Abstract
Elucidation of the metabolic pathways of triacylglycerol (TAG) synthesis is critical to the understanding of chronic metabolic disorders such as obesity, cardiovascular disease, and diabetes. sn-Glycerol-3-phosphate acyltransferase (GPAT) and sn-1-acylglycerol-3-phosphate acyltransferase (AGPAT) catalyze the first and second steps in de novo TAG synthesis. AGPAT6 is one of eight AGPAT isoforms identified through sequence homology, but the enzyme activity for AGPAT6 has not been confirmed. We found that in liver and brown adipose tissue from Agpat6-deficient (Agpat6−/−) mice, N-ethylmaleimide (NEM)-sensitive GPAT specific activity was 65% lower than in tissues from wild-type mice, but AGPAT specific activity was similar. Overexpression of Agpat6 in Cos-7 cells increased an NEM-sensitive GPAT specific activity, but AGPAT specific activity was not increased. Agpat6 and Gpat1 overexpression in Cos-7 cells increased the incorporation of [14C]oleate into diacylglycerol (DAG) or into DAG and TAG, respectively, suggesting that the lysophosphatidic acid, phosphatidic acid, and DAG intermediates initiated by each of these isoforms lie in different cellular pools. Together, these data show that “Agpat6−/− mice” are actually deficient in a novel NEM-sensitive GPAT, GPAT4, and indicate that the alterations in lipid metabolism in adipose tissue, liver, and mammary epithelium of these mice are attributable to the absence of GPAT4
Supplementary key words: triacylglycerol, phospholipid, lipodystrophy, acyl-coenzyme A, steatosis, sn-l-acylglycerol-3-phosphate O-acyltransferase-deficient mice
The regulation of triacylglycerol (TAG) synthesis and metabolism plays an important role in whole body energy homeostasis in mammals, and dysregulation of TAG synthesis and oxidation pathways has been implicated in the pathogenesis of obesity, lipodystrophy, cardiovascular disease, insulin resistance, and type 2 diabetes (1). De novo TAG synthesis begins with the formation of lysophosphatidic acid (LPA) through the acylation of the sn-1 position of glycerol-3-phosphate by sn-glycerol-3-phosphate acyltransferase (GPAT) (2). The second enzyme in de novo TAG synthesis, sn-1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), acylates LPA to form phosphatidic acid (PA). Phosphatidic acid phosphohydrolase (also called lipin) converts PA to diacylglycerol (DAG) (3, 4), and diacylglycerol acyltransferase acylates DAG to form TAG. PA and DAG are also precursors for the glycerophospholipids.
Three mammalian GPAT isoforms have been cloned (5–7). GPAT1 and GPAT2 are located in the outer mitochondrial membrane, whereas GPAT3 is located in the endoplasmic reticulum. GPAT1 is resistant to inactivation by sulfhydryl agents like N-ethylmaleimide (NEM), prefers palmitoyl-CoA as a substrate compared with oleoyl-CoA, and plays a regulatory role in liver TAG synthesis (2). GPAT2 is NEM-sensitive, has no preference for palmitoyl-CoA, and is highly expressed in rodent testis (8, 9). Recently, the gene product previously called AGPAT8 in the National Center for Biotechnology Information database was shown to have NEM-sensitive GPAT activity and to lack AGPAT activity; this gene product was renamed GPAT3 (7). Because small interfering RNA knockdown of Gpat3 in 3T3-L1 adipocytes reduces total GPAT activity by ∼60% (7, 10), it is likely that GPAT3 is important for TAG synthesis in white adipose tissue (WAT). However, GPAT3 is unlikely to play a major role in TAG synthesis in liver, where its mRNA expression is very low (7, 10). In liver, GPAT1 accounts for 30–50% of the total GPAT activity (2). The remaining hepatic GPAT activity is NEM-sensitive and microsomal, suggesting that one or more additional microsomal NEM-sensitive GPAT isoforms must exist.
Eight genes with predicted amino acid sequence similarities to GPAT1 have been named AGPAT isoforms 1–8. AGPAT1 and -2 are well characterized and have high AGPAT activity (11, 12). Mutations in AGPAT2 cause generalized human congenital lipodystrophy, with absent abdominal and subcutaneous adipose tissue, systemic insulin resistance, and hypertriglyceridemia (13). Compared with AGPAT1 and -2, the AGPAT activities reported for AGPAT3 to -5 are extremely low (14); no enzyme activity has been reported for AGPAT6 or -7.
