Abstract
This paper illustrates how epidemiologic principles can be used to investigate relationships between genetic susceptibility and other risk factors for disease. Five plausible models are described for relationships between genetic and environmental effects, and an example of a simple mendelian disorder that fits each model is given. Each model leads to a different set of predictions about disease risk in individuals with the genetic susceptibility alone, the risk factor alone, both, or neither. The risk predictions for the different models are described, and research designs for testing them are discussed.
Keywords: human genetics, epidemiology, risk
INTRODUCTION
Genetic models for complex diseases usually subsume environmental effects under the concepts of “reduced penetrance” (genetically susceptible individuals who are unaffected) and “sporadics” (genetically nonsusceptible individuals who are affected). These concepts imply etiologic relationships that are seldom made explicit. This paper illustrates how epidemiologic methods can be used to investigate relationships between genetic susceptibility and other risk factors. Five plausible models are described for relationships between genetic and environmental effects, and an example of a simple mendelian disorder that fits each model is given. Then, predictions about disease risk in families are given for each model, and research designs for testing the models are discussed.
MODELS
The diseases under investigation are assumed to have multiple genetic and nongenetic causes. For simplicity, only the causes involving a single discrete, measurable risk factor and genetic susceptibility are discussed. The risk factor itself may have multiple causes, some of which may involve genetic factors other than the susceptibility genes under study. For illustration, consider the relationship between breast cancer, age at first full-term pregnancy, and genetic susceptibility. Late age at first pregnancy (>30 years) is known to be associated with increased breast cancer risk [Kelsey, 1979] and may have either nongenetic or genetic origins (e.g., genetic syndromes involving infertility). Several investigators have demonstrated that susceptibility to breast cancer is increased in some families by an autosomal dominant gene [Newman et al., 1988; Williams and Anderson, 1984; Go et al., 1983], but the genetic form of the disease appears to affect only a minority of patients (approximately 4%) [Newman et al., 1988]. Some women, even in high-risk families, appear to inherit susceptibility from their mothers and transmit it to their daughters without ever becoming affected themselves. This “reduced penetrance” suggests that expression of the dominant susceptibility allele is influenced by environmental factors. What, specifically is the relationship between late first pregnancy and genetic susceptibility? Does the susceptibility gene cause endocrine changes similar to those resulting from delayed pregnancy? Alternatively, does the susceptibility gene operate through a mechanism separate from the effects of delayed pregnancy? If so, does late pregnancy have an effect on gene expression, such as exacerbating it or being required for it?
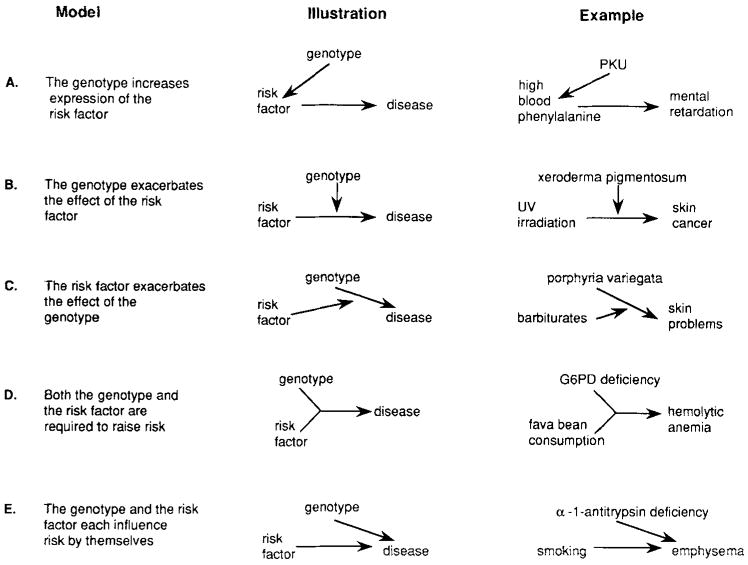
The five models to be discussed are illustrated in Figure 1. These illustrations are not path diagrams, but are similar to diagrams used for biochemical pathways. An arrow from one factor (gene, risk factor, or disease) to another indicates that the first factor has a causal influence on the second. An arrow from a factor to an arrow indicates that the factor influences the relationship between the two other factors. When two arrows merge (as in Fig. 1D), it indicates that two factors must both be present to influence disease risk. The five included models are not intended to be exhaustive. Other relationships between risk factors and genetic susceptibility are possible, and may be testable by using the approach described.
