Abstract
Mammalian immune responses to LPS exposure are typified by the robust induction of NF-κB and IFN-β responses largely mediated by TLR4 signal transduction pathways. In contrast to mammals, Tlr4 signal transduction pathways in non-tetrapods are not well understood. Comprehensive syntenic and phylogenetic analyses support our hypothesis that zebrafish tlr4a and tlr4b genes are paralogous rather than orthologous to human TLR4. Furthermore, we provide evidence to support our assertion that the in vivo responsiveness of zebrafish to LPS exposure is not mediated by Tlr4a and Tlr4b paralogs because they fail to respond to LPS stimulation in vitro. Zebrafish Tlr4a and Tlr4b paralogs were also unresponsive to heat-killed Escherichia coli and Legionella pneumophila. Using chimeric molecules in which portions of the zebrafish Tlr4 proteins were fused to portions of the mouse TLR4 protein, we show that the lack of responsiveness to LPS was most likely due to the inability of the extracellular portions of zebrafish Tlr4a and Tlr4b to recognize the molecule, rather than to changes in their capacities to transduce signals through their Toll/IL-1R (TIR) domains. Taken together, these findings strongly support the notion that zebrafish tlr4a and tlr4b paralogs have evolved to provide alternative ligand specificities to the Tlr immune defense system in this species. These data demonstrate that intensive examination of gene histories when describing the Tlr proteins of basally-diverging vertebrates is required in order to obtain fuller appreciation of the evolution of their function. These studies provide the first evidence for the functional evolution of a novel Tlr.
Keywords: Comparative Immunology/Evolution, Signal Transduction, Other Animals
Introduction
The landmark discovery of a human homolog to the Drosophila Toll protein (1), which later became known as TLR4 (2), dramatically transformed the immunology paradigm, forcing a re-evaluation of our understanding of the immune system to include concepts related to innate immunity. Since this initial breakthrough, additional vertebrate TLR genes have been identified and their fundamental importance to immunity has gathered fuller appreciation. The TLR family of pathogen-recognition receptors forms a central pillar in the immune defense of all vertebrates, from fishes to mammals. TLR proteins function as sentinels against infection, participating from the earliest innate immune responses to the eventual transition to adaptive responses (3). Upon recognition of conserved pathogen-associated molecular patterns, TLR proteins trigger signal transduction events resulting in the activation of cytokines, antimicrobial peptides and other immune response genes.
Mammalian TLR4s function as central proteins in LPS receptor complexes (4-8). LPS is a conserved glycolipid common to Gram-negative bacteria. LPS, facilitated by its association with LPS binding protein (LBP) (9, 10), interacts with CD14 at its N-terminal binding pocket (11), and then is transferred to TLR4, which is complexed with MD2. MD2 is a glycoprotein responsible for TLR4 localization and responsiveness to LPS (12, 13). The TLR4-MD2 complex homodimerizes and recruits intracellular adaptor proteins to the Toll-IL-1R (TIR)3 domain of TLR4. Bifurcated signaling processes, which have been termed MYD88-dependent and MYD88–independent, are triggered, resulting in a pro- and anti-inflammatory immune responses (reviewed in 14).
There has been an increased effort to identify tlr genes from lower vertebrates, particularly the fishes (15); however, the classification of Tlr proteins in basally-diverging vertebrates has tended to rely solely upon phylogenies. The failure to investigate the gene histories and functions of tlr genes has led to the false presumption that sequence homology equates to functional conservation. In truth, the way these Tlr proteins are induced and the transduction pathways through which they signal are poorly understood in basally-diverging vertebrates. The data presented herein demonstrate that zebrafish are responsive to LPS but through a mechanism that is independent of the prototypical TLR4-MD2 multiprotein LPS receptor complexes seen in mammals. Using a series of chimeric mouse-zebrafish and zebrafish-mouse Tlr4 constructs, we demonstrate that the failure of zebrafish Tlr4 proteins to respond to LPS is due to differences in the extracellular domains and not to the transmembrane or intracellular domains. We observe that the transmembrane and intracellular domains of the zebrafish Tlr4a and Tlr4b can mediate a positive NF-κB response when fused to extracellular mouse TLR4 domains, contradicting a recent statement that Tlr4a and Tlr4b TIR domains negatively-regulated NF-κB activation (16). Further, we show through analyses of conserved syntenies that zebrafish tlr4a and tlr4b are not orthologous to human TLR4 but rather are paralogous. Our findings confound the role these tlr4 paralogs play in zebrafish immunity and support the hypotheses that alternative LPS induction pathways predominate in fishes and that zebrafish Tlr4 proteins offer alternative, LPS-independent ligand specificities.
Materials and Methods
Nomenclature Conventions
Nomenclature rules for zebrafish, chicken, mouse, and human genes and proteins differ. Gene and protein names are presented according to the respective nomenclature conventions (species, gene, protein: zebrafish, tlr4, Tlr4; human and chicken, TLR4, TLR4; and mouse, Tlr4, TLR4; see http://zfin.org/zf_info/nomen.html#2).
DNA Constructs
A full-length zebrafish homolog of human TLR4, designated tlr4a, was identified through BLAST sequence analyses of zebrafish genome and available Tlr4a TIR domain sequence data (17). Based upon predicted sequence, we designed primers tlr4a F1 (5′-CTGCGAGTTCTTGATCTGTCAAGGT-3′) and tlr4a R1 (5′-ACTGTTGTTGCATTGTATGCTGA-3′). Total RNA was extracted from the livers, kidneys, and spleens of adult zebrafish with TRIzol and reverse transcribed with random hexamers using the ImPromII reverse transcription system (Promega), as described previously (18). The tlr4a gene was cloned by PCR from this cDNA pool with Phusion polymerase (New England Biolabs, Beverly, MA) under the following reaction conditions: 1 cycle at 98°C; 38 cycles at 98°C for 10 s, 58°C for 15 s, 72°C for 90 s; 1 cycle at 72°C for 5 min. A full-length zebrafish tlr4b clone (cDNA clone MGC:85690; IMAGE:6961784) was purchased from ATCC (Manassas, VA). Mouse Tlr4 (mTlr4), human MD2, human CD14, and the NF-κB luciferase reporter construct pBIIx-luc were generous gifts from R. Medzhitov (Yale University, New Haven, CT). The zebrafish expression vector frm2bl (19) was a gift from P. Gibbs (University of Miami, Miami, FL).
For luciferase assays, full-length tlr4a, tlr4b, mTlr4 were subcloned in frm2bl, resulting in the removal of blue-variant egfp gene. Deletion of the leucine-rich repeat (LRR) of TLR proteins has been shown to result in the creation of constitutively active mutants (1, 8, 20). Due to a truncated N-terminus, the Tlr4aΔLRR deletion construct was created by amplification with primers Tlr4aCAF and Tlr4aCAR (supplementary Table S1A, Figure 7A). The deletions of the LRR from Tlr4b were accomplished through PCR sewing. The 5′ portion of tlr4b was amplified with Tlr4bCASignalF and Tlr4bCASignalR; the 3′ portion of tlr4b was amplified with Tlr4bCATM&IntraF and Tlr4bCATM&IntraR. Through primer design and to facilitate splicing, 5′ amplicons contained 24 bases of 3′ sequence, and 3′ amplicons contained 24 bases of 5′ sequences. To create the ΔLRR deletion constructs, amplicons were combined, denatured, and spliced in the presence of Phusion polymerase (New England Biolabs), which filled in the overlapping PCR product. Following overlap extension, tlr4b outer primers Tlr4bCASignalF and Tlr4bCATM&IntraR were added to the reactions, and PCR amplification of the Tlr4bΔLRR deletion mutant construct was performed. Both the Tlr4aΔLRR and Tlr4bΔLRR products were expressed from frm. Similarly, a full-length myd88 amplicon (Accession No. NM_212814 [http://www.ncbi.nlm.nih.gov/nuccore/56118597]) was subcloned in frm for overexpression studies.
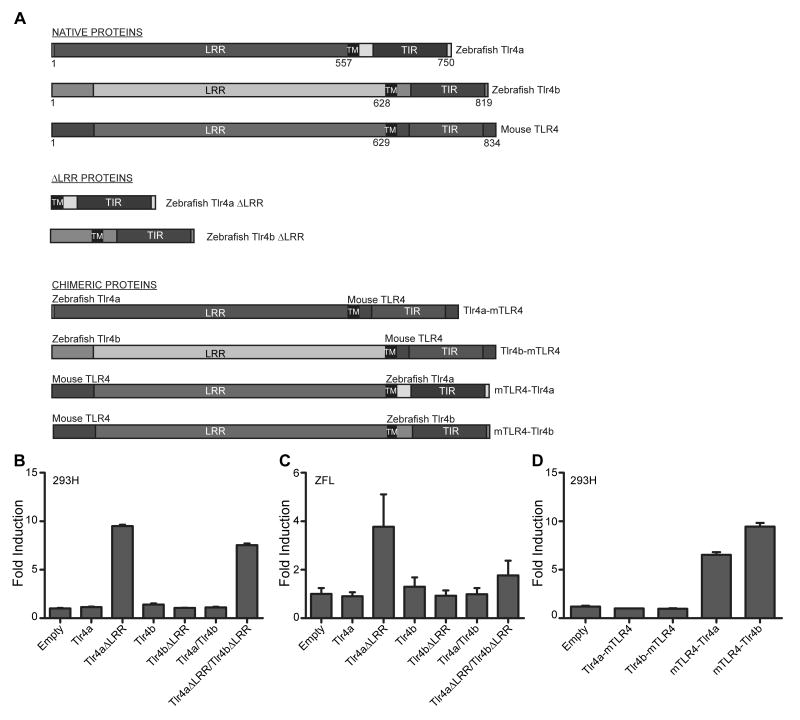
Figure 7. The lack of LPS responsiveness is due to the extracellular domains of Tlr4a and Tlr4b.