We previously characterized mice with a targeted deletion in Agpat6 (Agpat6−/− mice; NP_848934) (10, 15). These mice have a subdermal lipodystrophy, resistance to high-fat diet-induced obesity, and a 50% reduction of TAG content in liver, adipose tissue, and mammary epithelium, suggesting an important role for AGPAT6 in TAG synthesis in these tissues. AGPAT6 shares amino acid sequence homology with other glycerolipid acyltransferases at conserved motifs I–V (Table 1) and is closely related to GPAT1 and -2 in motifs I–IV, which were identified as important for acyltransferase catalysis and glycerol-3-phosphate binding domains (16). AGPAT6 shares 66% amino acid homology with “AGPAT8,” recently identified as GPAT3 (7), and confocal microscopy shows that AGPAT6 localizes to the endoplasmic reticulum (15). These observations raised the possibility that AGPAT6, like AGPAT8, is in fact a GPAT enzyme. Here, we demonstrate that expression of this enzyme in Cos-7 cells increases GPAT, but not AGPAT, activity. Furthermore, tissues from Agpat6−/− mice show reduced GPAT activity but normal AGPAT activity. Our studies thus establish that Agpat6 encodes the primary microsomal NEM-sensitive GPAT activity in liver, GPAT4.
TABLE 1.
Acyltransferase motifs in Escherichia coli and mouse AGPAT and GPAT isoforms
| Gene | Motif I | Motif II | Motif III | Motif IV | |
|---|---|---|---|---|---|
| Function | Catalysis | Glycerol-3-phosphate binding | Glycerol-3-phosphate binding | Catalysis | Motif V |
| AGPAT1 (NP_061350) | 104 HQSSLD | 140 GIIFIDR | 173 FPEGTRNH | 200 VPIIPIVNSSY | 231 VLPPVSTEGLTPDD |
| AGPAT2 (NP_080488) | 98 HQSLID | 137 GVYFINR | 170 YPEGTRND | 197 VPIIPVVYSSF | 227 VLDAVPTNGLTDAD |
| AGPAT6 (GPAT4a) (NP_061213) | 248 HTSPID | 286 PHVWFER | 320 FPEGT1CIN | 343 ATVYPVAIKYD | 385 VWYLPPMTREKDED |
| AGPAT8 (GPAT3b) (NP_766303) | 229 HRTRVD | 268 VHIFIHR | 301 FPEGTCLT | 324 GTIYPVAIKYN | 366 VWYMPPMTREEGED |
| E. coli GPAT (plsB) (BAE78043) | 306 HRSHMD | 349 GAFFIRR | 383 FVEGGRSR | 417 ITLIPIYIGYE | |
| GPAT1c (NP_032175) | 230 HRSHID | 272 GGFFIRR | 313 FlEGTRSR or | 347 ILVIPVGISYD | |
| GPAT2d (NP_001074558) | 202 HKSLLD | 231 TCSPALR or 235 ALRALLR or 247 LGGFLPP(5) | 291 GSPGRLSA or 285 F1EEPPGS(5) | 320 ATLVPVAIAYD |
AGPAT, sn-1-acylglycerol-3-phosphate O-acyltransferase; GPAT, sn-glycerol-3-phosphate acyltransferase. Acyltransferase motifs are as described (16), except for motif V, which is as described in Ref. 15. Boldface residues are highly conserved across these GPAT family members, and underlined amino acids are important for GPAT1 activity as determined by mutagenesis (2, 16). Cysteines that may be targets for N-ethylmaleimide are boldface and italicized.
Identified in GenBank as AGPAT6, but it has only GPAT activity as reported here.
Identified in GenBank as AGPAT8, but it has only GPAT activity (7).
Previously called mtGPAT or mtGAT.
Also identified as xGPAT1 (5).