Fig. 1.
Five hypothetical relationships between genetic susceptibility to disease and risk factors for disease identified in epidemiologic studies. The genetic susceptibility may be either polygenic or due to a dominant, recessive, or X-linked major locus. The risk factor may be only one of many factors associated with disease risk, and may itself have either genetic or nongenetic origins.
In Model A, the genetic susceptibility does not cause disease directly, but acts by increasing the level of expression of the risk factor. In this case, the genetic basis of disease is equivalent to the genetic basis of the risk factor, but the risk factor may have other, nongenetic causes. An example of this model is the relationship between the recessive gene for phenylketonuria (PKU), blood levels of phenylalanine, and mental retardation. Individuals who are homozygous for the PKU gene lack the enzyme necessary to convert phenylalanine to tyrosine, resulting in a buildup of blood levels of phenylalanine [Tourian and Sidbury, 1983]. These high blood levels, if uncorrected, cause mental retardation. The indirect nature of the effect of the homozygous PKU genotype is illustrated by the prevention of mental retardation if blood phenylalanine is maintained at a low level through dietary intervention. Further, intrauterine exposure to high blood levels of phenylalanine has been shown to cause mental retardation in individuals who lack the high-risk genotype—heterozygous offspring of homozygous PKU mothers [Mabry et al., 1966].
In Model B, the risk factor has a direct effect on disease susceptibility, and the genetic susceptibility exacerbates this effect. The genetic susceptibility has no effect in the absence of the risk factor, but the risk factor can act by itself to cause disease. An example of this mechanism is the relationship between xeroderma pigmentosum, ultraviolet radiation, and skin cancer. Individuals with xeroderma pigmentosum have a genetic defect in an enzyme required for repair of DNA damage induced by ultraviolet radiation [Cleaver and Bootsma, 1975] and are therefore unusually susceptible to sun-induced skin cancer.
Model C is the converse of the second. Here the genetic susceptibility has a direct effect, and the risk factor exacerbates this effect. The risk factor has no effect in the absence of the genetic susceptibility, but the genetic susceptibility can raise risk by itself. Porphyria variegata [Kappas et al., 1983] is an autosomal dominant genetic disease that fits this model. Affected individuals have skin problems of varying severity, including unusual sensitivity and tendency to blister easily. Upon exposure to barbiturates, however, they experience acute attacks that may lead to paralysis and/or death. Neither the skin problems nor the effects of barbiturates occur in individuals without the gene.
In Model D, neither the genetic susceptibility nor the risk factor can influence disease risk by itself, but risk is increased when both are present. An example of this model is the relationship between the Mediterranean form of glucose-6-phosphate dehydrogenase (G6PD) deficiency, fava bean consumption, and hemolytic anemia [Beutler, 1983]. G6PD-deficient individuals who consume fava beans develop severe hemolytic anemia, but the disease does not develop either in individuals without G6PD deficiency who eat fava beans or in G6PD-deficient individuals who avoid eating fava beans.
In Model E, either the genetic susceptibility or the risk factor can influence disease risk by itself, and the combined effect of the two may be different from the effect of each acting alone. An example of this model is the relationship between alpha-1-antitrypsin deficiency, smoking, and emphysema [Gadek and Crystal, 1983]. The disease occurs with elevated frequency in both smokers and individuals with alpha-1-antitrypsin deficiency, but smokers who also have the enzyme deficiency have even more dramatically elevated risk.
RISK PREDICTIONS
Each of these five models leads to different predictions about disease risk in the four groups defined by presence or absence of the risk factor and predisposing genotype. Table I shows the pattern of relative risk for each model, using as the reference group individuals who have neither the genetic predisposition nor the risk factor (relative risk denoted by “1” in the Table).
TABLE I.
Expected Patterns of Relative Risk for Five Models of Gene-Environment Interaction, by Presence of Risk Factor and Genetic Susceptibility
| Genetic susceptibility | ||||
|---|---|---|---|---|
| Present: Risk factor |
Absent: Risk factor |
|||
| Model | Present | Absent | Present | Absenta |
| A | +b | 1 | + | 1 |
| B | + +b | 1 | + | 1 |
| C | + + | + | 1 | 1 |
| D | + | 1 | 1 | 1 |
| E | ? | + | + | 1 |
Reference group; relative risk denoted by “1.”