(A) Schematic of native and chimeric Tlr4 constructs created by PCR sewing of DNA sequence encoding the mTLR4 extracellular domain to the transmembrane and intracellular domains of zebrafish Tlr4a and Tlr4b or the zebrafish Tlr4a and Tlr4b extracellular domains to the transmembrane and intracellular domains of mTLR4. The extracellular domain, including the LRR region, the transmembrane (TM) domain, and the intracellular domains, including the Toll-IL1 receptor (TIR) domain, for the native and chimeric proteins are labeled accordingly. Numbers below each of the native proteins indicate the N-terminus of the protein (left), the N-terminal portion of the transmembrane domain (middle), and the C-termimus of the protein (right). (B,C) Plasmids encoding Tlr4aΔLRR and Tlr4bΔLRR proteins were overexpressed in 293H (B) and ZFL (C), and the relative induction of an NF-κB-reporter was measured relative to an empty vector control. Native Tlr4a and Tlr4b proteins were overexpressed for comparison. To determine if Tlr4aΔLRR and Tlr4bΔLRR synergize, each was overexpressed in a manner such that the total amount of plasmid transfected did not exceed that which was transfected for the individual constructs (i.e. 200 ng of each, 400 ng total). (D) Chimeric Tlr4 constructs were overexpressed in 293H cells, along with CD14, MD2, pBIIx-luc, and pRL-CMV. Cells were exposed to LPS (10 μg · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM.
Zebrafish-mouse and mouse-zebrafish chimeric Tlr4 constructs were created by PCR sewing using primers listed in supplementary Table S1A and illustrated in Fig. 7A. The tlr4a-mTlr4 chimeric construct, which consists of the zebrafish extracellular domain fused to the mouse transmembrane and TIR domains, contains the nucleotide sequence encoding the first 557 amino acids of zebrafish Tlr4a and the final 207 amino acids of mouse TLR4. The tlr4b-mTlr4 chimeric construct, which is a fusion of the zebrafish Tlr4b extracellular domain and the mouse TLR4 transmembrane and TIR domains, contains nucleotide sequences encoding the first 628 amino acids of zebrafish Tlr4b linked to the final 207 amino acids of mouse TLR4. mTlr4-tlr4a and mTlr4-tlr4b contain nucleotide sequences encoding the extracellular 629 amino acids of mTLR4 fused to the transmembrane- and TIR domain-containing 194 amino acids of Tlr4a or 192 amino acids of Tlr4b, respectively. For luciferase assays, full-length tlr4a, tlr4b, and mTlr4, as well as all four zebrafish-mouse or mouse-zebrafish chimeric constructs, were subcloned in the frm2bl zebrafish expression vector, as previously described (21).
Phylogenetic Reconstruction
Representatives from a broad range of Tlr proteins were aligned with AlignX (Vector NTI, Invitrogen), a multiple sequence alignment program based upon the ClustalW algorithm. Aligned sequences corresponding to the extracellular, LRR regions of these Tlr proteins were extracted and used in the phylogenetic reconstruction. Molecular phylogenetic analysis was performed using PHYLIP, software version 3.6b, (distributed by the author J. Felsenstein, University of Washington, Seattle at http://evolution.gs.washington.edu/phylip.html), using the same methodologies previously described (21).
Genetic mapping
We designed radiation hybrid mapping primers for tlr4a (designed from AY389447) and tlr4b (designed from NM_212813) (supplementary Table S1B). These primers were used to amplify DNAs from the LN54 radiation hybrid mapping panel (22). Strain distribution patterns were submitted to http://mgchd1.nichd.nih.gov:8000/zfrh/beta.cgi to obtain the map location. Strain distributions for tlr4a were: 000002011000101111010000000020000111110000000000001000101011111100100100011110110102011010001, and for tlr4b were 000000011000101111010000000010000111100000000000001000101001111100100100011110110100012010001.
Paralogy mapping
To identify paralogons in the human genome, we created a pipeline that modifies the methods of Dehal and Boore (23) using data downloaded from ENSEMBL (http://www.ensembl.org/Homo_sapiens/index.html) (24). The pipeline starts with a blastn search for each human chromosome 10 (Hsa10) gene against the human genome. The pipeline then scores the top five results, keeping all of the reciprocal best hits (RBHs). It then looks at the location of each RBH and labels the genes as “paralogous duplications” if they are on a different chromosome or more than 1 Mb away on the same chromosome, or tandem duplications if the two sequences are closer than 1 Mb. The distant paralogs constitute paralog groups. For each of the paralog groups, the pipeline blastps each individual gene in the group against the genome of the urochordate Ciona intestinalis, whose lineage diverged from the vertebrate lineage before the amplification of the vertebrate genome (25). All of the human genes must BLASTP to the same C. intestinalis gene, and the C. intestinalis gene must BLASTP back as one of the top X hits (assuming X human genes in the paralogy group). Non-tandem gene paralogs that perform as such are considered “verified” and are plotted on a display of human chromosomes (Fig. 5).
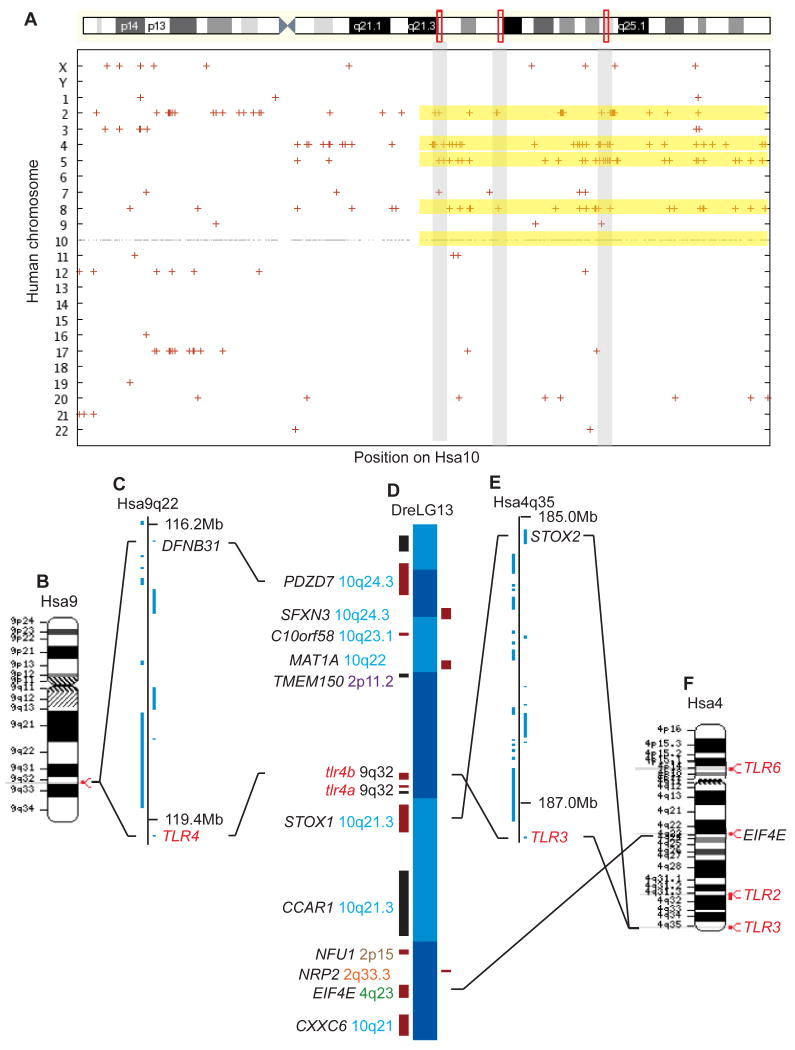
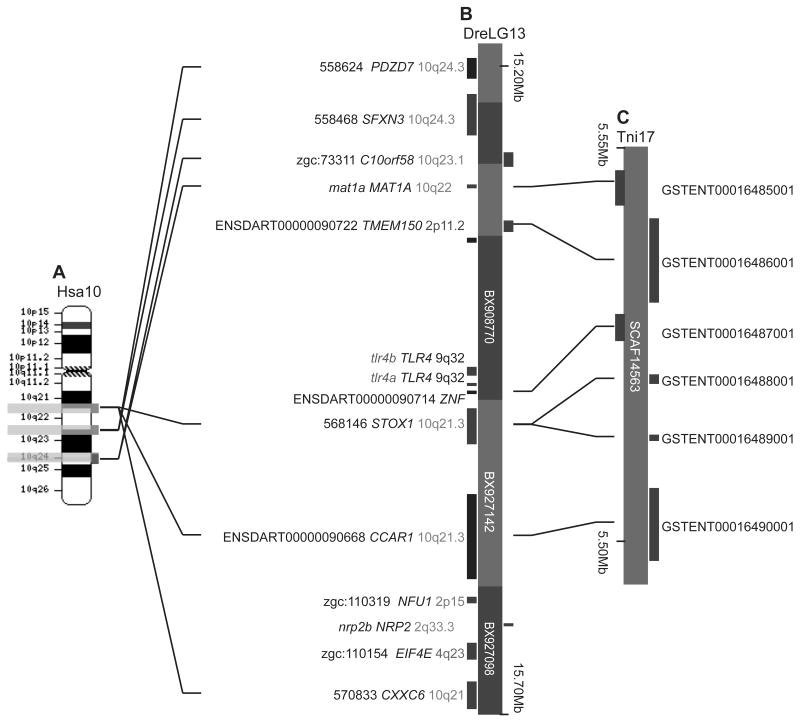
Figure 5. Paralogons for the zebrafish chromosome segment containing tlr4 genes.
(A) Paralogies for human chromosome 10, which is pictured at the top with the locations of several orthologs of genes neighboring zebrafish tlr4 genes (orthologs to STOX1, CCAR1, and CXXC6 [left red rectangle], PDZD7 and SFXN3 [middle red rectangle], and C10ORF58 and MAT1A [right red rectangles]). Yellow bars represent regions of Hsa2, 4, 5, and 8 paralogous to the corresponding portion of Hsa10q. (B) Hsa9, the location of TLR4. (C) A 3.3 Mb portion of Hsa9q22 containing TLR4 and the closest human paralog to PDZD7. (D) A 0.5 Mb portion of zebrafish LG13 containing the tlr4 genes. (E) A 2.2 Mb segment of Hsa4q35 containing STOX2, the closest paralog to stox1, and TLR3. (F) EIF4E, the human ortholog of the zebrafish tlr4 neighbor zgc:110154, resides on Hsa4q with TLR2 and TLR3. These data would be expected according to the hypothesis that the zebrafish tlr4 genes are paralogs, not orthologs, of the human TLR4 gene.
Zebrafish Care and Maintenance
AB strain zebrafish were maintained at 28°C in re-circulating systems (flow rate=150 liters/min) designed by Aquatic Habitats (Apopka, FL) in accordance with procedures and guidelines outlined by the Institutional Animal Care and Use Committee.