Methods
Agpat6−/− mice
Agpat6−/− mice were generated from a gene-trap cell line identified in BayGenomics, a gene-trapping resource (10, 15). This strain was back-crossed to C57BL/6J for at least four generations. The mice were housed with a 12 h light/12 h dark cycle and fed a chow diet containing 4.5% fat (Ralston Purina, St. Louis, MO). All animal protocols were approved by the institutional animal care and use committee. Tissues were harvested from 6 month old mice, snap-frozen in liquid nitrogen, and stored at −80°C. A previous characterization of the Agpat6−/− mice revealed that male and female mice had similar phenotypes in adipose tissue and liver (10, 15), and both sexes were used in the studies described here.
Cloning of Agpat6/Gpat4
Agpat6/Gpat4 was amplified by PCR from mouse liver RNA at 57.5°C with Pfx50 (Invitrogen) using a forward primer (5′-caccggatccacgtgcgtcctccaccat) containing a BamHI restriction site (boldface) and a reverse primer (5′-ccctctagactacttgtcgtcatcgtccttgtagtcggaccggctgcggtcct) containing a Flag epitope sequence (underlined) and an XbaI restriction site (boldface). The amplification product was digested with BamHI and XbaI and ligated into pcDNA3.1 mammalian expression vector (Invitrogen).
Nucleofection of Cos-7 cells with Agpat6/Gpat4
Cos-7 cells were grown in DMEM high glucose (Gibco) with 10% FBS and 1% penicillin/streptomycin. The cells were grown to confluence and passaged at 1 day before nucleofection. Cells were harvested, and 4.0 × 106 cells were nucleofected with 12 μg of Agpat6/Gpat4, Gpat1, Agpat2, or empty vector pcDNA3.1 plasmid DNA in 100 μl of Nucleofector™ Kit V Solution using the Nucleofector™ system (AMAXA Biosystems). Cells were then transferred to 100 mm cell culture dishes containing RPMI, 10% FBS, and 1% penicillin/streptomycin and harvested 18 h later. HepG2 cells were nucleofected (AMAXA Kit V program H-22) with pcDNA3.1 (vector) or pcDNA3.1-GPAT4-Flag and allowed to reattach to the dishes. After 12 h, the medium (DMEM-H, 10% FBS, and 1% penicillin/streptomycin) was removed and replaced with medium containing 100 μM oleate complexed with 0.5% BSA. Cells were harvested at 12 h after adding oleate.
Membrane isolation and measurement of GPAT activity
Liver or adipose tissue was homogenized in medium I + DTT (250 mM sucrose, 10 mM Tris, pH 7.4, 1 mM EDTA, and 1 mM DTT) with 10 up-and-down strokes in a Teflon-glass motor-driven homogenizer. Homogenates were centrifuged at 100,000 g for 1 h to obtain total membrane fractions. The membrane pellet was rehomogenized in medium I + DTT and stored in 100 μl aliquots at −80°C. GPAT specific activity was assayed at 23° C for 10 min in a 200 −μl reaction mixture containing 75 mM Tris-HCl, pH 7.5, 4 mM MgCl2, 2 mg/ml BSA (essentially FA-free), 1 mM DTT, 8 mM NaF, 300 μM [3H]glycerol-3-phosphate, and 80 μM palmitoyl-CoA (17). The reaction was initiated by adding 2.5–7.5 μg (WAT), 5–15 μg [brown adipose tissue (BAT)], or 10–30 μg (liver) of total membrane protein to the assay mixture after incubating the membrane protein on ice for 15 min in the presence or absence of 1 mM NEM. These assays measured initial rates. The reaction was stopped by adding 1% perchloric acid, and the products were extracted into chloroform (18). NEM-sensitive activity is calculated as the total GPAT activity minus the GPAT activity that is not inhibited by NEM. For nucleofected Cos-7 cells, culture dishes were scraped with medium I + DTT and the samples were homogenized and centrifuged as described above to obtain total membrane fractions. GPAT specific activity was measured as above with 5–15 μg of total Cos-7 membrane protein. To identify the products from the GPAT assay, the chloroform extracts were spotted on a Silica H thin-layer chromatography plate (Analtech) together with authentic standards (Avanti) and developed in CHCl3/pyridine/formic acid (88%) (50:30:7, v/v). The plates were exposed to iodine vapor, and the bands corresponding to LPA, PA, and DAG were scraped into scintillation vials containing Cytoscint (ICN) and counted. To determine acyl-CoA preference, 60 μM of each of several acyl-CoA species and 10–20 μg of protein were used in the GPAT assay.
