Abstract
Inward-rectifier potassium (K+) channels conduct K+ ions most efficiently in one direction, into the cell. Kir2 channels control the resting membrane voltage in many electrically excitable cells and heritable mutations cause periodic paralysis and cardiac arrhythmia. We present the crystal structure of Kir2.2 from chicken, which, excluding the unstructured amino and carboxyl termini, is 90% identical to human Kir2.2. Crystals containing rubidium (Rb+), strontium (Sr2+), and europium (Eu3+) reveal binding sites along the ion conduction pathway that are both conductive and inhibitory. The sites correlate with extensive electrophysiological data and provide a structural basis for understanding rectification. The channel’s extracellular surface, with large structured turrets and an unusual selectivity filter entryway, might explain the relative insensitivity of eukaryotic inward rectifiers to toxins. These same surface features also suggest a possible approach to the development of inhibitory agents specific to each member of the inward-rectifier K+ channel family.
In 1949 Bernard Katz introduced the term “anomalous rectification” to distinguish the K+ currents he observed in frog skeletal muscle from the “delayed rectification” K+ currents of the squid axon action potential (1, 2). Today we know that “delayed rectifiers” are a subset of the large family of voltage-dependent K+ (Kv) channels, whereas “anomalous rectifiers” are members of a different family of channels more commonly known as inward-rectifier K+ (Kir) channels (3). The name “inward rectifier” refers to a fundamental ion-conduction property exhibited to a greater or lesser degree by all members of the family: Given an equal but opposite electrochemical driving force, K+ conductance into the cell far exceeds conductance out of the cell. Thus, Kir channels are analogous to one-way conductors, or diodes, in solid-state electronic devices.
Electrophysiological experiments have shown that inward rectification is a consequence of voltage-dependent pore blockage by intracellular multivalent cations, especially Mg2+ and polyamines (4–8). At internal negative (hyperpolarizing) membrane voltages the blocking ions are cleared from the pore so that K+ conducts, whereas at internal positive (depolarizing) membrane voltages the blocking ions are driven into the pore from the cytoplasm so that K+ conduction is blocked. As a result, Kir channels are conductive when an excitable cell is at rest and non-conductive during excitation. This property is thought to foster energy efficiency because it enables Kir channels to regulate the resting membrane potential, but not dissipate the K+ gradient during an action potential (3).
A central mechanistic question is why are Kir channels blocked by intracellular multivalent cations? Mutational studies have identified several amino acids that confer sensitivity to blocking ions (9–19), but a structural description of these sites has remained elusive. Structures of prokaryotic Kir channels, because of their low sequence similarity to eukaryotic Kir channels, do not contain the specific amino acids that are known to underlie blockage and rectification (20, 21).
Another longstanding puzzle in eukaryotic Kir channel studies is their relative insensitivity to natural toxins that typically inhibit other K+ channels (22–24). Snake, spider and scorpion venoms, for example, contain numerous toxins against various Kv channels and Ca2+-activated K+ channels (25–27). By contrast, Kir channel toxins are rare, and no specific toxins against Kir2 channels have been discovered.
Eukaryotic Kir channels as a molecular family
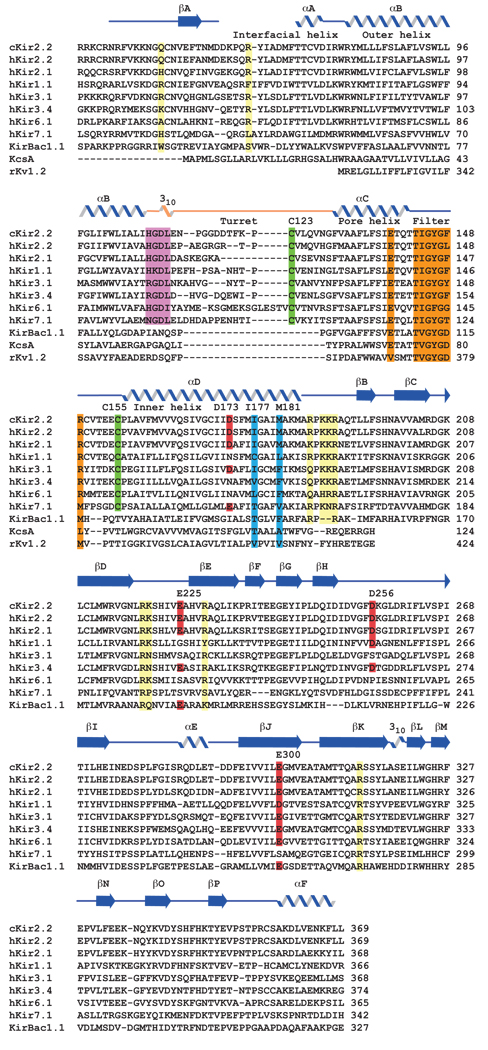
The eukaryotic Kir channels contain several amino acid sequence motifs and conserved amino acids that are essential to their functional properties (Fig. 1). For example, in most other K+ channels the selectivity filter comprises the “canonical” filter sequence TXGYGDX, where X represents an aliphatic amino acid (Fig. 1). The corresponding sequence in eukaryotic Kir channels is TXGYGFR, with F sometimes replaced by another amino acid. In light of the structural importance of DX in the canonical sequence, the amino acids FR signify a marked variation on the filter sequence. Eukaryotic Kir channels also contain an absolutely conserved pair of cysteine residues flanking the pore region, which is the reentrant peptide segment that forms the pore-helix and selectivity filter of K+ channels. Between the outer helix (the first transmembrane segment) and pore region the “turret”, though varied among inward rectifiers, contains the sequence HGDL that could be considered a signature of eukaryotic Kir channels. Finally, through extensive studies combining electrophysiology and mutagenesis several acidic amino acids (D and E) are known to be critical to inward rectification (9–19), and motifs containing basic amino acids (e.g. PKKR) are critical to phosphatidylinositol 4,5-biphosphate (PIP2) activation of Kir channels (28–35). These positions are highlighted on the sequences in Fig. 1.
Fig. 1.