RNA Isolation and cDNA Synthesis from Zebrafish Following Static Immersion in LPS or SHRV
Zebrafish aged 20-30 d post-fertilization (dpf) were exposed to 10 μg·mL-1 LPS (E. coli 055:B5) (Sigma, St. Louis, MO) for 24 h or 106TCID50 ·mL-1 SHRV for 5 h. Mock control exposures were also performed. At the end of the exposure duration, fish were returned to the isolated flow-through system for the remainder of the experiment. Three to five fish were collected at each of the following time points and preserved in TRIzol (Invitrogen) at -80°C until processed: 0, 6, 12, 24, 48, 72, and 96 h post-exposure (hpe). Total RNA was extracted from exposed and control fish by TRIzol method, according to manufacturer's recommendations. First-strand cDNA synthesis was performed with random hexamers using the ImpromII reverse transcription system (Promega), as described previously (18).
Cell Culture and Snakehead Rhabdovirus (SHRV) Propagation
Human embryonic kidney 293H cells (Invitrogen, Carlsbad, CA) were cultured at 37°C, 6% CO2 in DMEM (high glucose) supplemented with 10% heat-inactivated FBS. Epithelioma papulosum cyprini (EPC) cells were cultured at 28°C, 5% CO2 in MEM supplemented with 10% heat-inactivated FBS. Zebrafish liver (ZFL) cells were cultured at 28°C in LDF media (21). SHRV was propagated in EPC cells, and viral titers were quantified by determining the 50% tissue culture infective doses (TCID50) · mL-1 (18).
PCR and Quantitative RT-PCR
Total RNA was extracted from cultured ZFL cells by TRIzol method (Invitrogen) and reverse transcribed into first-strand cDNA with random hexamers using the ImpromII reverse transcription system (18). Primers were designed to amplify a 976 bp myd88 fragment, a 1971 bp ticam1 fragment, and a 726 bp tirap fragment from this ZFL cDNA pool (supplementary Table S1C). The amplicons were generated using the following PCR conditions: 98°C for 2 minutes followed by 40 cycles of 98°C for 10 s, 62°C for 15 s, and 72°C for 45 s. Products were subjected to a final 72°C extension for 5 min. Total RNA that had been mock reverse transcribed was used as negative control.
In comparing experimental versus control groups from the LPS- and SHRV-exposed groups, relative transcript levels of the il1, tnfa, ifn, and myxovirus (influenza virus) resistance (mx) genes were determined by quantitative RT-PCR (qRT-PCR) using Icycler iQ Detection system (BioRad, Hercules, CA), as described previously (18). qRT-PCR primer pairs used in these experiments are listed in supplementary Table S1C. Following a 3 min polymerase activation step, experimental conditions for tnfa were as follows: 95°C for 15 s, 56°C for 15 s, 72°C for 20 s, repeated for 40 cycles. il1, ifn, and mx qRT-PCR were performed under the following conditions, after the initial 3 min polymerase activation step: 95°C for 15 s, 54°C for 15 s, 72°C for 20 s, repeated for 40 cycles. All transcript levels were normalized to beta-actin-1 (bactin1). bactin1 qRT-PCR was performed at 94°C for 30 s, 53°C for 30 s, 72°C for 30 s, repeated for 40 cycles. Representative data are presented as fold induction ± SEM.
Heat-killed Escherichia coli (HKEC) and Legionella pneumophila (HKLP)
Colonies of E. coli (29839; ATCC, Manassas, VA) were isolated on Luria-Bertani (LB) agar plates. An individual colony was cultured in LB broth until an OD600 of 2.4 was reached. Cells were pelleted and washed in Dulbecco's phosphate-buffered saline (PBS). Cells were pelleted again, resuspended in Dulbecco's PBS to a concentration of 1010 CFU · mL-1, and then heat-killed at 70°C for 15 min. HKLP was purchased from Invivogen (San Diego, CA) and resuspended to 109 CFU · mL-1 in endotoxin-free distilled water prior to use.
NF-κB Luciferase Reporter Assays
293H and ZFL cells were plated in 24-well plates (Corning, Corning, NY) so that they were 90-95% confluent on the day of transfection. To assay the responsiveness of Tlr4a and Tlr4b to LPS, HKEC, or HKLP in the presence of mammalian TLR4 pathway accessory proteins MD2 and CD14 (mTLR4 positive control), 293H or ZFL cells were transfected with 133 ng of a TLR4 construct (tlr4a, tlr4b, mTlr4, mTLR2 [for L. pneumophila experiment], or frm empty vector control), 133 ng of MD2, and 133 ng of CD14, along with 400 ng of pBIIx-luc NF-κB-luciferase reporter construct, and 10 ng pRL-CMV (Promega, Madison, WI) Renilla luciferase internal control construct using Lipofectamine 2000 (Invitrogen), according to manufacturer's protocol. For co-expression studies, 66.5 ng of tlr4a and 66.5 ng tlr4b were used. To determine the responsiveness of Tlr4a and Tlr4b to LPS in the absence of MD2 and CD14 (mTLR4 negative control), 293H or ZFL cells were transfected with 133 ng of a TLR4 construct (tlr4a, tlr4b, mTlr4, or frm empty vector control), 266 ng pcDNA3.1 empty vector, along with 400 ng of pBIIx-luc NF-κB-luciferase reporter construct, and 10 ng pRL-CMV Renilla luciferase internal control construct using Lipofectamine 2000. As described, when expressed in combination, 66.5 ng of both tlr4a and tlr4b were transfected. The responsiveness of the chimeric constructs to LPS was measured following transfection of 293H with 133 ng of the chimeric construct (tlr4a:mTlr4, tlr4b:mTlr4, mTlr4:tlr4a, or mTlr4:tlr4b), 133 ng of MD2, 133 ng of CD14, 400 ng of pBIIx-luc NF-κB-luciferase reporter construct, and 10 ng pRL-CMV Renilla luciferase internal control construct.
Twenty-four hours post-transfection, cells were exposed to one of the following treatments: ultrapure LPS (E.coli 0111:B4) (Invivogen, San Diego, CA) at a concentration 10 μg · mL-1 for 6 h; HKEC at a concentration of 106 CFU · mL-1 for 6 h; or HKLP at 108 CFU · mL-1 for 6 h. Mock exposures were simultaneously performed using respective diluents. Following exposure, cells were lysed, and firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay system (Promega).
Deletion of the LRR from TLR proteins has been shown to lead to their constitutive activation (1, 8, 20). To determine the capacity of Tlr4a and Tlr4b to constitutively activate an NF-κB-luciferase reporter, 293H and ZFL cells were transfected with 400 ng of frm plasmid encoding an individual Tlr4 construct (Tlr4a, Tlr4aΔLRR, Tlr4b, or Tlr4bΔLRR), 400 ng of pBIIx-luc NF-κB-luciferase reporter construct, and 10 ng pRL-CMV (Promega) Renilla luciferase internal control construct using Lipofectamine 2000, according to manufacturer's protocol. To test for synergism between Tlr4a and Tlr4b and between Tlr4aΔLRR and Tlr4bΔLRR, 293H and ZFL cells were transfected with 200 ng of Tlr4 and 200 ng Tlr4b or 200 ng of Tlr4aΔLRR and 200 ng of Tlr4bΔLRR, along with 400 ng of pBIIx-luc NF-κB-luciferase reporter construct, and 10 ng pRL-CMV Renilla luciferase internal control construct using Lipofectamine 2000, For each experiment, cells were lysed 24 h post-transfection, and luciferase activities were measured using the Dual-Luciferase Reporter Assay system.
An frm expression plasmid containing myd88 was used to assay its effect on NF-κB activation, using the aforementioned NF-κB-luciferase reporter. ZFL cells were transfected with 10 ng, 100 ng, or 400 ng of the frm-myd88 plasmid, along with 400 ng of pBIIx-luc NF-κB-luciferase reporter construct, and 10 ng pRL-CMV Renilla luciferase internal control construct using Lipofectamine 2000, according to manufacturer's protocol. Empty frm plasmid was used as a negative control and was also added when necessary to ensure that the total amount of frm plasmid was 400 ng (0 ng frm-myd88, 400 ng frm-empty; 10 ng frm-my88, 390 ng frm-empty; 100 ng frm-myd88, 300 ng frm-empty; and 400 ng frm-myd88, 0 ng frm-empty). Cells were lysed 24 h post-transfection, and luciferase activities were measured using the Dual-Luciferase Reporter Assay system.
Results
Zebrafish exposed by static immersion to LPS exhibit elevated il1 and tnfa transcript levels but no antiviral response
In mammals, exposure to the LPS present on Gram-negative bacteria triggers inflammatory immune responses through TLR4. TLR4 signaling initiates bifurcated signal transduction cascades leading to the upregulation of assorted immune-responsive genes. One part of this signaling cascade is MYD88-dependent and relies upon MYD88 and MAL (TIRAP) to initiate an early pro-inflammatory response typified by the increased expression of IL1 and TNFA transcripts (26). The other part of this signaling cascade is MYD88-independent and relies upon TIR domain-containing adaptor molecule-1 (TICAM1, also known as TRIF) and TICAM2 (TRAM) to initiate a later NF-κB and an antiviral type I IFN response (27). In zebrafish, myd88 and mal are present in the genome but ticam2 is not (21), and thus we hypothesize that if zebrafish Tlr4 proteins could recognize LPS, their signaling cascades would involve a pro-inflammatory component but not an antiviral response.