AGPAT assay
AGPAT specific activity was assayed at 37°C in a 100 μl mixture containing 100 mM HEPES-NaOH, pH 7.5, 200 mM NaCl, 10 mM EDTA, 8 mM NaF, 1 mg/ml BSA (fatty acid-free), 1 mM DTT, 5% (w/v) glycerol, 40 μM oleoyl-CoA, and 20 μM l-[3H]oleoyl-LPA (1 μCi; Perkin-Elmer NET-1100). The reaction was initiated by adding 5 μg of total membrane protein to the assay mixture. Fifteen microliters of the reaction mixture was removed at the indicated times up to 10 min and spotted directly onto a Silica H plate together with authentic standards. The radiolabeled PA product was separated from the radiolabeled LPA substrate in chloroform-pyridine-formic acid (50:30:7, v/v). Authentic lipid standards for LPA and PA were visualized by exposure to iodine vapor. Spots corresponding to PA were scraped into 500 μl of methanol-water (1:1), Cytoscint (ICN) was added, and radioactivity was quantified.
Cellular TAG content
Cells were transfected and plated on 100 mm plates as described above. Ten hours after cells were plated, the medium was removed and growth medium (RPMI, 10% FBS, 1% penicillin/streptomycin, and 0.5% BSA) with or without 100 μM oleic acid was added. After a 14 h incubation with fatty acid, cells were washed twice with ice-cold PBS and scraped in 750 μl of distilled water. Cells were homogenized in water, and lipids were extracted into CHCl3 (18), dried in a SpeedVac, and resuspended in 100 μl of isopropanol and 1% Triton X-100 at room temperature for 1 h. The TAG content of 10–45 μl of the lipid sample was determined using an enzymatic colorimetric method (Stanbio Laboratory).
Cell labeling and lipid analysis
Cos-7 cells were plated on 60 mm plates and grown to 75% confluence in DMEM-H with 10% FBS and 1% penicillin/streptomycin. Cells were transfected with 4 μg of Agpat6/Gpat4, Gpat1, or vector DNA using Fugene 6 (Roche) in a 3:1 reagent-plasmid ratio. Seventeen hours later, the medium was removed and labeling medium (DMEM-H, 10% FBS, 0.5% BSA, and 1 mM carnitine) containing a trace amount of [14C] oleic acid (750,000 dpm in 2 ml) was added. After 3 h of incubation with the labeling medium, it was removed and the cells were washed twice in ice-cold PBS containing 10 mg/ml fatty acid-free BSA. Cells were scraped in 2 ml of ice-cold methanol and 0.5 ml of distilled water. Lipids were extracted and concentrated as described above. Cellular lipids were resolved on LK5D silica plates by thin-layer chromatography using either a solvent system for neutral lipids consisting of hexane-ethylether-acetic acid (80:20:2, v/v) or a solvent system consisting of chloroform-methanol-acetic acid-water (50:37.5:3.5:1.5, v/v). All samples were chromatographed in parallel with pure lipid standards. The 14C-labeled lipids were detected and quantified with the Bioscan 200 Image System.
Immunoblotting
Cell total membrane proteins were separated on an 8% polyacrylamide gel containing 1% SDS, transferred to a polyvinylidene difluoride membrane (Bio-Rad), and incubated with monoclonal antibody against the FLAG epitope (Clone M2; Sigma). For chemiluminescence detection, the immunoreactive bands were visualized by incubating the membrane with horseradish peroxidase-conjugated goat anti-mouse IgG and PicoWest reagents (Pierce) according to the manufacturer's instructions.
Gene expression analyses
Total RNA was extracted from mouse tissues with TRIzol (Invitrogen). Two micrograms of RNA was reverse-transcribed with oligo(dT) and random primers (Invitrogen). Five percent of the resulting cDNA was used for each real-time RT-PCR. Real-time RT-PCR was performed in duplicate with Quantitect SYBR Green PCR mix (Qiagen) in an iCycler Real-Time Detection System (Bio-Rad) (10, 19). Data presented were derived from starting quantity values normalized by the square root of the product of values obtained for the housekeeping genes β2-microglobulin and TATA box binding protein or to 18S rRNA Primer sequences used in this study have been described previously (19) or are shown in Table 2. Primers for Gpat3 were cited previously as Agpat8 (10).
