammalian vector digested with various restriction enzymes. The undigested pVP-16 vector (lane a), pVP-16 digested with HindIII and SalI (lane b), pVP-16-GS-2 clone digested with PstI and XbaI (lane c), pVP-16-GS-6 clone digested with HindIII and PstI (lane d), pVP-16-GS-7 clone digested with Hind III and SalI (lane e), pVP-16-GS-12 clone digested with HindIII and SalI (lane f), pVP-16-GS-18 clone digested with HindIII and EcoRI (lane g), and pVP-16-Col99 clone digested with HindIII and SalI (lane h). The DNA molecular weight ladder has been included for comparison (lane i). The authenticity of the clones was further confirmed by sequencing.
3.4. Effect of anti-UBAP2L antibody on human sperm-zona binding
The strongest reacting clone (GS-2 clone), which showed 97% homology both at the nucleotide and amino acid sequences with human ubiquitin associated protein-2 like (UBAP2L), was selected for further analysis. The commercially available antibody to UBAP2L was obtained and examined for its effect on human sperm-zona binding in the hemizona assay. Anti-UBAP2L antibody significantly (p < 0.001) inhibited binding of sperm to hemizonae in a concentration-dependent manner compared to control immunoglobulins or BSA control (Table 3). Control immunoglobulins did not affect sperm-zona binding (p < 0.05).
Table 3.
Effect of UBAP2L antibodies on human sperm-zona binding.
| Sperm treated with | Concentration (µg/100µl) |
Number of sperm tightly bound to* | Hemizona index (HZI) |
|
|---|---|---|---|---|
| Hemizona incubated with treated sperm |
Hemizona incubated with control sperm |
|||
| 1. Anti-UBAP2L antibody | 7 | 1.2 ± 0.3 | 7.0 ± 0.1* | 17.14** |
| 15 | 0.3 ± 0.04 | 6.3 ± 0.2 | 4.76** | |
| 2. Control immunoglobulins | 7 | 7.2 ± 0.2 | 6.1 ± 0.6 | 114.29 |
| 15 | 6.8 ± 0.1 | 7.1 ± 0.5 | 95.77 | |
| 3. BSA control | 15 | 7.3 ± 0.2 | 7.2 ± 0.6 | 101.39 |
Values are mean ± S.E.M.
P < 0.001; all others were non-significant (P > 0.05) versus BSA control.
4. Discussion
In humans, ZP3 has been identified as the primary receptor for binding to human sperm (Gupta et al., 2006; Dean, 2006). Both sugar residues (O- and N- linked, preferably N-linked) as well as peptide moieties of ZP3 have been proposed as mediators of human sperm-egg recognition, attachment and binding, and acrosomal exocytosis (Chapman et al., 1998; Chakravarty et al., 2005; Gupta et al., 2006). cDNA encoding ZP3 has been cloned and sequenced (Harris et al., 1994; 1999; Gupta et al., 2006). The expression of ZP3 in our system was confirmed using anti-ZP3 antibody in the Western blot procedure. It showed a band of ~53 kD in the expression vector culture extract. This was expected since ZP3 (~37 kD) was expressed along with a binding domain of 147 aa.
Using the Y2H system, six clones were obtained from the human testis library that reacted with ZP3. We were able to PCR-amplify these clones from the human testis cDNA library in λgt10 (Clontech) using gene-specific primer further confirming their expression in the human testis (data not shown). These clones showed homology with several published sequences, several of them have been shown to be expressed in the testis and/or involved in sperm fertilization of the oocyte. Ubiquitin-proteasome pathway has been implicated in sperm-ZP interaction in several species (Sutovsky et al., 1999; 2004; Sakai et al., 2004). Sperm acrosome associated 3/lysozyme is present on the acrosome of the human sperm, expressed primarily in the testis, and the antibodies to it block binding of human sperm to zona-free hamster oocytes (Mandal et al., 2003). Various olfactory receptor family members are not only expressed in the nostrum but also in the testis and on mature sperm (Parmentier et al., 1992; Vandergaeghen et al., 1993; Spehr et al., 2004). The sperm olfactory receptor has been hypothesized to play a role in sperm chemotaxis (Vosshall, 2004). Protocadherin is a calcium dependent cell-cell adhesion molecule (Yagi, 2008). MICAL like 1 proteins interact with Rab proteins and are involved in junction complexes (Yamamura et al., 2007).
The six interacting clones were then confirmed for interaction with ZP3 using mammalian chinese hamster ovarian (CHO) system. This system glycosylates the peptides and may mimic the in vivo pattern observed in humans. The six clones were found to interact with ZP3 with various degrees in the mammalian system as observed in Y2H system. These findings may indicate that glycosylation of the polypeptide moieties do not significantly modify the sperm-zona interaction patterns. Both specific sugar residues as well as peptide moieties have been proposed as mediators of sperm-egg recognition, attachment and binding, and acrosomal exocytosis in various mammalian species, including humans (Chapman et al., 1998; Chakravarty et al., 2005; Gupta et al., 2006).