Key residues in eukaryotic Kir channels. Sequence alignment of chicken Kir2.2 (GI:118097849), human Kir2.2 (GI:23110982), human Kir2.1 (GI:8132301), human Kir1.1 (GI:1352479), human Kir3.1 (GI:1352482), human Kir3.4 (GI:1352484), human Kir6.1 (GI:2493600), human Kir7.1 (GI:3150184), KirBac1.1 (GI:33357898), KcsA (GI:39654804), and rat Kv1.2 (GI:73536156). For all the Kir sequences only the core region corresponding to the expressed protein and atomic structure of Kir2.2 is included in the alignment. For Kv1.2 only the transmembrane pore region is shown. Secondary structure elements are indicated above the sequences and the turret is colored orange. Residues discussed in the text are highlighted in red (acidic residues), green (two disulfide-bonded cysteines), cyan (the inner helix bundle activation gate), purple (conserved residues among the turrets of eukaryotic Kir channels), orange (the selectivity filter and E139), and yellow (critical residues for channel-PIP2 interactions). Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
The Kir2.2 channel from chicken is 90% identical to the human ortholog (excluding the N and C termini) and contains all of the sequence characteristics of a strong inward rectifier (36). Figure S1 shows that the chicken Kir2.2 channel expressed in Xenopus oocytes indeed functions as a strong rectifier. In oocyte two-electrode voltage-clamp recordings with 98 mM KCl in the bath solution inward currents are much larger than outward currents (fig. S1B). In on-cell and excised gigaseal patch recordings channel activity is observed at hyperpolarizing (negative internal) membrane voltages but not at depolarizing (positive internal) voltages (fig. S1C). The single-channel conductance measured near −80 mV is ~40 pS, which is very similar to the values reported for the guinea pig and mouse Kir2.2 channels (37, 38) (fig. S1D, inset). The sharp transition between channel conductance and nonconductance as a function of membrane voltage is characteristic of a strong rectifier (36). Upon patch excision from the oocyte surface some outward current is observed at voltages slightly positive to the reversal potential because the concentration of intracellular blockers is decreased (fig. S1C, blue trace). However, the current still decreases with further depolarization (negative conductance) as channels become blocked in a voltage-dependent manner; this behavior reflects the inherent difficulty in washing away trace yet still active concentrations of polyamine molecules due to their very high affinity for the pore in strong rectifiers (39, 40). Several minutes after patch excision the currents decrease (fig. S1C, red trace). This “run-down” reflects altered channel regulation mediated by kinases, phosphatases and lipid signaling (34, 36, 41, 42).
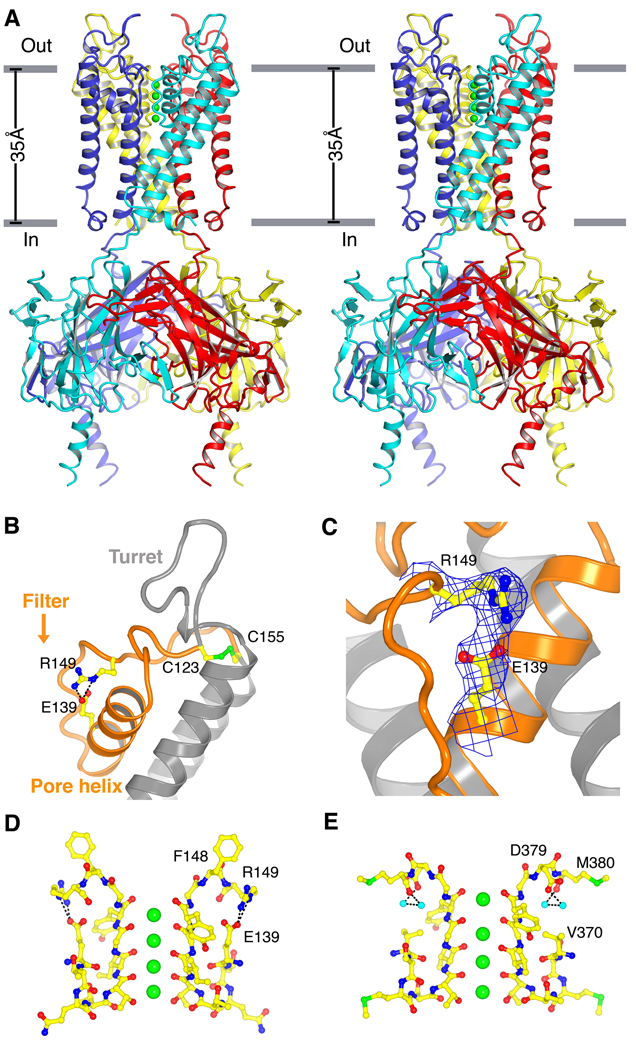
To obtain diffracting crystals, we removed the intrinsically disordered N- and C-terminal regions. The electrophysiological recordings shown in fig. S1 were made using a similar construct with N- and C-terminal truncations, confirming that the crystal structure corresponds to a functional channel unit with strong rectifying properties. The Kir2.2 model, consisting of the cytoplasmic domain and transmembrane channel, was refined at 3.1 Å to a free R-factor of 0.27. A ribbon diagram in stereo shows the transmembrane pore (above) and the cytoplasmic pore (below) (Fig. 2A). Lateral openings between the transmembrane and cytoplasmic pores, at the level of the lipid membrane headgroup layer, contain many arginine and lysine residues. The high density of positive charges makes it unlikely that K+ ions would pass through these openings (fig. S2). The structure is therefore consistent with mutagenesis studies, which support the conclusion that the ion pathway extends across the full length of the transmembrane and cytoplasmic pores (9–19). The overall architecture is similar to that of prokaryotic Kir channels but with a notable difference: The Kir2.2 channel contains prominent, highly structured turrets on the extracellular face of the channel. These surround as if to protect the pore entryway.
Fig. 2.