TABLE 2.
Primer sequences used in real-time PCR experiments
| Gene | Forward | Reverse |
|---|---|---|
| β2-Microglobulin | 5′-cagcatggctcgctcggtgac | 5′-cgtagcagttcagtatgttcg |
| Gpat1 | 5′-agcaagtcctgcgctatcat | 5′-ctcgtgtgggtgattgtgac |
| Gpat2 | 5′-aagaaagaggtacagcgtatcc | 5′-gtggagagccctcctgcacag |
| Gpat3 | 5′-gtacatgcctcccatgactag | 5′-gatccgttgcccacgatcatc |
| 18S rRNA | 5′-accgcagctaggaataatgga | 5′-gcctcagttccgaaaacca |
Statistics
Data are shown as means ± SEM. Significance was established using Student's t-test (unpaired, two-tailed). Differences were considered significant at P < 0.05.
Results
Agpat6−/− mouse tissues have reduced NEM-sensitive GPAT activity
To determine whether tissues from Agpat6−/− mice have reduced GPAT activity, we measured GPAT activity in total membrane fractions from liver, BAT, and gonadal WAT in the presence and absence of NEM. Total GPAT activity was 49 % lower in liver from Agpat6−/− compared with wild-type mice (P < 0.001) (Fig. 1A). NEM-resistant activity (GPAT1) was similar in liver from knockout and wild-type mice (Fig. 1B), but the NEM-sensitive activity was reduced by 65% (P < 0.0001) (Fig. 1C). AGPAT activity was unchanged in Agpat6−/− mouse liver (Fig. 1D). In Agpat6−/− BAT, total GPAT specific activity was reduced by 50% (P< 0.01) and NEM-sensitive specific activity was reduced by 65% (P< 0.001) (Fig. 2A). However, GPAT specific activities were identical in gonadal WAT from wild-type and Agpat6−/− mice (Fig. 2C). AGPAT activity was unchanged in Agpat6−/− mouse WAT and BAT (Fig. 2B, D). Thus, liver and BAT tissue from Agpat6−/− mice exhibit normal AGPAT activity but reduced NEM-sensitive microsomal GPAT activity. Based on these results, we henceforth refer to the gene and enzyme affected in these mice as Gpat4 and GPAT4, respectively.
Fig. 1.
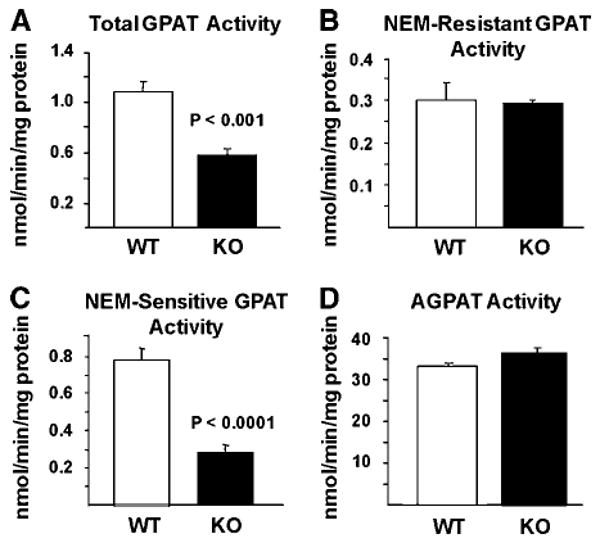
N-Ethylmaleimide (NEM)-sensitive sn-glycerol-3-phosphate acyltransferase (GPAT) specific activity is reduced in liver from sn-1-acylglycerol-3-phosphate O-acyltransferase-deficient (Agpat6−/−) mice. GPAT and AGPAT specific activity was measured in liver total particulate preparations from female mice (n = 6 for each genotype) as described in Methods. A: Total GPAT activity. B: NEM-resistant GPAT activity. C: NEM-sensitive GPAT activity. D: Total AGPAT activity. KO, knockout (Agpat6−/−); WT, wild type. Data are shown as means ± SEM.
Fig. 2.