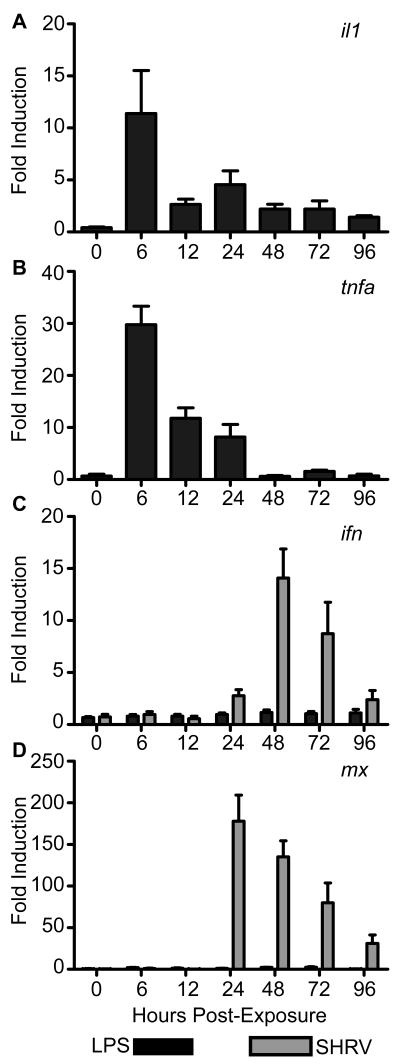
To test our hypothesis, zebrafish aged 20-30 dpf were exposed by static immersion to 10 μg·ml-1 LPS for 24 h. Whole body samples were collected at specific time points following this initial exposure and stored in TRIzol. Previous studies have shown that zebrafish exposed to bacterial and viral pathogens by static immersion exhibit potent pro-inflammatory immune responses (18, 28). Robust increases in il1 and tnfa transcript levels have been measured as part of the antibacterial response (28). Similarly, remarkable elevations in ifn and mx transcript levels are measured as part of the antiviral response (18). Upon exposure to LPS, our studies showed that zebrafish exhibited elevated levels of il1 transcripts, as measured by qRT-PCR, relative to controls (Fig. 1A). Changes were noted as early as 6 hpe, with an observed 11.4-fold increase in il1 expression relative to controls. Expression of il1 increased in the LPS-exposed groups and remained elevated through 96 hpe. Similarly, tnfa expression (Fig. 1B) was upregulated most during the initial 24 hpe, peaking at 6 hpe when a 29.8-fold induction was observed. By 48 hpe, tnfa expression levels had returned to control levels.
Figure 1. Expression of immune-responsive genes upon exposure to known TLR4 agonist LPS.
Zebrafish aged 20-30 dpf were exposed to 10 μg·mL-1 LPS for 24 h or 106TCID50 ·mL-1 SHRV for 5 h. qRT-PCR was performed on first-strand cDNA reverse transcribed from total RNA collected from zebrafish that had been sacrificed at the indicated times. Changes in gene expression, measured as fold induction ± SEM, were recorded for (A) il1, (B) tnfa, (C) ifn, and (D) mx at 0, 6, 12, 24, 48, 72, and 96 hpe. Elevated levels of transcription were observed for il1 (A) and tnfa (B) upon LPS exposure, but not for ifn (C) and mx (D).
In contrast to the changes observed in il1 and tnfa transcript levels following exposure to LPS, expression of the antiviral gene ifn (Fig. 1C) exhibited no significant changes relative to the mock-exposed controls. In contrast, zebrafish exposed to SHRV did exhibit the expected potent antiviral responses, with increases in ifn levels initially observed at 24 hpe (2.8-fold induction). Expression of ifn peaked at 48 hpe with 14.1-fold induction, and then gradually declined through 96 hpe, when 2.4-fold induction was observed. Similarly, expression levels of mx (Fig. 1D), an antiviral protein responsive to Ifn stimulation (29), were minimally induced by LPS exposure, particularly when compared to SHRV-exposed fish. LPS-exposure led to a very modest, biphasic induction of mx expression, with an initial peak of 1.9-fold over control at 6 hpe and a second peak of 2.5-fold over control at 72 hpe. In stark contrast, SHRV exposure induced very strong expression of mx, with peak expression of 178-fold over control observed at 24 hpe. Levels of mx remained strongly elevated relative to the controls through 96 hpe, when a 31.1-fold induction was recorded. The data indicate that, in contrast to mammals (30, 31), zebrafish do not exhibit a potent antiviral response to LPS stimulation, as is observed upon SHRV exposure.
Identification of zebrafish tlr4 homologs
Sequence encoding the TIR domain of zebrafish tlr4a, originally described by Meijer et al. (17), was used to search genomic databases in an effort to identify a putative full-length clone based upon an in silico prediction of the entire coding sequence. We amplified a 2297 bp clone of tlr4a containing a 2253 nt open reading frame by PCR and deposited the sequence in GenBank (Accession No. EU551724 [http://www.ncbi.nlm.nih.gov/nuccore/190341704]). A full-length zebrafish tlr4b clone had previously been amplified (Accession No. NM_212813 [http://www.ncbi.nlm.nih.gov/nuccore/47086406]), but its role has not been directly investigated.
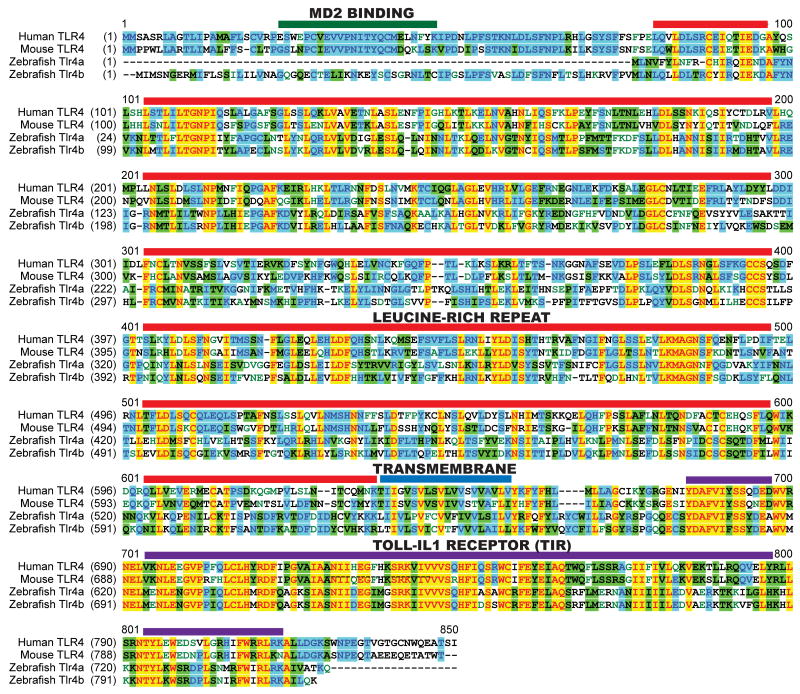
The zebrafish tlr4a gene encodes a 750 amino acid protein, with 38% identity and 57% similarity to 839 amino acid human TLR4 protein and 36% identity and 56% similarity to 835 amino acid mouse TLR4 protein. An in-frame stop codon was observed just upstream of the tlr4a start codon and accounts for the truncated N-terminus. The zebrafish tlr4b gene encodes a slightly larger 819 amino acid protein, with 38% identity and 55% similarity to human TLR4 and 35% identity and 54% similarity to mouse TLR4. Zebrafish Tlr4a and Tlr4b share 67% identity and 78% similarity with each other. A ClustalW-based AlignX amino acid alignment of human TLR4, mouse TLR4, zebrafish Tlr4a, and zebrafish Tlr4b reveals sequence differences in the extracellular protein domains that may have functional consequences (Fig. 2). For example, in humans, the region between Glu24 and Lys47 has been shown to be essential for MD2 binding (32, 33). Close examination of our amino acid alignment reveals strong conservation of this region between mouse TLR4 and human TLR4. In contrast, zebrafish Tlr4a does not possess a homologous MD2 binding region, and zebrafish Tlr4b exhibits only 35% identity in this area. This difference, combined with the fact that an MD2 homolog is not observed in the zebrafish (34), strongly indicates that this structural difference may have significant effects on the mechanisms of signaling that zebrafish Tlr4a and Tlr4b use. Human TLR4 requires MD2 and the glycosylations that MD2 facilitates at Asn526 or Asn575 of TLR4 to be expressed on the cell surface (35). Under these conditions, human TLR4 can translocate to the cell surface and be LPS-responsive. Analysis of homologous residues in the zebrafish reveals that glycosylation sites with the canonical sequence Asp-Xxx-Ser/Thr (where Xxx represents any amino acid) are not present in either Tlr4a or Tlr4b. The significance of these differences is in question, however, due to the lack of MD2 in the zebrafish genome and the potential for compensatory adaptations through evolution. Indeed, when we stably expressed zebrafish Tlr4a-GFP and Tlr4b-GFP in ZFL cells, fluorescence appeared on the cell surface (Fig. S1), suggesting that Tlr4a and Tlr4b are transported to the cell surface through an MD2-independent mechanism.
Figure 2. Amino acid alignment of human, mouse and zebrafish TLR4 homologs.
Amino acid sequences for human TLR4 (Accession No. U93091 [http://www.ncbi.nlm.nih.gov/nuccore/2429102]), mouse TLR4 (Accession No. Q9QUK6 [http://www.ncbi.nlm.nih.gov/protein/20140894]), zebrafish Tlr4a (Accession No. ACE74929 [http://www.ncbi.nlm.nih.gov/protein/190341705]), and zebrafish Tlr4b (Accession No. AAH68358 [http://www.ncbi.nlm.nih.gov/protein/46249703]) were aligned using AlignX, a ClustalW-based algorithm that is part of the VectorNTI software package. Dashes indicate gaps in aligned sequence. The red overlines indicate the LRR domain; the blue overline indicates the predicted transmembrane domain; and the purple overlines identify the TIR domain. The color scheme of the alignment is the default output of the AlignX software: black on white indicates a non-similar residue for which no consensus sequence could be determined; blue on cyan denotes a consensus residue from a block of similar residues at the aligned position; black on green indicates a consensus residue with equal to or greater than 50% identity at the aligned position; red on yellow denotes a consensus residue with 100% identity to the other residues at the aligned position; and green on white describes a residue with weak similarity to the consensus residue at a given position.
In contrast to the extracellular domains, the intracellular portions of Tlr4a and Tlr4b, particularly the TIR domains, are more similar to mammalian TLR4 proteins. Alignment of the intracellular portions of the proteins reveal 50% identity and 66% similarity of residues for both zebrafish Tlr4a and Tlr4b when compared to human TLR4. In this intracellular region of the polypeptide, zebrafish Tlr4a and Tlr4b share 91% identity and 96% similarity with each other. This sequence homology is significant because it supports the notion that Tlr4a and Tlr4b differ from mammalian TLR4 proteins at the point of ligand recognition rather than at the level of signal transduction through adaptor proteins like Myd88.