Structure of Kir2.2. (A) Stereoview of a ribbon representation of the Kir2.2 tetramer from the side with the extracellular solution above. Four subunits of the channel are uniquely colored. Approximate boundaries of the lipid bilayer are shown as gray bars. (B) A close-up view of the pore-region of a single subunit (in ribbon representation) with the turret, pore helix and selectivity filter labeled. Side chains of residues E139, R149 and a pair of disulfide-bonded cysteines (C123 and C155) are shown as sticks and colored according to atom type: carbon, yellow; nitrogen, blue; oxygen, red; and sulfur, green. Ionized hydrogen bonds are indicated by dashed black lines. The region flanked by the two disulfide-bonded cysteines is colored orange. (C) Electron density (blue wire mesh, 2Fo-Fc, calculated from 50 to 3.1Å using phases from the final model and contoured at 1.0 σ) is shown for the side chains of E139 and R149 [sticks, colored the same scheme as in (B)] forming a salt bridge. (D and E) K+ selectivity filter of the Kir2.2 channel (D) compared with that of the Kv1.2-Kv2.1 paddle chimera channel [(E), PDB ID 2R9R]. For clarity, only two of the four subunits [sticks, colored with the same scheme as in (B)] are shown. K+ (green spheres), water molecules (cyan spheres), and hydrogen bonds between R149 and E139 (Kir, dashed black lines), or between D379, M380 and waters (Kv, dashed black lines) are shown.
The selectivity filter
At a detailed structural level Kir2.2 is quite different from prokaryotic Kir channels owing to minimal (<20%) sequence conservation. The cysteine pair that is absolutely conserved among eukaryotic Kir channels creates a circularized pore region through covalent linkage of the segment preceding the pore helix (C123) to the segment following the selectivity filter (C155) (Fig. 2B). The existence of a disulfide bond was correctly predicted on the basis of mutagenesis studies: Mutation of the corresponding cysteines in Kir2.1 led to the absence of currents even though expressed protein was detectable by Western blot analysis (43, 44). Application of 10 mM dithiothreitol (DTT) or reduced glutathione to the outside of cells expressing the wild-type channels did not affect currents. From these two observations it was concluded that a disulfide bridge must be essential for proper folding, but apparently not for function (43, 44). The structure provides an alternative interpretation. The disulfide bridge is buried beneath the protein surface at the level of the membrane interface. Furthermore, the Kir2.2 channel was purified and crystallized in the presence of 20 mM DTT and 3 mM TCEP [(tris(2-carboxyethyl)phosphine)], and yet the disulfide bridge remained intact. It is therefore possible that the disulfide bridge remains intact upon exposure to moderate concentrations of DTT and that the bridge may be important for channel function.
The pore region is further stapled together by an ionized hydrogen bond between R149 in the filter sequence TXGYGFR and E139 (Fig. 2, B and C). The Glu O-ε to Arg N-η distance is 2.4 Å, compatible with an energetically strong interaction. Mutations altering this interaction are known to alter channel function (45, 46). On the basis of studies with concatenated subunits the salt bridge was thought to be inter-subunit, but the crystal structure shows that this interaction ties together two segments of the pore-region within a single subunit (46).
Despite the presence of substantially different protein contacts surrounding the selectivity filter, the main-chain structure of the filter in Kir2.2 is the same as in other K+ channels (47). For example, the main chain root mean square deviation between Kv1.2 and Kir2.2 is 0.4 Å, which is within the margin of certainty to discriminate atomic positions with 3.1 Å diffraction data (Fig. 2, D and E) (20, 48, 49). One structural difference near the filter could possibly account for important pharmacological differences between Kir and other K+ channels. In the canonical filter sequence the Asp (D) residue in the filter sequence is buried, creating a flat surface surrounding the filter opening. By contrast, in Kir channels the Phe (F) residue at the corresponding position projects directly into aqueous solution, creating four protrusions on the perimeter where the filter opens to the extracellular solution.
The cavity and gates
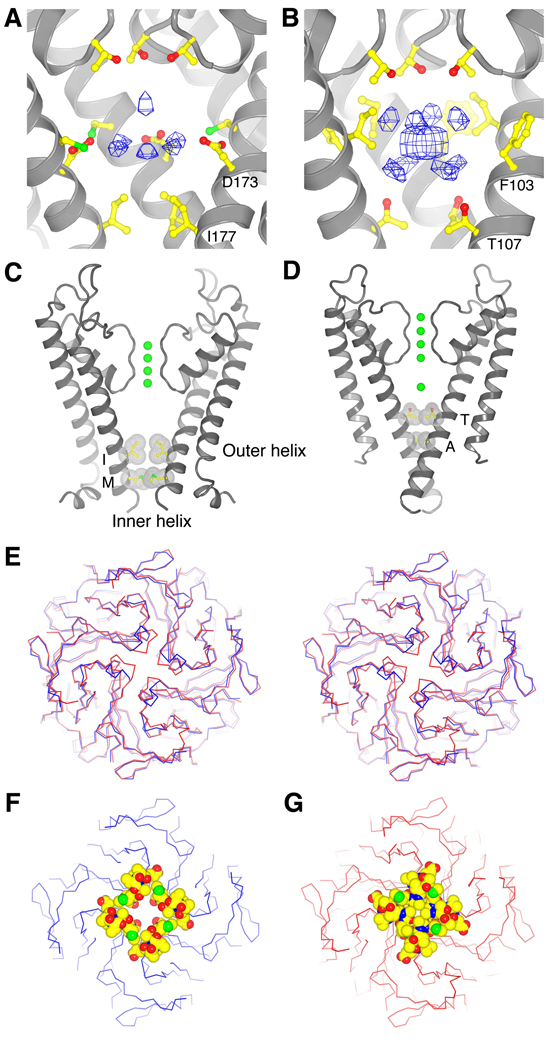
The pore lining on the intracellular side of the selectivity filter is mainly hydrophobic in nearly all K+ channels. Eukaryotic Kir channels are an exception in which the central region of the pore – known as the central cavity – contains four polar amino acids (one from each subunit) projecting toward the ion pathway (Fig. 3. A and B). In Kir2.2 and other strong rectifiers these polar amino acids are Asp (D173), whereas in weak rectifiers such as Kir1.1 and Kir6.1 they are Asn (Fig. 1). On the basis of electrophysiological studies, Asp residues in the central cavity of strong rectifiers are hypothesized to influence the affinity of Mg2+ and polyamines by an electrostatic mechanism (12, 18).
Fig. 3.