“+” indicates relative risk above 1, “+ +” indicates a greater elevation in relative risk, “?” indicates unpredictable relative risk.
In Model A, the effect of the risk factor is the same regardless of whether it is caused by genetic susceptibility or by another factor (such as intrauterine exposure to phenylalanine in the PKU example). If the predisposing genotype has incomplete penetrance, or if its effect can be prevented (such as with dietary intervention in PKU), then some individuals with the genetic susceptibility will lack the risk factor, and will not have increased risk. Thus, four predictions can be made from this model:
The risk factor has an effect regardless of whether or not the genetic susceptibility is present.
The relative risk for the risk factor is the same in the presence and absence of the genetic susceptibility.
There is no effect of the genetic susceptibility within either the group with the risk factor or the group without the risk factor.
The risk factor is more prevalent among those with the genetic susceptibility than among those without it.
In Model B, individuals who have only the risk factor have increased risk, but risk is even higher for those with both the risk factor and the genetic susceptibility (denoted by + + in the Table). Because there is no direct effect of the genetic susceptibility, individuals who have only the predisposing genotype (and who lack the risk factor) do not have increased risk. From this model, the following predictions can be made:
The risk factor has an effect regardless of the presence or absence of the genetic susceptibility.
The relative risk for the risk factor is larger in the presence than in the absence of the genetic susceptibility.
There is no effect of the genetic susceptibility in the absence of the risk factor.
There is an effect of the genetic susceptibility in the presence of the risk factor.
In Model C, individuals who lack the genetic predisposition do not have increased risk regardless of whether or not they have the risk factor, but individuals with the genetic predisposition have higher risk in the presence than in the absence of the risk factor. The predictions from this model are:
There is no effect of the risk factor in the absence of the genetic susceptibility.
There is an effect of the risk factor in the presence of the genetic susceptibility.
There is an effect of the genetic predisposition both in the presence and the absence of the risk factor.
The relative risk for the genetic predisposition is larger in the presence than in the absence of the risk factor.
In Model D, risk is increased only in individuals with both the predisposing genotype and the risk factor. Thus the following predictions can be made:
There is no effect of the risk factor in the absence of the genetic susceptibility.
There is an effect of the risk factor in the presence of the genetic susceptibility.
There is no effect of the genetic predisposition in the absence of the risk factor.
There is an effect of the genetic predisposition in the presence of the risk factor.
In Model E, risk is increased both in individuals with the risk factor who lack the genetic predisposition and in individuals with the genetic predisposition who lack the risk factor. In Table I, risks in these two groups are depicted as equal (denoted by +), but they may differ depending upon their relative effects. Risks in individuals with both the genetic susceptibility and the risk factor may be higher, lower, or the same as in those with only one factor (denoted by ? in the Table). This model would predict:
There is an effect of the risk factor in the absence of the genetic susceptibility.
There is an effect of the genetic susceptibility in the absence of the risk factor.
The effect of the risk factor in the presence of the genetic susceptibility depends upon the relationship (synergistic, antagonistic, etc.) between the risk factor and the genetic susceptibility.
The effect of the genetic susceptibility in the presence of the risk factor depends upon the relationship between the risk factor and the genetic susceptibility.
RESEARCH DESIGNS
To discriminate among these five models, subjects must be classified by both the presence or absence of the predisposing genotype and the presence or absence of the risk factor. Classification according to the presence of the high-risk genotype presents problems, because in most cases the relevant genetic susceptibility cannot be measured. Classification according to risk factor status is usually less problematic. Nondifferential misclassification of either genotype or risk factor will blur the distinctions among the models shown in Table I. Distinctions among the models can also be blurred by contributions of other loci or risk factors, especially if their effects vary across the subgroups shown in Table I.
The following designs can be used for testing the models.