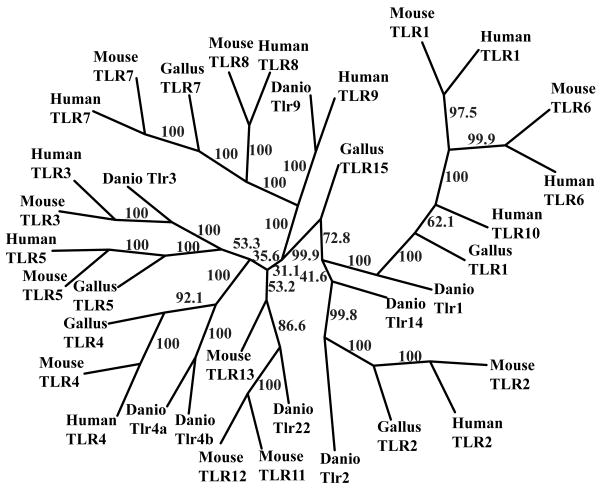
Phylogenetic analyses of the extracellular and transmembrane portions from a broad representation of the TLR proteins predict that zebrafish Tlr4a and Tlr4b form a monophyletic group with mouse, human, and chicken TLR4s well separated from other TLRs (Fig. 3). These findings are supported by other analyses (15) and bolster the notion that tlr4a and tlr4b share a common evolutionary history with other TLR4 genes.
Figure 3. Zebrafish Tlr4 proteins form a clade with human, mouse, and Gallus TLR4s.
A broad representation of TLR proteins were aligned with AlignX. Extracellular domains were extracted and subjected to molecular phylogenetic analysis using PHYLIP, software version 3.6b. The TLR4 proteins from human, mouse, Gallus, and zebrafish form a monophyletic group with 100% bootstrap support.
Genomics of the origin of tlr4a and tlr4b
To identify the location of tlr4a and tlr4b in the zebrafish genome, we designed mapping primers and located the genes on the LN54 radiation hybrid panel (22). Results placed tlr4a and tlr4b at a distance of 4.71 cR and 2.63 cR, respectively, from Z9049, showing that these two genes lie very close together on linkage group (LG) 13. The zv6 assembly of the zebrafish genome agrees with our mapping data, and places tlr4a and tlr4b as nearest neighbors separated by about 7.2kb and transcribed in the same direction from the same DNA strand.
The hypothesis that tlr4a and tlr4b are co-orthologs of the human TLR4 gene predicts that they should share conserved syntenies. To test this proposition, we examined the orthologies of loci surrounding the zebrafish tlr4 genes using the best reciprocal BLAST hit as the criterion for orthology (36). Results showed that of the 12 annotated sequences surrounding the tlr4 genes (Fig. 4B), seven were orthologous to genes in three small regions on the long arm of human chromosome 10 (Hsa10q, Fig. 4A). Others were orthologous by this criterion to human genes in Hsa2, Hsa4, but none were from Hsa9, the location of TLR4. This situation is not as predicted if tlr4a and tlr4b occupy a chromosome segment conserved with the region surrounding TLR4 in the human genome.
Figure 4. Conserved syntenies surrounding zebrafish tlr4 genes.
(A) Human chromosome 10, showing three regions that contain orthologs of the indicated tlr4 neighbors of zebrafish tlr4 genes. In each of the three regions, the indicated genes are either adjacent to each other or are within two genes of each other. (B) A 0.5 megabase region of zebrafish LG13 containing tlr4a and tlr4b. Labels such as 558624 PDZD7 10q24.3 indicate the annotated designation of the zebrafish sequence, the abbreviation of the human ortholog, and the location of the human ortholog. Results show that the tlr4-containing portion of the zebrafish genome is orthologous to parts of human chromosome 10. (C) 50kb of pufferfish chromosome 17, demonstrating co-linear orthologies with the corresponding segment of the zebrafish genome, with an exception for the absence of the two tlr4 genes. The most parsimonious explanation is that the tlr4 gene region was deleted from the pufferfish lineage after the divergence of the zebrafish and pufferfish lineages.
To investigate whether this situation is similar among teleost fish, we examined the genome of the green-spotted pufferfish Tetraodon nigroviridis (Tni). Results showed that a series of six genes in pufferfish were orthologous to genes to the right and left of the zebrafish tlr4 genes, but there was no tlr4 sequence in this portion of the pufferfish genome (Fig. 4C), and in fact, the pufferfish genome database has no ortholog of TLR4. The closest sequence to human or zebrafish Tlr4 genes in T. nigroviridis is GSTENT00026075001, which is orthologous to human TLR7 on HsaXp22. Furthermore, neighbors of GSTENT00026075001 are also orthologous to HsaXp22 genes, confirming orthology by conserved synteny. Two possible hypotheses can explain these results. First, the zebrafish tlr4 genes may have been translocated into their current location in the middle of Hsa10q orthologs after the divergence of the zebrafish and pufferfish lineages and, in addition, the teleost tlr4 gene was deleted from the pufferfish lineage, a hypothesis requiring two steps. Second and alternatively, the teleost tlr4 gene was in a location orthologous to the zebrafish location in the last common ancestor of zebrafish and pufferfish and a single event deleted it from the pufferfish lineage. Because the latter hypothesis requires a single step, it is preferred by parsimony. The loss of the tlr4 genes from the pufferfish genome and their retention in the zebrafish genome leads us to speculate that there may be functional redundancies in the pufferfish that make tlr4 genes dispensable. As a corollary, their presence in the zebrafish genome is suggestive of a potential loss of this functional redundancy in the zebrafish through the sub- or neo-functionalization of the Tlr4 or other Tlr proteins.
To discover the mechanisms that could have caused the zebrafish tlr4 genes to be surrounded by genes primarily orthologous to genes in Hsa10q rather than in Hsa9q, the location of the TLR4 gene, we analyzed paralogons in the human genome. To determine the regions of the human genome that are paralogous to Hsa10q, we wrote software to identify the most similar paralog in the human genome to each gene on Hsa10 in a manner similar to that of Dehal and Boore (23). Fig. 5A shows that Hsa10q genes have paralogy mainly with Hsa8p, Hsa5q, Hsa4, and Hsa2p. Significantly, the genes surrounding tlr4 genes in zebrafish that are not orthologous to genes on Hsa10q are highly similar to genes on Hsa2p or Hsa4q, suggesting that they may be paralogs, rather than orthologs of the human genes. This raises the hypothesis that the zebrafish tlr4 genes are paralogs, rather than orthologs, of human TLR4. To explore this hypothesis, we determined the closest human paralogs of the human orthologs of the genes surrounding zebrafish tlr4 genes. The closest human paralog to PDZD7 is DFNB31, which is located about 3 Mb from the human TLR4 gene (Fig. 5B, 5C, and 5D). Likewise, the closest human paralog of STOX1, whose apparent ortholog is the nearest neighbor of zebrafish tlr4a, is about 2 Mb from the human TLR3 gene (Fig. 5D, 5E, and 5F). EIF4E, the human ortholog of the zebrafish sequence zgc:110154, is found on Hsa4q along with TLR2 and TLR3 (Fig. 5F). Human paralogs of the other neighbors of tlr4 genes are almost exclusively on Hsa2p (CXXC6/MGC22014, MAT1A/MAT2A), Hsa5 (SFXN3/SFXN1, MAT1A/MAT2B, EIF4E/EIF4E1B), and Hsa8 (CXXC6/KIAA1967). This evidence from the analysis of conserved syntenies suggests that the zebrafish tlr4 genes are paralogs, not orthologs, of the human TLR4 gene despite their position in the phylogenetic analysis.
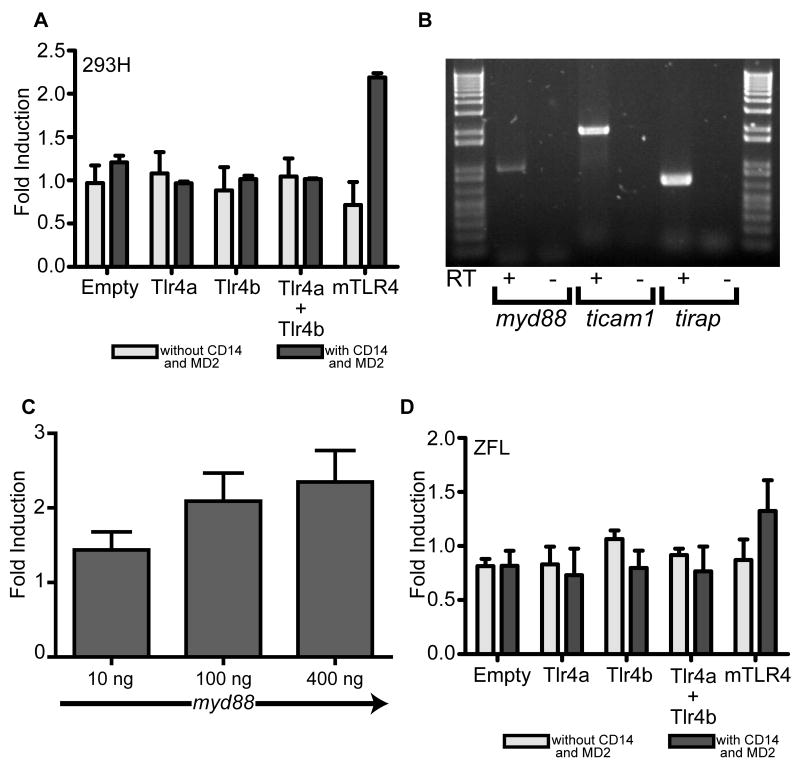
Zebrafish Tlr4a and Tlr4b are unresponsive to LPS stimulation
Due to the significant sequence divergence observed between zebrafish Tlr4 proteins and mammalian TLR4 proteins, particularly in the extracellular domain, we sought to determine whether zebrafish Tlr4a and Tlr4b, alone or in combination as a potentially heterodimeric complex, were capable of activating an NF-κB-luciferase reporter, even though it was not anticipated that MD2 could interact with these proteins. In 293H cells and in the presence of CD14 and MD2, zebrafish Tlr4a alone, Tlr4b alone, and Tlr4a combined with Tlr4b exhibited no induction (1.0-fold) of the NF-κB-luciferase reporter (Fig. 6A) when compared to uninduced controls. In contrast, mTLR4 overexpression, when combined with CD14 and MD2, exhibited 2.2 ± 0.05-fold activation. In the absence of CD14 and MD2, similar results were observed. Tlr4a exhibited 1.1 ± 0.25-fold activation; Tlr4b was induced 0.88 ± 0.27-fold; and the combined Tlr4a + Tlr4b were activated 1.0 ± 0.21-fold. Without MD2 and CD14 co-expression, mTLR4 showed 0.72 ± 0.26-fold activation.