The cavity and gates. (A and B) Electron density in the cavity of the Kir2.2 channel [(A), Fo-Fc omit map, calculated from 50 to 3.1Å using phases from the final model and contoured at 2.0 σ] and of the KcsA channel [(B), PDB ID 1K4C, Fo-Fc omit map, calculated from 50 to 3.1Å using phases from the final model and contoured at 2.8 σ]. The channels are shown as ribbon representations with the subunit closest to the viewer removed. Only the side chains facing the cavity are shown (sticks). (C and D) Comparison of the transmembrane inner helix bundle activation gate of Kir2.2 (C) with the KcsA structure [(D), PDB ID 1K4C]. For clarity, only two of the four subunits (gray ribbon) are shown. Side chains of the residues in the bundle crossing are shown as sticks [colored with the same scheme as in (B)] and van der Waals surfaces (gray dots). K+ ions are shown as green spheres. Inner and outer helices are indicated. (E) Superposition of the chicken Kir2.2 cytoplasmic domain (blue α-carbon trace) and the mouse Kir2.1 cytoplasmic domain (red α-carbon trace, PDB ID 1U4F) in stereo viewed from the extracellular side. (F and G) Comparison of the apex (G loop) of the cytoplasmic pores of Kir2.2 (F) and mouse Kir2.1 (G), with the same view as (E). The cytoplasmic domains are shown as α-carbon traces, with residues 303 to 309 (Kir2.2) and 302 to 308 (Kir2.1) shown as CPK models (carbon, yellow; nitrogen, blue; oxygen, red; and sulfur, green).
Beneath the central cavity, residues I177 and M181 on the inner helices form two hydrophobic seals that close off the pore leading to the cytoplasm (Fig. 3C). Kir2.2 is therefore physically shut at the “activation gate” (50). Amino acids corresponding to positions 177 and 181 are also large and hydrophobic in most other eukaryotic Kir channels, but not in many other K+ channels (Fig. 1). For example, in KcsA, Kv channels and prokaryotic Kir channels, the position corresponding to 177 usually contains a small and sometimes polar amino acid, typically Val or Thr. In KcsA both seal positions contain small amino acids (Fig. 3D). Because of the large hydrophobic residues at positions 177 and 181, the inner helices of Kir2.2 do not come as close together in the closed conformation as in KcsA (Fig. 3, C and D).
Figure 3E shows the cytoplasmic domain tetramer from the Kir2.2 channel superimposed onto the domain from Kir2.1, which was solved by crystallography in the absence of a transmembrane channel (11). Over most of the domain these structures are nearly identical. This observation supports the expectation (based on 80% sequence identity) that Kir2.2 should represent an excellent model for the complete Kir2.1 channel. In addition to the activation gate formed by the transmembrane inner helices, Kir channels have been proposed to have a second gate (G loop) at the apex of the cytoplasmic domain tetramer (11, 51). The G loop is physically open in Kir2.2 and closed in the Kir2.1 domain (Fig. 3, F and G). The differences in conformation are due to local movements of the G loop rather than to rigid body motions of the cytoplasmic domains. Local G loop movements contrast observations on the cytoplasmic domain of Kir3.1, in which the G loop opening appears associated with rigid body movements of domains in the tetramer (20).
Ion binding sites for conduction and inward rectification
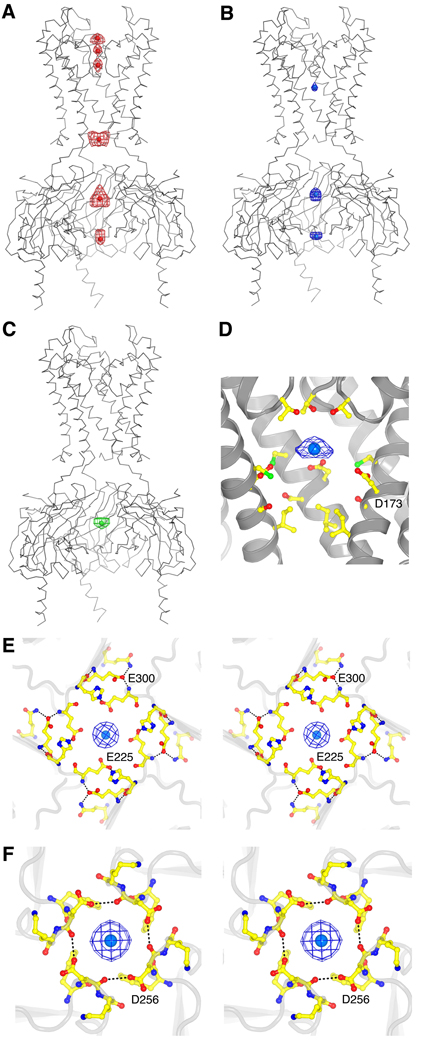
Figure 4, A to F shows the locations of ions in difference Fourier maps from crystals containing Rb+, Sr2+, and Eu3+. Rb+ is a K+ analog that conducts curret. Density for this ion is observed at multiple sites in the selectivity filter and at three positions within the pore on the intracellular side of the selectivity filter, but is absent in the central cavity (Fig. 4A). The three occupied intracellular positions are as follows: immediately internal to the activation gate in the transmembrane pore, in the cytoplasmic pore internal to the G loop, and at the entryway to the cytoplasmic pore. We refer to the two sites in the cytoplasmic pore as the upper and lower rings of charges, respectively (fig. S3). The presence of multiple sites along the pore occupied by conducting ions is a prerequisite for strong voltage-dependent block by intracellular cations that cannot pass through the selectivity filter (12, 52–57).
Fig. 4.