Antibody to UBAP2L protein significantly inhibited human sperm-zona binding in the hemizona assay. The other molecules need to be tested for their effect on sperm-zona binding.
5. Conclusion
In conclusion, we found at least six genes/proteins that interact with human ZP3 in the Y2H and mammalian systems. Antibody to one of these molecules (UBAP2L) that was examined further also inhibited sperm-zona binding when tested in vitro in the hemizona assay. Further studies are needed to confirm these preliminary findings. Our data also indicate that Y2H is a valid system to identify novel genes encoding proteins that interact with a protein of interest (Suter et al., 2008; Bao et al., 2009). To our knowledge, this is the first study using the Y2H system to identify sperm proteins that interact with human oocyte ZP3 and/or other zona pellucida components. Although we identified six proteins reacting with the human zona pellucida using Y2H, it does not rule out that there are additional proteins that react with ZP3. Novel proteins identified using this system may find applications in elucidating the fertilization cascade, development of a new generation of non-steroidal contraceptives, and specific diagnosis and treatment of human infertility. Further studies are needed to examine their structure-function relationship and molecular mechanism of action.
Acknowledgement
We thank Sarah Wesson and Sarah Davis kindly for the excellent technical and typing assistance. We also sincerely thank members, especially Don Zeh, of Andrology and In Vitro Fertilization (IVF) laboratories for technical assistance. This work was supported in part by grant (HD24425) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen JB, Walberg MW, Edwards MC, Elledge SJ. Finding prospective partners in the library: the two-hybrid system and phage display find a match. Trends in Biochem. Sci. 1995;20:511–516. doi: 10.1016/s0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- Bao L, Redondo C, Findlay JB, Walker JH, Ponnambalam S. Deciphering soluble and membrane protein function using yeast systems. Mol. Membr. Biol. 2009;26:127–135. doi: 10.1080/09687680802637652. [DOI] [PubMed] [Google Scholar]
- Chakravarty S, Suraj K, Gupta SK. Baculovirus expressed recombinant human zona pellucida glycoprotein-B induces acrosomal exocytosis in capacitated spermatozoa in addition to zona pellucida glycoprotein –C. Mol. Hum. Reprod. 2005;11:365–372. doi: 10.1093/molehr/gah165. [DOI] [PubMed] [Google Scholar]
- Chapman NR, Kessopoulou E, Andrews P, Hornby D, Barratt CR. The polypeptide backbone of recombinant human zona pellucida glycoprotein-3 initiates acrosomal exocytosis in human spermatozoa in vitro. Bioch. J. 1998;330:839–845. doi: 10.1042/bj3300839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J. The enigma of sperm-egg recognition in mice. In: Gupta SK, Koyama K, Murray FF, editors. Gamete Biology: Emerging Frontiers in Fertility and Contraceptive Development. Nottingham, United Kingdom: Nottingham University Press; 2006. pp. 359–365. [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Guarente L. Strategies for the identification of interacting proteins. Proc. Natl. Acad. Sci. USA. 1993;90:1639–1641. doi: 10.1073/pnas.90.5.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Chakravarty S, Suraj K, Bansal P, Ganguly A, Jain MK, Bhandari B. Structural and functional attributes of zona pellucida glycoproteins. In: Gupta SK, Koyama K, Murray FF, editors. Gamete Biology: Emerging Frontiers in Fertility and Contraceptive Development. Nottingham, United Kingdom: Nottingham University Press; 2006. pp. 203–216. [Google Scholar]
- Harris JD, Hibler DW, Fontenot GK, Hsu KT, Yurewicz EC, Sacco AG. Cloning and characterization of zona pellucida genes and cDNAs from a variety of mammalian species: the ZPA, ZPB and ZPC gene families. DNA Sequence. 1994;4:361–393. doi: 10.3109/10425179409010186. [DOI] [PubMed] [Google Scholar]
- Harris JD, Seid CA, Fontenot GK, Liu HF. Expression and purification of recombinant human zona pellucida proteins. Protein Expression and Purification. 1999;16:298–307. doi: 10.1006/prep.1999.1060. [DOI] [PubMed] [Google Scholar]
- Kadam AL, Fateh M, Naz RK. Fertilization antigen (FA-1) completely blocks human sperm binding to human zona pellucida: FA-1 antigen may be a sperm receptor for zona pellucida in humans. J. Reprod. Immunol. 1995;29:19–30. doi: 10.1016/0165-0378(95)00928-e. [DOI] [PubMed] [Google Scholar]