Figure 6. Zebrafish Tlr4a and Tlr4b are unresponsive to LPS stimulation.
Zebrafish Tlr4a and Tlr4b, alone and in combination, were overexpressed in 293H (A) cells, in the presence or absence of human CD14 and human MD2. mTLR4 was similarly overexpressed to serve as a positive control. Each was expressed in the presence or absence of human CD14 and MD2 in order to facilitate the mTLR4 positive and negative control, respectively. In the absence of MD2 and CD14, pcDNA3.1 empty vector was overexpressed to maintain an equal amount of plasmid in the transfection. pBIIx-luc (to measure NF-κB reporter activity) and pRL-CMV (to normalize luciferase activity data) were also co-transfected. Following transfection, cells were exposed to ultrapure LPS (10 μg · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM. (B) RT-PCR reveals expression of myd88, ticam1 (trif), and tirap (mal) in ZFL cells. Total RNA was extracted from ZFL cells and reverse transcribed to cDNA (+). A mock reverse transcription, in which the reverse transcriptase was excluded, was also performed (-). PCR was performed using gene-specific primers for myd88, ticam1 (trif), and tirap (mal) (see supplementary Table S1C). (C) Full-length zebrafish myd88 was subcloned in frm2bl so that the egfp was replaced. ZFL cells were transfected with 10, 100, and 400 ng of myd88 plasmid, along with empty frm (no egfp) so that the total frm plasmid transfected was 400 ng. In addition, 400 ng of pBIIx-luc and 10 ng of pRL-CMV were co-transfected. Forty-eight hours post-transfection, cells were lysed, and luciferase activities were measured. The data are presented as normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM over empty vector control. (D) As in (A), zebrafish Tlr4a and Tlr4b, alone and in combination, were overexpressed in ZFL cells, in the presence or absence of human CD14 and human MD2. mTLR4 was overexpressed as a positive control. pBIIx-luc and pRL-CMV were also co-transfected. Following transfection, cells were exposed to ultrapure LPS (10 μg · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB-luciferase reporter activity ± SEM.
We have previously shown that ZFL cells were useful for measuring NF-κB activities in response to the overexpression of constitutively-activated tlr3, traf6, irak4, (20) and ticam1 (trif) (21). We also demonstrate that ZFL cells express full-length myd88, tirap (mal), and ticam1 (Fig. 6B). Further, ectopic expression of Myd88 leads to dose-dependent activation of an NF-κB-luciferase reporter (Fig. 6C). Despite the presence of these intact signaling cascades in ZFL cells, none of the Tlr4 constructs tested exhibited significant activation of the NF-κB-luciferase reporter when cells were stimulated with LPS (Fig. 6D). When overexpressed in combination with CD14 and MD2, Tlr4a exhibited 0.73 ± 0.25-fold activation when compared to uninduced controls; Tlr4b was induced 0.79 ± 0.16-fold; Tlr4a and Tlr4b together was activated 0.77 ± 0.23-fold; and mTLR4 was induced 1.3 ± 0.29-fold. Likewise, in the absence of CD14 and MD2, Tlr4a showed 0.83 ± 0.17-fold activation; Tlr4b exhibits 1.1 ± 0.08-fold induction; Tlr4a and Tlr4b combined is activated 0.92 ± 0.06-fold; and mTLR4 is induced 0.87 ± 0.19-fold. Based upon these data, we conclude that in ZFL cells, ectopic expression of zebrafish or mammalian TLR4 proteins followed by stimulation with LPS is not enough to activate a robust NF-κB response.
The failure of zebrafish Tlr4a and Tlr4b to respond to LPS is due to differences in their extracellular domains
We wanted to determine whether the lack of responsiveness by zebrafish Tlr4a and Tlr4b to LPS stimulation was due to differences in the extracellular domain or the transmembrane and intracellular domains, or both. Previous experiments have shown that invertebrate Toll proteins (37) and vertebrate Tlr proteins (1, 8, 20) can be made constitutively-active by the removal of the LRR regions. We sought to determine whether the removal of the LRRs from the zebrafish Tlr4a and Tlr4b proteins would result in their constitutive activation (Fig. 7A). In 293H cells, overexpression of the Tlr4aΔLRR led to a 9.5 ± 0.1-fold activation of the NF-κB-luciferase reporter (Fig. 7B). Similarly, a 3.8 ± 1.3-fold activation was observed in ZFL cells (Fig. 7C). Overexpression of the full-length Tlr4a failed to activate the NF-κB-luciferase reporter (1.1 ± 0.1-fold in 293H cells; 0.9 ± 0.2-fold in ZFL cells) (Fig. 7B,C). In stark contrast, overexpression of the Tlr4bΔLRR in 293H and ZFL cells failed to activate NF-κB-luciferase reporter expression (1.1 ± 0.02-fold in 293H cells; 0.9 ± 0.2-fold in ZFL cells) (Fig. 7B,C). Full-length Tlr4b similarly failed to activate the NF-κB-luciferase reporter (1.4 ± 0.1-fold in 293H cells; 1.3 ± 0.4-fold in ZFL cells). We tested for potential synergism between the Tlr4a and Tlr4b proteins by co-expression of the full-length or ΔLRR constructs in 293H and ZFL cells. Cells were transfected with half the amount of individual plasmid (200 ng) so that the total amount of plasmids (400 ng) did not change and comparisons between individual plasmids and combined plasmids could be made. In both 293H and ZFL cells, we did not observe any synergism resulting from the co-transfection. No antagonism could be detected either. In 293H cells, the NF-κB-luciferase reporter was activated 7.5 ± 0.2-fold (Fig. 7B), and in ZFL cells, the reporter was activated 1.8 ± 0.6-fold (Fig. 7C). Combined overexpression of full-length Tlr4a and Tlr4b resulted in no activation in either 293H or ZFL cells (Fig. 7B,C). In light of 91% identity and 96% similarity between Tlr4a and Tlr4b across their TIR domains, these data were both surprising and concerning, particularly in light of recent findings by Sepulcre et al. (16). Our findings indicate that the overexpression of the Tlr4b TIR domain alone may not give an accurate representation of Tlr4b function.
To further investigate the roles of the LRR, transmembrane, and TIR domains and address this question about the lack of LPS responsiveness, we created chimeric Tlr4 proteins consisting of portions of the zebrafish Tlr4a or Tlr4b protein fused to a portion of the mTLR4 protein (Fig. 7A) through PCR sewing of genes: zebrafish Tlr4a or Tlr4b extracellular domains fused to mTLR4 transmembrane and intracellular domains (Tlr4a-mTLR4 or Tlr4b-mTLR4 respectively) and mTLR4 extracellular domain fused to zebrafish Tlr4a or Tlr4b transmembrane and intracellular domains (mTLR4-Tlr4a or mTLR4-Tlr4b, respectively). Each of these constructs was overexpressed in 293H (Fig. 7D), and the NF-κB-luciferase reporter activities were measured following exposure to LPS for 6 h. In 293H cells, stimulation of Tlr4a-mTLR4 and Tlr4b-mTLR4 chimeric proteins with LPS failed to activate NF-κB-luciferase reporter activity (1.0 ± 0.01-fold and 0.98 ± 0.05-fold, respectively) (Fig. 7D). As a result, it is apparent that the extracellular portions of the zebrafish Tlr4a and Tlr4b fail to recognize LPS. In contrast, the expression construct encoding both mTLR4-Tlr4a and mTLR4-Tlr4b chimeric proteins exhibited robust activation upon stimulation with LPS (6.5 ± 0.27-fold and 9.5 ± 0.38-fold). These data support the notion that the transmembrane and intracellular domains of zebrafish Tlr4a and Tlr4b retain a capacity to interact with components of the intracellular downstream TLR signal transduction pathway (in human 293H cells, MYD88 and MAL and/or TICAM2 and TICAM1) and positively regulate NF-κB. These data directly contradict findings by Sepulcre et al. (16), who determined through overexpression of the TIR domain alone, rather than the entire protein, that Tlr4a and Tlr4b were negative regulators of NF-κB. Taken together, these data indicate that there may be ligand(s) other than LPS that are recognized by Tlr4a and Tlr4b. It is also possible that Tlr4a and Tlr4b may function as co-receptors for each other or for other zebrafish Tlr proteins in a manner similar to the role mammalian TLR1 and TLR6 play in supporting the alternative ligand specificities of TLR2.