Ion binding sites. (A to C) Electron density (wire mesh) of Rb+ [(A), Fo-Fc map calculated to 4.0 Å, contoured at 3.5 σ for density in the filter and 2.0 σ for density elsewhere], Sr2+ [(B), 10 mM, Fo-Fc map calculated to 3.3 Å, contoured at 1.5 σ for density in the cavity and 3.0 σ for density elsewhere] and Eu3+ [(C), 10 mM, anomalous difference map calculated to 6.0 Å, contoured at 2.8 σ] inside the Kir2.2 channel ion conduction pathway. Kir2.2 is represented as a gray α-carbon trace with the transmembrane domain and cytoplasmic domain closest to the viewer removed for clarity. The ions are shown as spheres and colored red (Rb+), blue (Sr2+), and green (Eu3+). (D) Electron density (200 mM Sr2+, Fo-Fc map calculated from 50 to 3.8 Å, contoured at 2.5 σ, blue wire mesh) of Sr2+ (blue-green spheres) in the cavity of Kir2.2. The channel is shown as a ribbon with the subunit closest to the viewer removed. Only the side chains facing the cavity are shown (sticks). (E) Stereoview of the ion binding site near the upper ring of charges in the cytoplasmic domain of Kir2.2, viewed from the extracellular side. Residues E225, H227, E300, and Q311 are shown as sticks, and hydrogen bonds between them are indicated as dashed black lines. Electron density (200 mM Sr2+, Fo-Fc map calculated from 50 to 3.8 Å, contoured at 4.5 σ) of Sr2+ (blue-green spheres) is shown as blue wire mesh. (F) Stereoview of the ion binding site at the lower ring of charges in the cytoplasmic domain of Kir2.2, viewed from the intracellular side. Residues F255, D256, and K257 are shown as sticks, and hydrogen bonds between D256 from different subunits are indicated as dashed black lines. Electron density (200 mM Sr2+, Fo-Fc map calculated from 50 to 3.8 Å, contoured at 4.5 σ) of Sr2+ (blue-green spheres) is shown as blue wire mesh.
Crystals of Kir2.2 were grown in the presence of 650 mM Rb+ and yet electron density for Rb+ is not observed in the cavity (Fig. 4A). This finding is noteworthy because under similar conditions a strong monovalent cation peak is observed in the cavity of KcsA (47, 58). Native crystals of Kir2.2, grown in the presence of 150mM K+ and 500mM Na+, show a weak electron density peak at the cavity center with additional peaks on the perimeter, apparently bridging toward the D173 side chain (Fig. 3A). We cannot discern whether these peaks represent a disordered ion, multiple ions, or a low occupancy K+ (or Na+) in the center, perhaps surrounded by water molecules hydrogen bonded to the Asp carboxylate. We can conclude, however, that the central cavity in Kir2.2, at least in the closed conformation, has cation attractive properties that are different from those of KcsA.
The divalent cation Sr2+ should behave as an electron dense mimic of Mg2+, a biologically important metal ion inhibitor of eukaryotic Kir channels (7, 8). In Fo-Fc Fourier maps from crystals with 10 mM Sr2+, 500 mM Na+ and 150 mM K+, density peaks due to Sr2+ are observed at three sites inside the pore intracellular to the selectivity filter: in the cavity, at the upper ring and at the lower ring of charges (fig. S3 and Fig. 4B). The magnitude of the Sr2+ peak is small in the cavity (3.4 σ) compared to the peaks at the upper (9.6σ) and lower (7.2 σ) rings of charges. Separate experiments with crystals containing 200 mM Sr2+ support the notion that the weak cavity peak is indeed due to Sr2+, which is present apparently at relatively low occupancy. Detailed views of these sites are shown (Fig. 4, D to F). They each consist of planar rings of acidic amino acids arranged on the pore’s perimeter. All three sites exhibit a preference for Sr2+: 10 mM Sr2+ outcompetes 150 mM K+. This selectivity is likely to be electrostatic in origin. The sites are too wide (10.5, 8.9, and 9.3 Å diameter for the cavity, upper and lower ring of charges) to mediate direct coordination of an ion at the center. Presumably ions at the center of these sites interact through bridging water molecules. Because each site has the potential to contain multiple negatively charged carboxyl groups, the resulting strong electric field is expected to create a good match for a multivalent cation. Crystals containing the lanthanide Eu3+, which we assume to be trivalent (59), provide support for this hypothesis. An anomalous difference Fourier map shows that Eu3+ binds at only one site, the upper ring of charges (Fig. 4C). This site appears to be more electronegative than the others because it contains two concentric rings of acidic amino acids, E225 and E300.
Mutagenesis studies have identified several amino acids that, when mutated, affect the affinity of Mg2+ and polyamines in strong rectifiers. D173 in the cavity, E225 and E300 forming the upper ring of charges, and D256 forming the lower ring of charges are among those known to be important (9–19). The weak Sr2+ peak in the cavity might seem incompatible with the large influence that mutations of the cavity Asp (D173) have on Mg2+ affinity. However, the channel in the crystal is not in an applied electric field: In an electric field imposed by a depolarized (positive inside) membrane we expect that the distribution of blocker occupancies among the multiple sites will change. Specifically, we expect the blocking cations to be driven deeper into the pore toward the cavity. In correlating the crystallographic with electrophysiological data, it is most notable that the amino acids forming the Sr2+ sites in the crystal are the same amino acids that are known to affect blockage and rectification in electrophysiology experiments (36). Beyond providing a structural basis with which to explain past electrophysiological studies, the Kir2.2 structure also suggests many new experiments. For example, most studies on the mechanism of rectification have focused on electrostatic interactions between the positively charged blocker and negatively charged groups on the protein. But hydrophobic interactions between methylene groups of polyamine molecules and hydrophobic residues in the channel may be important. In particular, we might anticipate that when the pore opens polyamines could interact strongly with the large hydrophobic amino acids at positions 177 and 181 when the leading amino group of the polyamine reaches into the central cavity (Fig. 3C) (54).
Since the earliest investigations of strong inward rectifiers two important properties have been noted: a sharp transition from a conductive state to a non-conductive (blocked) state over a very narrow voltage range, and a dependence of the transition on the extracellular K+ concentration (60–63). Specifically, the voltage at which the transition occurs shifts to more depolarizing values as extracellular K+ concentration is increased. Both properties, the sharp transition (i.e. strong voltage dependence) and its dependence on extracellular K+, have been attributed to the simple notion that conducting ions and blocking ions compete for sites in the pore (12, 52–57, 64–66). The crystallographic data presented here support this conclusion. We observe in the crystal Rb+ binding at the same sites that can bind multivalent blocking ions. Therefore a high extracellular K+ (or Rb+) concentration should favor occupation of the sites by conducting ions, and a more depolarizing voltage should be required to drive blocking ions into the pore from the cytoplasm to replace the conducting ions. Moreover, as blocking ions enter the pore from the intracellular side, the displaced conducting ions must move through the selectivity filter to the extracellular side; that is, movements of blocking and conducting ions must be coupled. Such coupling would have energetic consequences because movement of an ion across the membrane voltage difference constitutes work. Hence, a blocking ion entering the pore will exhibit a voltage dependence that results from a combination of its own charge and the charge of the displaced ions. This can be the origin of strong voltage-dependent block, which can be the origin of a biologically important property of strong rectifiers – their diode property of a sharp transition from a conductive to a nonconductive state as a function of membrane voltage (12, 52–55, 64).