Studies Employing Genetic Markers
This “ideal” design for testing the models can be applied only to diseases for which linkage to a genetic marker has been demonstrated (e.g., X-linked manic depression [Baron et al., 1987], familial polyposis coli [Leppert et al., 1987], and multiple endocrine neoplasia type 2A [Simpson et al., 1987]). In this case, even though the susceptibility gene itself has not been isolated or characterized, information from the linked marker can be used to classify individuals according to their probabilities of carrying the disease gene. In the absence of linkage disequilibrium, this approach can be used only if genetic marker data are available for families since different alleles at the marker locus will segregate with the disease gene in different families. Each individual within a family can be assigned a probability that he or she has the susceptibility genotype, and a threshold value can be used to dichotomize the probabilities. This procedure can be used in a group of families as long as the genotype assignment is made within families and genetic homogeneity can be assumed. Then, subjects can be classified also by presence or absence of environmental risk factors, and disease risks can be compared in the resulting four groups. The power of this approach depends on the informativeness of the linked marker, the tightness of linkage to the disease locus, and the availability of family members for testing. Under optimal conditions, it can provide very accurate information about genotypes at the susceptibility locus.
For diseases in which linkage disequilibrium—or population association with a genetic marker—has been demonstrated (e.g., insulin-dependent diabetes and HLA [Svejgaard et al., 1980]), genotype assignments can be made without family linkage data. In this case it can be assumed that individuals with the associated marker are more likely, and those without the marker less likely, to carry the susceptibility allele. Naturally, for this approach the degree of misclassification of disease genotypes depends upon the strength of association with the marker.
When genetic marker data are not available, family history data can be used as a rough indicator of genetic susceptibility, on the assumption that individuals with a positive family history are more likely to have the genetic predisposition than are those without. Presence or absence of a positive family history is influenced by a variety of factors, including family size, the relatives’ age distribution, and the genetic distance of included relatives to the proband [Susser and Susser, 1989], leading to great potential misclassification of disease genotypes. As a result of this nondifferential misclassification, the relative risk for the genetic susceptibility will be underestimated, or biased toward the null hypothesis. More serious problems arise in family history studies when there is differential misclassification. In case-control studies this can result from a difference between cases and controls in the accuracy of family history information (such as a greater awareness of affected relatives for cases). In cohort studies, it can result from an association between family history and the rate of disease detection (such as more intensive screening in the presence of a positive family history).
Cohort Studies
In a cohort study, in which subjects are ascertained on the basis of presence or absence of a risk factor, subjects can be stratified also by presence or absence of a positive family history, and disease incidence can be studied in the resulting four groups. This approach requires a very large sample size to achieve adequate statistical power, since the proportion of individuals with a positive family history is usually small.
Case-Control Studies
Two different approaches can be used in this type of study. First, cases and controls can be classified by both family history and a risk factor, and odds ratios can be calculated for family history alone, the risk factor alone, and both family history and the risk factor, in each case compared with absence of both family history and the risk factor. Second, the case-control sampling scheme can be converted to a cohort type of analysis [Susser and Susser, 1989], with the at-risk cohorts defined as relatives of cases (positive family history) and relatives of controls (negative family history). Then, each group of relatives can be stratified by presence or absence of the risk factor, and once again disease occurrence can be studied in the resulting four groups. Information on risk factors in relatives is seldom available, because it is difficult to obtain from interviews with the cases and controls. Thus, interviews with the relatives themselves may be required for valid collection of the relatives’ histories of such factors as smoking and diet.
A special case of genotype misclassification occurs in the case-control sampling scheme where relatives of the cases and controls comprise the study population. Some relationships between risk factor and genetic susceptibility predict associations between the probability of the high-risk genotype and the risk factor status of cases and controls. For example, it can be shown that in Models B and D the probability of the high-risk genotype is higher for cases than for controls only among individuals with the risk factor. Cases and controls without the risk factor have equal probabilities of carrying the high-risk genotype. Consequently, with this design the predictions in Table I should be tested within strata defined by the risk factor status of the cases and controls [Ottman et al., 1990].
EXAMPLES
In a case-control design, Brinton et al. [1982] examined the odds of breast cancer by family history and age at first pregnancy (<20 years vs. 30 + years). The results were most consistent with the Model E—the odds ratios, compared with a negative family history and early first pregnancy, were: 2.7 for late first pregnancy alone, 2.4 for positive family history alone, and 5.0 for late first pregnancy and positive family history.
In a classical study of lung cancer, smoking, and family history, Tokuhata and Lilienfeld [1963] examined lung cancer mortality in the relatives of cases and controls. Smoking status of living and deceased relatives was ascertained by mailing postcards to the relatives or their next of kin. The relative risks, compared with nonsmoking relatives of controls, were 5.3 for smoking relatives of controls, 4.0 for nonsmoking relatives of cases, and 13.6 for smoking relatives of cases. Again, the results were most consistent with Model E.