Zebrafish Tlr4a and Tlr4b are unresponsive to stimulation by heat-killed E. coli and L. pneumophila
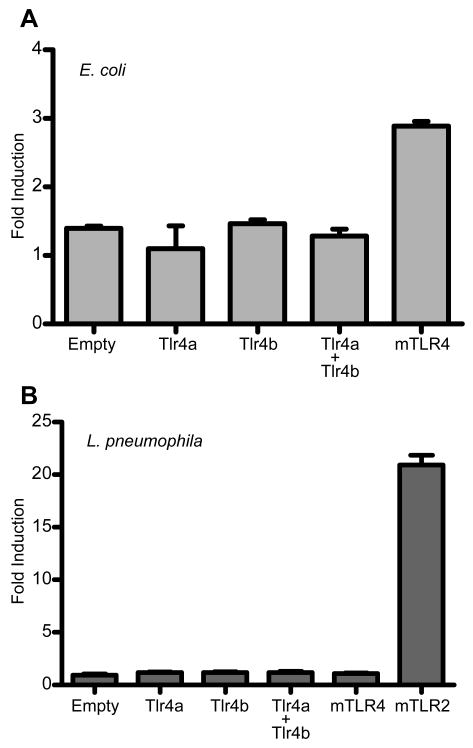
The failure of Tlr4a and Tlr4b to respond to LPS leads to speculation regarding their ligand specificities. To begin to address this question, we attempted to stimulate Tlr4a and Tlr4b with HKEC and HKLP. Both HKEC and HKLP can present an assortment of ligands, in addition to LPS, to the receptors (Fig. 8). For HKEC stimulation, empty vector, Tlr4a, Tlr4b, Tlr4a and Tlr4b in combination, and mTLR4 were overexpressed in 293H cells along with CD14, MD2, and the NF-κB-luciferase reporter construct. Cells were then stimulated for 6 h and reporter activities were measured (Fig. 8A). Cells transfected with empty vector exhibited a 1.4 ± 0.03-fold induction of reporter activity, indicating that the 293H cells possess a modest, intrinsic propensity for NF-κB activation by HKEC. Cells transfected with tlr4a exhibited no significant induction of NF-κB activation (1.1 ± 0.33-fold activation). Similar to the empty vector control, cells overexpressing Tlr4b exhibited modest activation of the reporter (1.5 ± 0.06-fold). In combination, stimulation of Tlr4a and Tlr4b led to a 1.3 ± 0.10-fold induction of NF-κB activity. In contrast, stimulation of mouse TLR4 resulted in a 2.9 ± 0.07-fold-induction of NF-κB activity. HKEC has been shown to stimulate a wide variety of mammalian TLRs, particularly TLR2, TLR5, TLR9, and possibly TLR4 (38-42). Because of this diverse interaction potential, we sought to determine if zebrafish Tlr4a and Tlr4b could be stimulated by HKLP. We overexpressed Tlr4a and Tlr4b alone or in combination, along with an empty vector control, a mTLR4 negative control, and a mTLR2 positive control. Cells were exposed to HKLP for 6 h and then NF-κB-luciferase reporter activities were recorded (Fig. 8B). The cells expressing mTLR4 exhibited 1.1 ± 0.05 activation of the NF-κB-luciferase reporter upon exposure to HKLP. The cells expressing mTLR2 were robustly activated by HKLP, with a 20.9 ± 0.92 induction of reporter expression. None of the Tlr proteins exhibited a capacity to be stimulated by HKLP (empty vector: 1.1 ± 0.05-fold; Tlr4a: 0.9 ± 0.05-fold; Tlr4b: 1.2 ± 0.07-fold; and Tlr4a + Tlr4b: 1.0 ± 0.08-fold.). These data further support the idea that Tlr4a and Tlr4b possess alternative ligand specificities, including the possibility that they do not function as stand-alone, homodimeric receptors.
Figure 8. Zebrafish Tlr4a and Tlr4b are unresponsive to stimulation by heat-killed E. coli or L. pneumophila.
Constructs encoding zebrafish Tlr4a and Tlr4b, alone and in combination, were overexpressed in 293H cells along with CD14, MD2, pBIIx-luc, and pRL-CMV. For HKEC exposure, empty vector was transfected as a negative control, and plasmid encoding mTLR4 was transfected as a positive control. For HKLP exposure, empty vector and plasmid encoding mTLR4 were transfected as negative controls, and plasmid encoding mTLR2 was transfected as a positive control. Following transfection, cells were exposed to (A) HKEC (106 CFU · mL-1) or (B) HKLP (108 CFU · mL-1) for 6 h and then lysed. Firefly and Renilla relative luminescence units were recorded. The data are presented as the normalized mean fold induction of NF-κB luciferase reporter activity ± SEM.
Discussion
Zebrafish Tlr pathways consist of evolutionarily-conserved and –divergent parts. We and others have previously shown that many, but not all, of the components present in the mammalian TLR signal transduction pathways are present in the zebrafish (17, 20, 21, 29, 43-45). For example, it appears that Myd88 (45) and Ticam1 (21) play essential roles in mediating anti-bacterial and anti-viral immunity, as they do in mammals. At this point, however, it has not been determined how the Tlr proteins identified in zebrafish respond to their environments. The temptation to presume that phylogenetic similarities equate to functional conservation needs to be tempered in light of the data we present herein. The data show that while zebrafish do respond to LPS stimulation, this activation is not dependent upon mechanisms homologous to the prototypical LPS receptor complexes observed in mammals, which require CD14 and involve TLR4 and MD2 as its core components. Our findings have been bolstered by Sepulcre et al. (16), who demonstrated, using a novel in vivo assay, that the knockdown of Tlr4a and Tlr4b expression, through a morpholino-mediated translation blocker and a morpholino-mediated splice blocker, did not disrupt the immune response by zebrafish to LPS exposure. Future studies will be directed towards identifying the role the zebrafish Tlr4 proteins play in the immune system. The failure of Tlr4a and Tlr4b to respond to LPS, HKEC, and HKLP allows for broad speculation regarding their roles. It may be possible that they respond to heretofore unidentified pathogen-associated molecular patterns of bacterial, fungal, protistan, helminthic, and/or viral origins. It is also possible that Tlr4a and Tlr4b contribute to the ligand-recognizing diversity of other zebrafish Tlr proteins by functioning as co-receptors. This particular possibility is reminiscent of what is observed with mammalian TLR1 and TLR6, which heterodimerize with TLR2 to broaden the diversity and specificity of TLR2 signaling to include responses to diacyl (TLR2:TLR6) and triacyl (TLR2:TLR1) lipoproteins (46, 47). In zebrafish, Tlr4a and Tlr4b may lack the capacity to function as “stand-alone” receptors and thus may operate in this manner. It is also possible that the zebrafish Tlr4s may behave as “decoy” receptors, modulating the activation of other TLR pathways although the capacity for the mTLR4-Tlr4a and mTLR4-Tlr4b chimeric proteins to be stimulated by LPS (Fig. 7) more than likely precludes this notion (48).
Another caveat that may need explanation in order to ascertain the true function of Tlr4a and Tlr4b relates to the role adaptor proteins play in their activation. In the zebrafish genome, orthologs of CD14 and MD2 are not observed (34). A homolog of MD1 (also known as LY86) is present, but MD1 protein is typically associated with the non-TLR CD180 (also known as RP105) receptor. MD1 facilitates the role CD180 plays as a negative regulator of TLR4 signaling. Phylogenetic analyses reveal that despite the absence of a TIR domain, CD180 and TLR4 are more closely related to each other than any other TLR is to TLR4 (34, 49). The relative phylogenetic closeness of CD180 to TLR4 makes it possible that perhaps in the zebrafish, Cd180 and Tlr4a or Tlr4b share Md1 in order to function. Future experiments aimed at establishing a definitive role for Cd180 and the Tlr4s in the zebrafish immune system will be designed to address this specific issue. In addition, a zebrafish gene showing identity with mammalian LBP also has strong similarity to the mammalian BPI (34), an LPS binding protein with antimicrobial activity (50). The unclear origins of this gene and the lack of functional data confound the role of this LBP/BPI protein homolog in both LPS recognition (associated with mammalian LBP function) and antimicrobial activity (associated with mammalian BPI function). With this in mind, it has become apparent that strong consideration for additional adaptor molecules in any zebrafish Tlr signaling complex must be considered to correctly and comprehensively characterize a particular Tlr function. In this regard, ectopic expression of tlr4a and tlr4b in ZFL cells, which we have used in previous Tlr pathway functional studies (20, 21, 44), may not allow for the full exploration of Tlr4a and Tlr4b function. It may be that heretofore unrecognized but essential adaptor proteins not present in these cells may be required for Tlr4a and Tlr4b to achieve functional competence. The derivation of alternative zebrafish cell lines, perhaps expressing Tlr4a and/or Tlr4b, and the utilization of gene knockdown strategies including morpholinos and/or RNAi may be required to address these functional questions in vitro. In addition, novel in vivo approaches, like the one utilized by Sepulcre et al. (16), may prove useful in the ultimate characterization of these receptors.
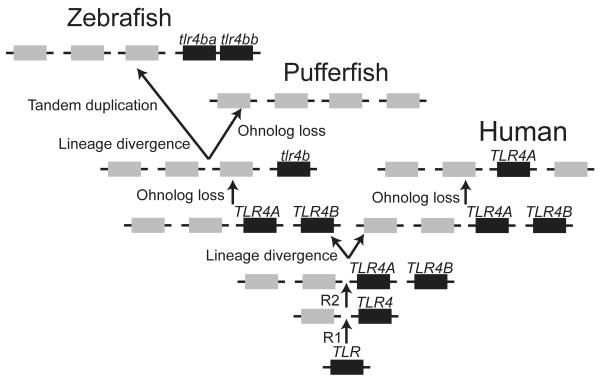
Results from the analysis of conserved syntenies are as expected by the hypothesis that the zebrafish tlr4 genes are paralogous, not orthologous to human TLR4 (Fig. 5). This contrasts with the data from phylogenetic analysis, which shows tlr4a and tlr4b falling clearly in the clade with mammalian and bird Tlr4 genes (Fig. 3). The following gene history model can account for these results (Fig. 9). It is likely that two rounds of whole genome duplication (R1 and R2) occurred at the base of the vertebrate radiation (23, 51). These events produced paralogons that appear today in the human genome, generally four copies of each paralogon, but occasionally five if a translocation subsequently divided one of the original four paralogons, as apparently happened with the Hsa2p, Hsa4, Hsa5q, Hsa8p, and Hsa10 paralogon shown in Fig. 5A. Paralogous genes derived from these genome duplications are called ohnologs (52, 53). After the R1 and R2 duplication events, genes were gradually lost, and sometimes, different lineages lost different ohnologs (reviewed in 54). Our historical model suggests that the last common ancestor of zebrafish and human had a precursor of vertebrate TLR4 genes present on at least two of the paralogous chromosomes from the R1 and R2 genome duplication events. The model imagines that the Hsa10q member of the paralogon lost the TLR4 ohnolog in the human lineage, and different member of the paralogon lost its TLR4 ohnolog in the zebrafish lineage. Thus, the gene in today's human genome that is most similar to tlr4a and tlr4b would be TLR4, as the phylogenetic analysis shows, although these two genes would not be orthologs (genes in two different species descended from a single gene in the last common ancestor of two species). Instead, the tlr4 genes of zebrafish and the TLR4 gene of humans are descended from a single gene in a distant common ancestor just after the R2 genome duplication event. A translocation is required to explain why TLR4 is on Hsa9, which otherwise has few genes paralogous to those in the Hsa10, Hsa2p, Hsa4, Hsa5q, Hsa8p paralogon. The loss of the tlr4 gene in the pufferfish lineage after it diverged from the zebrafish lineage, and a tandem gene duplication of tlr4 to form tlr4a and tlr4b after the divergence of the human and zebrafish lineage can help explain the current genomic situation. The strong conclusion from the analysis of conserved syntenies is that the zebrafish genes called tlr4a and tlr4b are not orthologs of the human TLR4 gene, but represent a reciprocal loss of ohnologs derived from the second round of genome duplication at about the time of the vertebrate radiation.