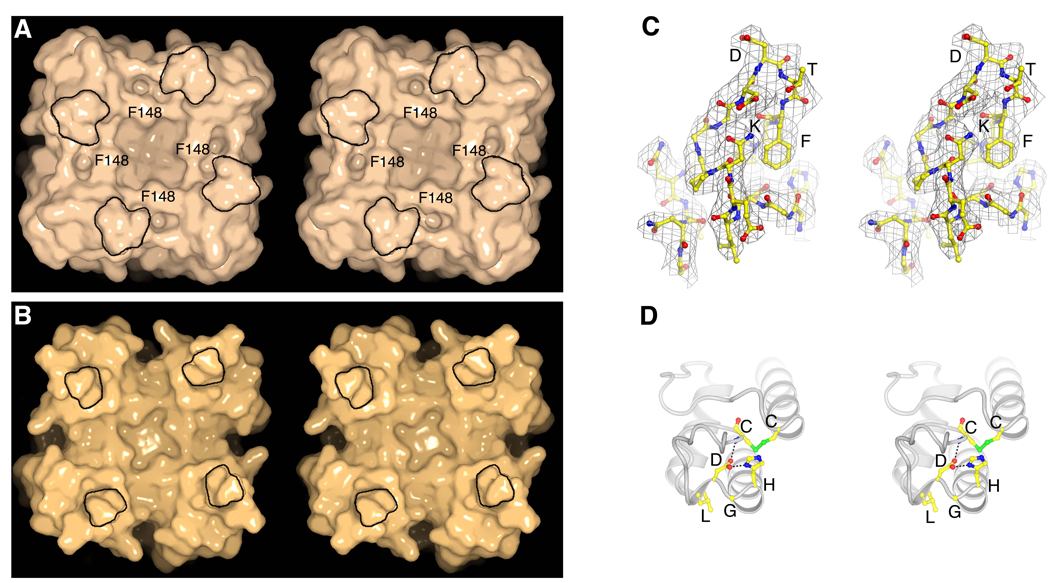
The extracellular pore entryway and pharmacology of Kir channels
Two aspects of the structure may account for the relative insensitivity of eukaryotic Kir channels, especially members of the Kir2 subfamily, to K+ channel toxins (22–24). The turrets in Kir2.2 are larger and come closer together, constricting the pore entryway compared to Kv1.2; and F148 in the sequence TXGYGFR creates four protrusions on the surface at the pore opening (Fig. 5, A and B). Thus, in Kv channels the entryway is wider and the pore opens onto intersecting grooves with a flat base, which form the docking surface for pore-blocking scorpion toxins (Fig. 5B). In Kir2.2 the entryway is constricted and the grooves are absent (Fig. 5A).
Fig. 5.
Unique structure of the extracellular entryway. (A and B) Surface representation of chicken Kir2.2 (A) and Kv1.2–Kv2.1 paddle chimera [(B), PDB ID 2R9R] in stereo, viewed from the extracellular side. The four protrusions formed by the top of the turrets are highlighted with a black perimeter and F148 in Kir2.2 is labeled. (C) Stereo representation of electron density (gray wire mesh) for the turret region (2Fo-Fc, calculated from 50 to 3.1Å using phases from the final model and contoured at 1.0 σ). The turret is shown as sticks (colored according to atom types), and residues corresponding to the highlighted protrusions in (A) are labeled. (D) A close-up view of the turret region in a single subunit in stereo. Side chains of those conserved residues among the turrets of eukaryotic Kir channels, as well as C155 are shown as sticks. Hydrogen bonds between H108, D110 and C123 are indicated as dashed black lines.
Though the shape of the eukaryotic Kir channel pore entryway might offer fewer opportunities for inhibitory protein-protein interactions, inhibition might occur by a somewhat different strategy. Inhibitors of Kir1.1 and Kir3.4 channels have been identified. A bee venom toxin, tertiapin, inhibits both of these channels (22). At 21 amino acids in length, tertiapin is smaller than most other venom toxins so it might fit between the turrets more effectively. Alternatively, the turrets themselves might form the binding site for tertiapin (67–69). At 57 amino acids δ-dendrotoxin from the green mamba snake is rather large and yet it inhibits Kir1.1 channels (23). Compared to tertiapin less is known about the binding site on the channel for δ-dendrotoxin, but one aspect of its inhibition is intriguing: The blocked state reduces single-channel conductance to about 10% rather than inhibiting it completely. δ-Dendrotoxin most likely binds to the turrets but is too large to fit tightly over the pore, which would imply that binding to the turret may be sufficient to alter the channel’s function.
The idea that binding to the turrets could alter function is not surprising when one considers that the turret in Kir2.2 is not a loop, but forms a highly ordered structure (Fig. 5C). The base of the turret is formed and pinned together by the HGDL sequence, which with only minor variation is found in all eukaryotic Kir channels (Fig. 1 and Fig 5D). H108 stabilizes D110 through a hydrogen bond. The Asp (D) itself is hydrogen bonded to the amide nitrogen of C123, which effectively holds the two ends of the turret together. L111 projects from the surface of a short 310 helix into the protein interior to make stabilizing hydrophobic interactions. Thus, the turrets are structurally important elements of the channel. Between the sequence HGDL and the first Cys of the disulfide bridge the turret sequence is highly variable among Kir channel subtypes. The Kir2.1 channel becomes sensitive to tertiapin if the variable sequence is mutated to be Kir3.4-like (68). Therefore, the turrets appear to be structures through which specific inhibition of Kir channel subtypes might be achievable through directed evolution of specific protein binding partners.