DISCUSSION
Relationships between environmental and genetic influences on disease susceptibility have been discussed previously [Haldane, 1946; MacMahon, 1968; Kidd and Matthysee, 1978; Khoury et al., 1987; King et al., 1984; Ottman et al., 1990], but little attention has been paid to epidemiologic research designs for testing them. Khoury et al. [1987, 1988] discussed such models in terms of the effect on the relative risk associated with environmental factors when genetic susceptibility is not considered, and the effect on the relative risk associated with genetic susceptibility when environmental risk factors are not considered. Ottman et al. [1990] have recently examined methods for control of familial environmental risk factors in studies of familial aggregation.
Although these models are usually called interactions between genotype and environment, they do not necessarily involve statistical interaction [Rothman et al., 1980]. Evaluation of the degree of statistical interaction involves examining the effect of each factor (risk factor or genetic susceptibility) at each level of the other. For example, disease risk in those with vs. without the risk factor can be examined within strata defined by presence or absence of the genetic susceptibility. If the risk difference is the same in these two strata, an additive model holds; if the risk ratio is the same in the two strata, a multiplicative model holds; and if neither is the same in the two strata, statistical interaction may be said to exist. From this point of view, Model A is noninteractive by definition, Models B, C, and D are interactive, and Model E can be either additive, multiplicative, or interactive.
Discovery of the relationships between susceptibility genes and environmental factors has important public health implications. Studies of the type presented here can elucidate these relationships, and provide a basis for well-informed public health recommendations.
Acknowledgments
This work was supported by NIH grants NS22050 and NS20656. The author thanks Drs. Mary-Claire King, Mervyn Susser, Dorothy Warburton, Melissa Austin, Stephanie Sherman, and R. Arlen Price for helpful comments.
References
- Baron M, Risch N, Hamburger R, Mandel B, Kushner S, Newman M, Drumer D, Belmaker R. Genetic linkage between X-chromosome markers and bipolar affective illness. Nature. 1987;326:289–292. doi: 10.1038/326289a0. [DOI] [PubMed] [Google Scholar]
- Beutler E. Glucose-6-phosphate dehydrogenase deficiency. In: Stanbury JB, Wyngaarden JB, Fredrickson DS, Goldstein JL, Brown MS, editors. The Metabolic Basis of Inherited Disease. 5. New York: McGraw-Hill; 1983. pp. 1629–1653. [Google Scholar]
- Brinton LA, Hoover R, Fraumeni JF., Jr Interaction of familial and hormonal risk factors for breast cancer. J Natl Cancer Inst. 1982;69:817–822. [PubMed] [Google Scholar]
- Cleaver JE, Bootsma D. Xerodema pigmentosum: Biochemical and genetic characteristics. Annu Rev Genet. 1975;9:19–38. doi: 10.1146/annurev.ge.09.120175.000315. [DOI] [PubMed] [Google Scholar]
- Gadek JAE, Crystal RG. Alpha-1-antitrypsin deficiency. In: Stanbury JB, Wyngaarden JB, Fredrickson DS, Goldstein JL, Brown MS, editors. The Metabolic Basis of Inherited Disease. 5. New York: McGraw-Hill; 1983. pp. 1450–1467. [Google Scholar]
- Go RCP, King M-C, Bailey-Wilson J, et al. Genetic epidemiology of breast and associated cancers in high risk families. I. Segregation analysis. J Natl Cancer Inst. 1983;71:455–462. [PubMed] [Google Scholar]
- Haldane JBS. The interaction of nature and nurture. Ann Eugen. 1946;13:197–205. doi: 10.1111/j.1469-1809.1946.tb02358.x. [DOI] [PubMed] [Google Scholar]