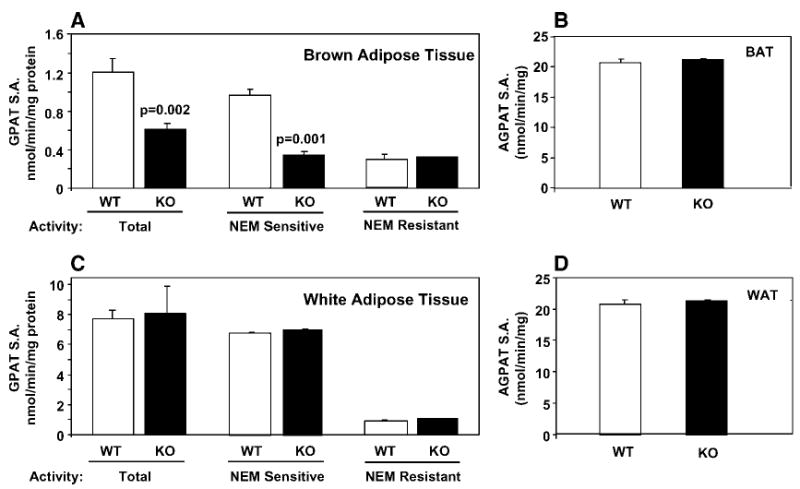
NEM-sensitive GPAT specific activity (S.A.) is reduced in brown adipose tissue (BAT) but not in gonadal white adipose tissue (WAT). Total, NEM-sensitive, and NEM-resistant GPAT specific activity was measured in BAT from male mice (A) and in gonadal WAT from female mice (C) (n = 6). AGPAT activity was measured in BAT from male mice (B) and in gonadal WAT from female mice (D) (n = 3). KO, knockout (Agpat6−/−); WT, wild type. Data are shown as means ± SEM.
Expression of Gpat1 to -3 mRNA transcripts in Agpat6−/− / Gpat4−/− mice
We previously demonstrated that BAT from Agpat6−/− / Gpat4−/− mice does not show altered expression of mRNA for Agpat1 to -5, -7, or -8 (10). To determine whether the deficiency of microsomal GPAT activity altered the mRNA expression of other GPAT isoforms, we measured the mRNA expression of Gpat1, -2, and -3 in liver, BAT, and inguinal WAT from Agpat6−/− / Gpat4−/− mice. Gpat1 expression was 2-fold higher in liver of the knockout mice (Table 3), but this increase in mRNA expression did not correspond to an increase in GPAT1 (NEM-resistant) activity (Fig. 1B). Agpat6−/− /Gpat4−/− and wild-type mice had similar expression levels for Gpat2 and Gpat3 in liver and for Gpat1, Gpat2, and Gpat3 in BAT and WAT.
TABLE 3.
mRNA levels of GPAT enzymes in wild-type and Agpat6−/− mouse tissues
| Liver | Brown Adipose Tissue | Inguinal White Adipose Tissue | ||||
|---|---|---|---|---|---|---|
| Enzyme | Wild Type | Agpat6−/− | Wild Type | Agpat6−/− | Wild Type | Agpat6−/− |
| Gpat1 | 0.20 ± 0.02 | 0.37 ± 0.1a | 0.73 ± 0.16 | 0.84 ± 0.23 | 0.07 ± 0.03 | 0.06 ± 0.05 |
| Gpat2 | 0.10 ± 0.04 | 0.08 ± 0.02 | 0.05 ± 0.002 | 0.03 ± 0.03 | 0.08 ± 0.08 | 0.11 ± 0.17 |
| Gpat3 | 0.64 ± 0.13 | 0.80 ± 0.19 | 1.41 ± 0.64 | 1.00 ± 0.19 | 2.05 ± 0.62 | 1.58 ± 1.24 |
P < 0.05 versus the wild type (n = 4).