- Yi Y, Manandhar G, Oko RJ, Breed WG, Sutovsky P. Mechanism of sperm-zona pellucida penetration during mammalian fertilization. In: Gupta SK, Koyama K, Murray FF, editors. Gamete Biology: Emerging Frontiers in Fertility and Contraceptive Development. Nottingham, United Kingdom: Nottingham University Press; 2006. pp. 385–408. [Google Scholar]
- Mandal A, Klotz KL, Shetty J, Jayes FL, Wolkowicz MJ, Bolling LC, Coonrod SA, Black MB, Diekman AB, Haystead TA, Flickinger CJ, Herr JC. SLLP1, a unique, intra-acrosomal, non-bacteriolytic, c lysozyme-like protein of human spermatozoa. Biol. Reprod. 2003;68:1525–1537. doi: 10.1095/biolreprod.102.010108. [DOI] [PubMed] [Google Scholar]
- Naz RK, Ahmad K. Molecular identities of human sperm proteins that bind human zona pellucida: nature of sperm-zona interaction, tyrosine kinase activity, and involvement of FA-1. Mol. Reprod. Dev. 1994;39:397–408. doi: 10.1002/mrd.1080390408. [DOI] [PubMed] [Google Scholar]
- Naz RK, Chauhan SC. Human sperm-specific peptide vaccine that causes long term reversible contraception. Biol. Reprod. 2002;67:674–680. doi: 10.1095/biolreprod67.2.674. [DOI] [PubMed] [Google Scholar]
- Naz RK, Zhu X, Kadam AL. Identification of human sperm peptide sequence involved in egg binding for immunocontraception. Biol. Reprod. 2000;62:318–324. doi: 10.1095/biolreprod62.2.318. [DOI] [PubMed] [Google Scholar]
- Parmentier M, Libert F, Schurmans S, Schiffmann S, Lefort A, Eggerickx D, Ledent C, Mollereau C, Gerard C, Perret J, et al. Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature. 1992;355:453–455. doi: 10.1038/355453a0. [DOI] [PubMed] [Google Scholar]
- Sakai N, Sawada MT, Sawada H. Non-traditional roles of ubiquitin-proteasome system in fertilization and gametogenesis. Int. J. Biochem. Cell Biol. 2004;36:776–784. doi: 10.1016/S1357-2725(03)00263-2. [DOI] [PubMed] [Google Scholar]
- Spehr M, Hatt H. hOR17-4 as a potential therapeutic target. Drug News Perspect. 2004;17:165–171. doi: 10.1358/dnp.2004.17.3.829014. [DOI] [PubMed] [Google Scholar]
- Suter B, Kittanakom S, Stagljar I. Two-hybrid technologies in proteomics research. Curr. Opin. Biotechnol. 2008;19:316–323. doi: 10.1016/j.copbio.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Manadhar G, McCauley TC, Caamano JN, Sutovsky M, Thompson WE, Day BN. Proteasomal interference prevents zona pellucida penetration and fertilization in mammals. Biol. Reprod. 2004;71:1625–1637. doi: 10.1095/biolreprod.104.032532. [DOI] [PubMed] [Google Scholar]
- Tulsiani DRP, Zeng H, Abou-Haila A. Biology of sperm capacitation: evidence for multiple signaling pathways. In: Gupta SK, Koyama K, Murray FF, editors. Gamete Biology: Emerging Frontiers in Fertility and Contraceptive Development. Nottingham University Press: Nottingham, United Kingdom; 2006. pp. 257–272. [Google Scholar]
- Vanderhaeghen P, Schurmans S, Vassart G, Parmentier M. Olfactory receptors are displayed on dog mature sperm cells. J Cell Biol. 1993;123:1441–1452. doi: 10.1083/jcb.123.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB. Olfaction: attracting both sperm and the nose. Curr. Biol. 2004;14:918–920. doi: 10.1016/j.cub.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Xu B, Hao Z, Jha JN, Digilio L, Urekar C, Kim YH, Pulido S, Flickinger CJ, Herr JC. Validation of a testis specific serine/threonine kinase [TSSK] family and the substrate of TSSK1 &2, TSKS, as contraceptive targets. In: Gupta SK, Koyama K, Murray FF, editors. Gamete Biology: Emerging Frontiers in Fertility and Contraceptive Development. Nottingham, United Kingdom: Nottingham University Press; 2006. pp. 87–101. [Google Scholar]
- Yagi T. Clustered protocadherin family. Dev. Growth Differ. 2008;50:S131–S140. doi: 10.1111/j.1440-169X.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- Yamamura R, Nishimura N, Nakatsuji H, Arase S, Sasaki T. The interaction of JRAB/MIKAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol. Biol. Cell. 2007;19:971–983. doi: 10.1091/mbc.E07-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi R. Mammalian fertilization. In: Knobil E, Neil JD, editors. Physiology of Reproduction. New York: Raven Press; 1994. pp. 189–317. [Google Scholar]