Summary
This paper presents the atomic structure of a eukaryotic Kir channel, Kir2.2, a strong inward rectifier. The sequence TXGYGFR gives rise to a K+ selectivity filter stabilized by disulfide bridges and salt bridges that distinguish eukaryotic Kir channels. Multiple ion binding sites on the intracellular side of the selectivity filter can be occupied by conducting ions but exhibit higher affinity for multivalent blocking ions. Thus, blocking ions entering from the cytoplasm must displace conducting ions through the pore. This situation is expected to give rise to strong voltage-dependent block and diode-like conduction properties. Structural features of the extracellular pore entryway offer an explanation for the relative insensitivity of Kir channels to venomous toxins and a possible approach to the development of selective Kir channel inhibitors.
Supplementary Material
Footnotes
Supporting Online Material
Materials and Methods
Figs. S1 to S3
Table S1
References
References and Notes
- 1.Hodgkin AL, Huxley AF, Katz B. Arch. Sci. Physiol. (Paris) 1949;3:129. [Google Scholar]
- 2.Katz B. Arch. Sci. Physiol. (Paris) 1949;3:285. [Google Scholar]
- 3.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 4.Fakler B, et al. Cell. 1995 Jan 13;80:149. doi: 10.1016/0092-8674(95)90459-x. [DOI] [PubMed] [Google Scholar]
- 5.Lopatin AN, Makhina EN, Nichols CG. Nature. 1994 Nov 24;372:366. doi: 10.1038/372366a0. [DOI] [PubMed] [Google Scholar]
- 6.Horie M, Irisawa H, Noma A. J Physiol. 1987 Jun;387:251. doi: 10.1113/jphysiol.1987.sp016572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda H, Saigusa A, Irisawa H. Nature. 1987 Jan 8–14;325:156. doi: 10.1038/325156a0. [DOI] [PubMed] [Google Scholar]
- 8.Vandenberg CA. Proc Natl Acad Sci U S A. 1987 Apr;84:2560. doi: 10.1073/pnas.84.8.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurata HT, Cheng WW, Arrabit C, Slesinger PA, Nichols CG. J Gen Physiol. 2007 Aug;130:145. doi: 10.1085/jgp.200709742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara Y, Kubo Y. J Gen Physiol. 2006 Apr;127:401. doi: 10.1085/jgp.200509434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pegan S, et al. Nat Neurosci. 2005 Mar;8:279. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 12.Guo D, Lu Z., a J Gen Physiol. 2003 Nov;122:485. doi: 10.1085/jgp.200308890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubo Y, Murata Y. J Physiol. 2001 Mar 15;531:645. doi: 10.1111/j.1469-7793.2001.0645h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Jan YN, Jan LY. Neuron. 1995 May;14:1047. doi: 10.1016/0896-6273(95)90343-7. [DOI] [PubMed] [Google Scholar]
- 15.Taglialatela M, Ficker E, Wible BA, Brown AM. EMBO J. 1995 Nov 15;14:5532. doi: 10.1002/j.1460-2075.1995.tb00240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wible BA, Taglialatela M, Ficker E, Brown AM. Nature. 1994 Sep 15;371:246. doi: 10.1038/371246a0. [DOI] [PubMed] [Google Scholar]
- 17.Stanfield PR, et al. J Physiol. 1994 Jul 1;478(Pt 1):1. doi: 10.1113/jphysiol.1994.sp020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Z, MacKinnon R. Nature. 1994 Sep 15;371:243. doi: 10.1038/371243a0. [DOI] [PubMed] [Google Scholar]
- 19.Fakler B, et al. FEBS Lett. 1994 Dec 19;356:199. doi: 10.1016/0014-5793(94)01258-x. [DOI] [PubMed] [Google Scholar]
- 20.Nishida M, Cadene M, Chait BT, MacKinnon R. EMBO J. 2007 Sep 5;26:4005. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo A, et al. Science. 2003 Jun 20;300:1922. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 22.Jin W, Lu Z. Biochemistry. 1998 Sep 22;37:13291. doi: 10.1021/bi981178p. [DOI] [PubMed] [Google Scholar]
- 23.Imredy JP, Chen C, MacKinnon R. Biochemistry. 1998 Oct 20;37:14867. doi: 10.1021/bi980929k. [DOI] [PubMed] [Google Scholar]
- 24.Lu Z, MacKinnon R. Biochemistry. 1997 Jun 10;36:6936. doi: 10.1021/bi9702849. [DOI] [PubMed] [Google Scholar]
- 25.Harvey AL, Robertson B. Curr Med Chem. 2004 Dec;11:3065. doi: 10.2174/0929867043363820. [DOI] [PubMed] [Google Scholar]
- 26.Swartz KJ, MacKinnon R. Neuron. 1995 Oct;15:941. doi: 10.1016/0896-6273(95)90184-1. [DOI] [PubMed] [Google Scholar]
- 27.Garcia ML, Hanner M, Knaus HG, Slaughter R, Kaczorowski GJ. Methods Enzymol. 1999;294:624. doi: 10.1016/s0076-6879(99)94035-1. [DOI] [PubMed] [Google Scholar]
- 28.Schulze D, Krauter T, Fritzenschaft H, Soom M, Baukrowitz T. J Biol Chem. 2003 Mar 21;278:10500. doi: 10.1074/jbc.M208413200. [DOI] [PubMed] [Google Scholar]
- 29.Zeng WZ, Liou HH, Krishna UM, Falck JR, Huang CL. Am J Physiol Renal Physiol. 2002 May;282:F826. doi: 10.1152/ajprenal.00300.2001. [DOI] [PubMed] [Google Scholar]
- 30.Lopes CM, et al. Neuron. 2002 Jun 13;34:933. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 31.Soom M, et al. FEBS Lett. 2001 Feb 9;490:49. doi: 10.1016/s0014-5793(01)02136-6. [DOI] [PubMed] [Google Scholar]
- 32.Shyng SL, Cukras CA, Harwood J, Nichols CG. J Gen Physiol. 2000 Nov;116:599. doi: 10.1085/jgp.116.5.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, He C, Yan X, Mirshahi T, Logothetis DE. Nat Cell Biol. 1999 Jul;1:183. doi: 10.1038/11103. [DOI] [PubMed] [Google Scholar]
- 34.Huang CL, Feng S, Hilgemann DW. Nature. 1998 Feb 19;391:803. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 35.Fan Z, Makielski JC. J Biol Chem. 1997 Feb 28;272:5388. doi: 10.1074/jbc.272.9.5388. [DOI] [PubMed] [Google Scholar]