Overexpression of Gpat4 in Cos-7 cells increases NEM-sensitive GPAT activity
To further investigate the enzyme activity encoded by Gpat4 in a heterologous cell system, we expressed Gpat4 cDNA tagged with a FLAG epitope in Cos-7 cells. Protein expression levels were monitored by Western blot using an anti-FLAG antibody (Fig. 3A). Cells transfected with the Gpat4 expression construct exhibited a 73% higher total GPAT activity and an 80% higher NEM-sensitive GPAT activity than cells transfected with vector (P< 0.0001 and P < 0.0001, respectively) (Fig. 3B). Overexpression of Gpat4 did not affect NEM-resistant GPAT activity, whereas overexpression of Gpat1 increased the NEM-resistant GPAT activity from a very low level to approximately one-third of total GPAT activity in the cell. Overexpression of Agpat2 in Cos-7 cells increased AGPAT activity by 3-fold compared with vector control cells but did not increase either NEM-sensitive or NEM-resistant GPAT activities (Fig. 3B, C). Cells transfected with Gpat4 or Gpat1 did not have increased AGPAT activity (Fig. 3C). The products of the GPAT assay were analyzed by thin-layer chromatography and showed a 2-fold increase in labeled LPA and a 70% increase in labeled PA in samples from Gpat4-transfected cells (Fig. 3D). Thus, overexpression of Gpat4 increased both NEM-sensitive GPAT activity and flux through the de novo glycerolipid synthetic pathway.
Fig. 3.
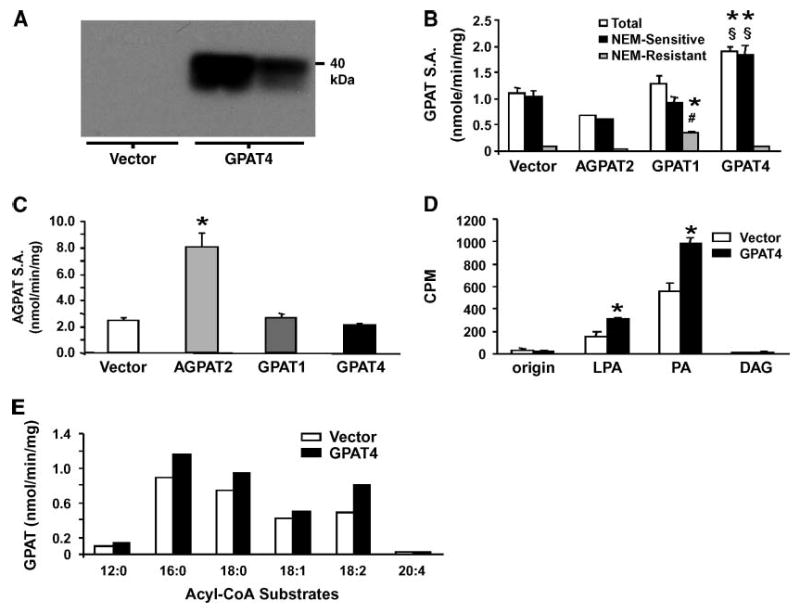
Overexpression of GPAT4 in Cos-7 cells increases NEM-sensitive GPAT activity. Cos-7 cells were transfected with pcDNA3.1 vector. Agpat2, Gpat1, or Gpat4. A: Immunoblot of Cos-7 cell total membrane fractions against the FLAG epitope (two independent transfections). B: GPAT specific activity (SA.) in Cos-7 total membrane fraction (vector, n = 13; GPAT4, n = 12; GPAT1, n = 4; AGPAT2, n = 2). * P < 0.0001 compared with vector; # P < 0.0001 compared with GPAT4; § P < 0.0001 compared with GPAT1. C: AGPAT specific activity in Cos-7 cell total membrane fraction (n = 3). * P < 0.01 compared with vector. D: Radiolabeled glycerolipid products from GPAT assays of Cos-7 cell total membranes were separated by TLC and quantified as described in Methods (average of two independent assays with three separate samples per assay). DAG, diacylglycerol; LPA, lysophosphatidic acid; PA, phosphatidic acid. * P < 0.05 compared with vector. E: Total GPAT activity in Cos-7 total membrane preparations with different acyl-CoA substrates (60 μM) (average of two independent experiments). Data are shown as means ± SEM.
To determine which acyl-CoA substrates GPAT4 prefers, we measured GPAT activity in Cos-7 cell total membranes from Gpat4-transfected cells in the presence of acyl-CoA substrates of varying chain length and saturation. Although able to use C12:0-CoA and C20:4-CoA, the highest GPAT4 activity was observed with acyl-CoA substrates having 16 or 18 carbons (Fig. 3E).