Figure 9. A model for the historical relationships of human and zebrafish TLR4-related genes.
Zebrafish tlr4 genes arose from a common ancestral TLR4 gene. Following two rounds of genome duplication, TLR4A and TLR4B appeared. With lineage divergence, there was reciprocal loss of TLR4 ohnologs, leading to the appearance of the TLR4A gene now present in the mammalian genome (known as TLR4) and tlr4b in the zebrafish/pufferfish ancestor. Following an additional ohnolog loss, the tlr4b gene disappeared from the pufferfish lineage. In contrast, the tlr4b gene was duplicated in the zebrafish lineage, leading to the appearance of tlr4ba (currently known as tlr4a) and tlr4bb (currently known as tlr4b).
Our conclusion about the historical origins of tlr4 and TLR4 raises questions regarding nomenclature. The names tlr4 and TLR4 suggest orthology, which is misleading. If both ohnologs had been retained in the human lineage, they might have been called TLR4A and TLR4B, analogous to the nomenclature for the tlr4 neighbors MAT2A and MAT2B, occupants of the Hsa2p and Hsa5q members of the paralogon, or EIF4E1 and EIF4E1B, occupants of the Hsa10q and Hsa5q members of the paralogon shown in Fig. 5A. The implication of orthology can be avoided where none exists by calling the zebrafish genes tlr4ba and tlr4bb, and the human gene TLR4A (Fig. 9).
Supplementary Material
Acknowledgments
The authors wish to thank Akshata Nayak for technical assistance. This is Maine Agricultural and Forest Experiment Station publication 3012.
Footnotes
The project described was supported in part by Grant Number R15AI049237-02 to C.S., C.R.L., and C.H.K. from the National Institute for Allergy and Infectious Disease (NIAID), Grant Number R01 RR10715 to J.H.P. from the National Center for Research Resources (NCRR), and Grant Number R15AI065509-01 to P.J.M. from NIAID. All institutes are components of the National Institutes of Health (NIH) and its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Abbreviations used in this paper: TIR, Toll/IL-1R; d post-fertilization (dpf); h post-exposure (hpe); mx, myxovirus (influenza virus) resistance gene; ZFL, zebrafish liver cells; SHRV, snakehead rhabdovirus; HKEC, heat-killed E. coli; HKLP, heat-killed L. pneumophila; TICAM, TIR domain-containing adaptor molecule.
Disclosures: The authors have no financial conflict of interest.
References
- 1.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 2.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci U S A. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 4.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 5.Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, Thompson P, Chan EK, Ledesma J, Roe B, Clifton S, Vogel SN, Beutler B. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 6.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 7.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 8.Rhee SH, Hwang D. Murine TOLL-like receptor 4 confers lipopolysaccharide responsiveness as determined by activation of NF kappa B and expression of the inducible cyclooxygenase. J Biol Chem. 2000;275:34035–34040. doi: 10.1074/jbc.M007386200. [DOI] [PubMed] [Google Scholar]
- 9.Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, Tobias PS, Ulevitch RJ. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 10.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 11.Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, Lee H, Lee JO. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J Biol Chem. 2005;280:11347–11351. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]
- 12.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 13.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 15.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci U S A. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sepulcre MP, Alcaraz-Perez F, Lopez-Munoz A, Roca FJ, Meseguer J, Cayuela ML, Mulero V. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- 17.Meijer AH, Gabby Krens SF, Medina Rodriguez IA, He S, Bitter W, Snaar-Jagalska B Ewa, Spaink HP. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. 2004;40:773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Phelan PE, Pressley ME, Witten PE, Mellon MT, Blake S, Kim CH. Characterization of snakehead rhabdovirus infection in zebrafish (Danio rerio) J Virol. 2005;79:1842–1852. doi: 10.1128/JVI.79.3.1842-1852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbs PD, Schmale MC. GFP as a Genetic Marker Scorable Throughout the Life Cycle of Transgenic Zebra Fish. Mar Biotechnol (NY) 2000;2:107–125. doi: 10.1007/s101269900014. [DOI] [PubMed] [Google Scholar]
- 20.Phelan PE, Mellon MT, Kim CH. Functional characterization of full-length TLR3, IRAK-4, and TRAF6 in zebrafish (Danio rerio) Mol Immunol. 2005;42:1057–1071. doi: 10.1016/j.molimm.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan C, Postlethwait JH, Lage CR, Millard PJ, Kim CH. Evidence for evolving Toll-IL-1 receptor-containing adaptor molecule function in vertebrates. J Immunol. 2007;178:4517–4527. doi: 10.4049/jimmunol.178.7.4517. [DOI] [PubMed] [Google Scholar]
- 22.Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, Hudson TJ, Zon LI, McPherson JD, Chevrette M, Dawid IB, Johnson SL, Ekker M. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci U S A. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catchen JM, Conery JS, Postlethwait JH. Inferring Ancestral Gene Order. In: Keith J, editor. Methods in Molecular Biology: Bioinformatics. Vol. 568. Humana Press; Totowa: 2008. [DOI] [PubMed] [Google Scholar]
- 25.Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- 26.Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, Shahab A, Zhao XD, Hibberd M, Wei CL, Lim B, Ng HH, Ruan Y, Chin KC. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-kappaB upon TLR4 activation. Mol Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 27.Covert MW, Leung TH, Gaston JE, Baltimore D. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309:1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 28.Pressley ME, Phelan PE, 3rd, Witten PE, Mellon MT, Kim CH. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev Comp Immunol. 2005;29:501–513. doi: 10.1016/j.dci.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Altmann SM, Mellon MT, Johnson MC, Paw BH, Trede NS, Zon LI, Kim CH. Cloning and characterization of an Mx gene and its corresponding promoter from the zebrafish, Danio rerio. Dev Comp Immunol. 2004;28:295–306. doi: 10.1016/j.dci.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 32.Nishitani C, Mitsuzawa H, Hyakushima N, Sano H, Matsushima N, Kuroki Y. The Toll-like receptor 4 region Glu24-Pro34 is critical for interaction with MD-2. Biochem Biophys Res Commun. 2005;328:586–590. doi: 10.1016/j.bbrc.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 33.Nishitani C, Mitsuzawa H, Sano H, Shimizu T, Matsushima N, Kuroki Y. Toll-like receptor 4 region Glu24-Lys47 is a site for MD-2 binding: importance of CYS29 and CYS40. J Biol Chem. 2006;281:38322–38329. doi: 10.1074/jbc.M606904200. [DOI] [PubMed] [Google Scholar]
- 34.Iliev DB, Roach JC, Mackenzie S, Planas JV, Goetz FW. Endotoxin recognition: in fish or not in fish? FEBS Lett. 2005;579:6519–6528. doi: 10.1016/j.febslet.2005.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohnishi T, Muroi M, Tanamoto K. MD-2 is necessary for the toll-like receptor 4 protein to undergo glycosylation essential for its translocation to the cell surface. Clin Diagn Lab Immunol. 2003;10:405–410. doi: 10.1128/CDLI.10.3.405-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- 37.Schneider DS, Hudson KL, Lin TY, Anderson KV. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 38.Newton CA, Perkins I, Widen RH, Friedman H, Klein TW. Role of Toll-like receptor 9 in Legionella pneumophila-induced interleukin-12 p40 production in bone marrow-derived dendritic cells and macrophages from permissive and nonpermissive mice. Infect Immun. 2007;75:146–151. doi: 10.1128/IAI.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akamine M, Higa F, Arakaki N, Kawakami K, Takeda K, Akira S, Saito A. Differential roles of Toll-like receptors 2 and 4 in in vitro responses of macrophages to Legionella pneumophila. Infect Immun. 2005;73:352–361. doi: 10.1128/IAI.73.1.352-361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, Skerrett SJ, Beutler B, Schroeder L, Nachman A, Ozinsky A, Smith KD, Aderem A. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J Exp Med. 2003;198:1563–1572. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawn TR, Smith KD, Aderem A, Skerrett SJ. Myeloid differentiation primary response gene (88)- and toll-like receptor 2-deficient mice are susceptible to infection with aerosolized Legionella pneumophila. J Infect Dis. 2006;193:1693–1702. doi: 10.1086/504525. [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi T, Kobayashi T, Gomi K, Suzuki T, Tokue Y, Watanabe A, Nukiwa T. Dendritic cells pulsed with live and dead Legionella pneumophila elicit distinct immune responses. J Immunol. 2004;172:1727–1734. doi: 10.4049/jimmunol.172.3.1727. [DOI] [PubMed] [Google Scholar]
- 43.Jault C, Pichon L, Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol Immunol. 2004;40:759–771. doi: 10.1016/j.molimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Altmann SM, Mellon MT, Distel DL, Kim CH. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J Virol. 2003;77:1992–2002. doi: 10.1128/JVI.77.3.1992-2002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Sar AM, Stockhammer OW, van der Laan C, Spaink HP, Bitter W, Meijer AH. MyD88 innate immune function in a Zebrafish embryo infection model. Infect Immun. 2006;74:2436–2441. doi: 10.1128/IAI.74.4.2436-2441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 47.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 48.Mantovani A, Locati M, Polentarutti N, Vecchi A, Garlanda C. Extracellular and intracellular decoys in the tuning of inflammatory cytokines and Toll-like receptors: the new entry TIR8/SIGIRR. J Leukoc Biol. 2004;75:738–742. doi: 10.1189/jlb.1003473. [DOI] [PubMed] [Google Scholar]
- 49.Divanovic S, Trompette A, Atabani SF, Madan R, Golenbock DT, Visintin A, Finberg RW, Tarakhovsky A, Vogel SN, Belkaid Y, Kurt-Jones EA, Karp CL. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat Immunol. 2005;6:571–578. doi: 10.1038/ni1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray PW, Flaggs G, Leong SR, Gumina RJ, Weiss J, Ooi CE, Elsbach P. Cloning of the cDNA of a human neutrophil bactericidal protein. Structural and functional correlations. J Biol Chem. 1989;264:9505–9509. [PubMed] [Google Scholar]
- 51.Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 52.Wolfe K. Robustness--it's not where you think it is. Nat Genet. 2000;25:3–4. doi: 10.1038/75560. [DOI] [PubMed] [Google Scholar]
- 53.Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 54.Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zoolog B Mol Dev Evol. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.