- Kappas A, Sassa S, Anderson KE. The porphyrias. In: Stanbury JB, Wyngaarden JB, Fredrickson DS, Goldstein JL, Brown MS, editors. The Metabolic Basis of Inherited Disease. 5. New York: McGraw-Hill; 1983. pp. 1301–1384. [Google Scholar]
- Kelsey J. A review of the epidemiology of human breast cancer. Epidemiol Rev. 1979;1:74–109. doi: 10.1093/oxfordjournals.epirev.a036215. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Adams MJ, Jr, Flanders WD. An epidemiologic approach to ecogenetics. Am J Hum Genet. 1988;42:89–95. [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, Stewart W, Beaty TH. The effect of genetic susceptibility on causal inference in epidemiologic studies. Am J Epidemiol. 1987;126:561–567. doi: 10.1093/oxfordjournals.aje.a114695. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Matthysee S. Research designs for the study of gene-environment interactions in psychiatric disorders. Arch Gen Psychiatry. 1978;35:925–932. doi: 10.1001/archpsyc.1978.01770320019001. [DOI] [PubMed] [Google Scholar]
- King M-C, Lee GM, Spinner NB, Thomson G, Wrensch MR. Genetic epidemiology. Annu Rev Pub Health. 1984;5:1–52. doi: 10.1146/annurev.pu.05.050184.000245. [DOI] [PubMed] [Google Scholar]
- Leppert M, Dobbs M, Scambler P, O’Connell P, Nakamura Y, Stauffer D, Woodward S, Burt R, Hughes J, Gardner E, Lathrop M, Wasmuth J, Lalouel JM, White R. The gene for familial polyposis coli maps to the long arm of chromosome 5. Science. 1987;238:1411–1413. doi: 10.1126/science.3479843. [DOI] [PubMed] [Google Scholar]
- Mabry CC, Denniston JC, Coldwell JG. Mental retardation in children of phenylketonuric mothers. N Engl J Med. 1966;275:1331–1336. doi: 10.1056/NEJM196612152752403. [DOI] [PubMed] [Google Scholar]
- MacMahon B. Gene-environment interaction in human disease. J Psychiatr Res. 1968;(6 Suppl):393–402. [Google Scholar]
- Newman B, Austin MA, Lee M, King M-C. Inheritance of human breast cancer: evidence for autosomal dominant transmission in high-risk families. Proc Natl Acad Sci USA. 1988;85:3044–3048. doi: 10.1073/pnas.85.9.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottman R, Susser E, Meisner M. Control for familial environmental risk factors in familial aggregation studies. 1990 doi: 10.1093/oxfordjournals.aje.a116085. (Submitted.) [DOI] [PubMed] [Google Scholar]
- Rothman KJ, Greenland S, Walker AM. Concepts of interaction. Am J Epidemiol. 1980;112:467–470. doi: 10.1093/oxfordjournals.aje.a113015. [DOI] [PubMed] [Google Scholar]
- Simpson WE, Kidd KK, Goodfellow PJ, McDermid H, Myers S, Kidd JR, Jackson CE, Duncan AMV, Farrer LA, Brasch K, Castiglione C, Genel M, Gertner J, Greenberg CR, Gusella JF, Holden JJA, White BN. Assignment of multiple endocrine neoplasia type 2A to chromosome 10 by linkage. Nature. 1987;328:528–30. doi: 10.1038/328528a0. [DOI] [PubMed] [Google Scholar]
- Svejgaard A, Platz P, Ryder LP. Insulin-dependent diabetes mellitis. In: Terasaki PC, editor. Histocompatibility. Los Angeles: Univ of California Press; 1980. pp. 638–656. [Google Scholar]
- Susser E, Susser M. Familial aggregation studies: A note on their epidemiologic properties. Am J Epidemiol. 1989;129:23–30. doi: 10.1093/oxfordjournals.aje.a115119. [DOI] [PubMed] [Google Scholar]
- Tokuhata GK, Lilienfeld AM. Familial aggregation of lung cancer in humans. J Natl Cancer Inst. 1963;30:289–312. [PubMed] [Google Scholar]
- Tourian A, Sidbury JB. Phenylketonuria and hyperphenylalaninemia. In: Stanbury JB, Wyngaarden JB, Fredrickson DS, Goldstein JL, Brown MS, editors. The Metabolic Basis of Inherited Disease. 5. New York: McGraw-Hill; 1983. pp. 270–286. [Google Scholar]
- Williams WR, Anderson DE. Genetic epidemiology of breast cancer: Segregation analysis of 200 Danish pedigrees. Genetic Epidemiology. 1984;1:7–20. doi: 10.1002/gepi.1370010104. [DOI] [PubMed] [Google Scholar]