- 36.Stanfield PR, Nakajima S, Nakajima Y. Rev.Physiol Biochem.Pharmacol. 2002;145:47. doi: 10.1007/BFb0116431. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi N, et al. J Biol Chem. 1994 Sep 16;269:23274. [PubMed] [Google Scholar]
- 38.Liu GX, et al. J Physiol. 2001 Apr 1;532:115. doi: 10.1111/j.1469-7793.2001.0115g.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Z. Annu Rev Physiol. 2004;66:103. doi: 10.1146/annurev.physiol.66.032102.150822. [DOI] [PubMed] [Google Scholar]
- 40.Nichols CG, Lopatin AN. Annu Rev Physiol. 1997;59:171. doi: 10.1146/annurev.physiol.59.1.171. [DOI] [PubMed] [Google Scholar]
- 41.Henry P, Pearson WL, Nichols CG. J Physiol. 1996 Sep 15;495(Pt 3):681. doi: 10.1113/jphysiol.1996.sp021625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fakler B, Brandle U, Glowatzki E, Zenner HP, Ruppersberg JP. Neuron. 1994 Dec;13:1413. doi: 10.1016/0896-6273(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 43.Cho HC, Tsushima RG, Nguyen TT, Guy HR, Backx PH. Biochemistry. 2000 Apr 25;39:4649. doi: 10.1021/bi992469g. [DOI] [PubMed] [Google Scholar]
- 44.Leyland ML, Dart C, Spencer PJ, Sutcliffe MJ, Stanfield PR. Pflugers Arch. 1999 Nov;438:778. doi: 10.1007/s004249900153. [DOI] [PubMed] [Google Scholar]
- 45.Dibb KM, et al. J Biol Chem. 2003 Dec 5;278:49537. doi: 10.1074/jbc.M307723200. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Yu M, Jan YN, Jan LY. Proc Natl Acad Sci U S A. 1997 Feb 18;94:1568. doi: 10.1073/pnas.94.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Nature. 2001 Nov 1;414:43. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 48.Long SB, Tao X, Campbell EB, MacKinnon R. Nature. 2007 Nov 15;450:376. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 49.Long SB, Campbell EB, Mackinnon R. Science. 2005 Aug 5;309:897. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 50.Jiang Y, et al. Nature. 2002 May 30;417:523. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 51.Pegan S, Arrabit C, Slesinger PA, Choe S. Biochemistry. 2006 Jul 18;45:8599. doi: 10.1021/bi060653d. [DOI] [PubMed] [Google Scholar]
- 52.Shin HG, Lu Z. J Gen Physiol. 2005 Apr;125:413. doi: 10.1085/jgp.200409242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin HG, Xu Y, Lu Z. J Gen Physiol. 2005 Aug;126:123. doi: 10.1085/jgp.200509296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo D, Ramu Y, Klem AM, Lu Z. J Gen Physiol. 2003 Apr;121:261. doi: 10.1085/jgp.200208771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo D, Lu Z. J Gen Physiol. 2000 Jun;115:799. doi: 10.1085/jgp.115.6.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spassova M, Lu Z. J Gen Physiol. 1998 Aug;112:211. doi: 10.1085/jgp.112.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson WL, Nichols CG. J Gen Physiol. 1998 Sep;112:351. doi: 10.1085/jgp.112.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y, MacKinnon R. Biochemistry. 2004 May 4;43:4978. doi: 10.1021/bi049876z. [DOI] [PubMed] [Google Scholar]
- 59.Cotton A, Wilkinson G. Wiley interscience [Google Scholar]
- 60.Noble D, Tsien RW. J Physiol. 1968 Mar;195:185. doi: 10.1113/jphysiol.1968.sp008454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leech CA, Stanfield PR. J Physiol. 1981;319:295. doi: 10.1113/jphysiol.1981.sp013909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagiwara S, Yoshii M. J Physiol. 1979 Jul;292:251. doi: 10.1113/jphysiol.1979.sp012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hodgkin AL, Horowicz P. J Physiol. 1959 Oct;148:127. doi: 10.1113/jphysiol.1959.sp006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo D, Lu Z. J Gen Physiol. 2001 May;117:395. doi: 10.1085/jgp.117.5.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliver D, Hahn H, Antz C, Ruppersberg JP, Fakler B. Biophys J. 1998 May;74:2318. doi: 10.1016/S0006-3495(98)77941-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spassova M, Lu Z. J Gen Physiol. 1999 Sep;114:415. doi: 10.1085/jgp.114.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Felix JP, et al. Biochemistry. 2006 Aug 22;45:10129. doi: 10.1021/bi060509s. [DOI] [PubMed] [Google Scholar]
- 68.Ramu Y, Klem AM, Lu Z. Biochemistry. 2004 Aug 24;43:10701. doi: 10.1021/bi049125x. [DOI] [PubMed] [Google Scholar]
- 69.Jin W, Klem AM, Lewis JH, Lu Z. Biochemistry. 1999 Oct 26;38:14294. doi: 10.1021/bi991206j. [DOI] [PubMed] [Google Scholar]
- 70.We thank P. Hoff and members of D. Gadsby’s laboratory (Rockefeller University) for assistance with oocyte preparation; R. Molday (University of British Columbia) for providing the anti-1D4 tag cell line; KR. Rajashankar and K. Perry at beamline 24ID-C (Advanced Photon Source, Argonne National Laboratory) and H. Robinson at beamline X29 (National Synchrotron Light Source, Brookhaven National Laboratory) for assistance at the synchrotron; members of the MacKinnon laboratory for assistance; and A. Banerjee, J. Butterwick, M. Whorton, and J. Chen (Purdue University) for comments on the manuscript. J.L.A. was a fellow of the Damon Runyon Cancer Research Foundation. R.M. is an Investigator in the Howard Hughes Medical Institute. The x-ray crystallographic coordinates and structure factor files have been deposited in the Protein Data Bank with accession ID 3JYC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.