Overexpression of Gpat4 increases the incorporation of oleate into DAG
In CHO cells (20) and hepatocytes (21), GPAT1 increases TAG synthesis and storage. To determine whether Gpat4 overexpression alters TAG synthesis in Cos-7 cells, we measured the cellular TAG content of Cos-7 cells transfected with Gpat4, Gpat1, and the vector control. Cells transfected with Gpat1 had increased cellular TAG content compared with vector control cells (P = 0.02) (Fig. 4A). Surprisingly, the content of TAG in cells transfected with Gpat4 was similar to that of control cells, even though NEM-sensitive GPAT activity increased by >60%. To determine whether adding exogenous fatty acids to the medium would affect TAG synthesis in Cos-7 cells transfected with Gpat4, we treated Cos-7 and HepG2 cells with 100 μM oleic acid for 14 h. Compared with Cos-7 cells without added oleate, cellular TAG content was >6-fold higher in cells transfected with the vector control, with Gpat1, or with Gpat4, although the cellular TAG content did not differ significantly from the vector control value. In HepG2 cells incubated with 100 μM oleate, nucleofection with Gpat4 increased both NEM-sensitive GPAT activity and TAG content by 20% (Fig. 4B). These small increases reflect the difficulty of transfecting cultured hepatoma cells, and further studies with adenovirus vectors will be required to fully understand the role of GPAT4 in liver.
Fig. 4.
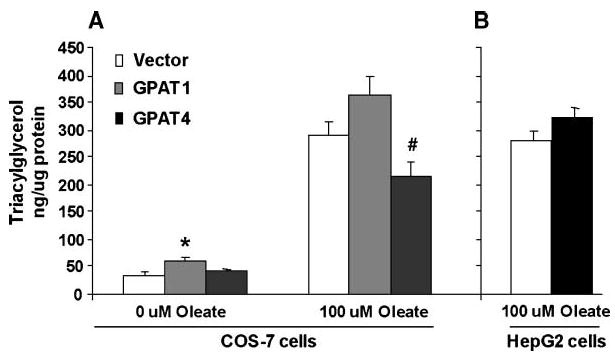
Overexpression of GPAT4 in Cos-7 and HepG2 cells. A: Cos-7 cells were transfected with Gpat1, Gpat4, or pcDNA3.1 for 10 h before a 14 h incubation with or without 100 μM oleic acid. In Cos-7 cells, GPAT activities increased as shown in Fig. 3B. In HepG2 cells, NEM-sensitive GPAT activity increased by 20% (P < 0.05) (0 μM oleate, n = 6–9; 100 μM oleate, n = 3). * P< 0.05 compared with vector; # P < 0.05 compared with GPAT1. B: HepG2 cells were transfected with Gpat4 or pcDNA3.1 for 12 h before a 12 h incubation with 100 μM oleic acid (n = 3). Data are shown as means ± SEM.
Because total cellular TAG content did not increase in Cos-7 cells that overexpressed Gpat4, we incubated Cos-7 cells with [14C] oleate to examine the role of GPAT4 in glycerolipid synthesis. In contrast to incubating cells with high concentrations of fatty acid, which are primarily incorporated into TAG, an incubation with trace (∼3 μM) oleate allows one to examine the relative incorporation into TAG, DAG, and phospholipids (20). As expected, transfection of Cos-7 cells with Gpat1 increased [14C] oleate incorporation into DAG and TAG by 2-fold (P < 0.0004) and 1.5-fold (P < 0.01), respectively (Fig. 5A, B). In contrast, overexpressing Gpat4 did not significantly alter label incorporation into TAG but increased [14C] oleate incorporation into DAG by 2-fold (P < 0.0002). The incorporation of [14C] oleate into total phospholipids and cholesteryl esters was not altered by overexpressing either Gpat1 or Gpat4 (Fig. 5C, D), but overexpression of Gpat4 increased the incorporation of [14C] oleate into phosphatidylinositol (P = 0.04) (Fig. 5E). Label incorporation into phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine did not change with overexpression of Gpat4, and overexpression of Gpat1 did not affect label incorporation into any phospholipid species (data not shown).
Fig. 5.
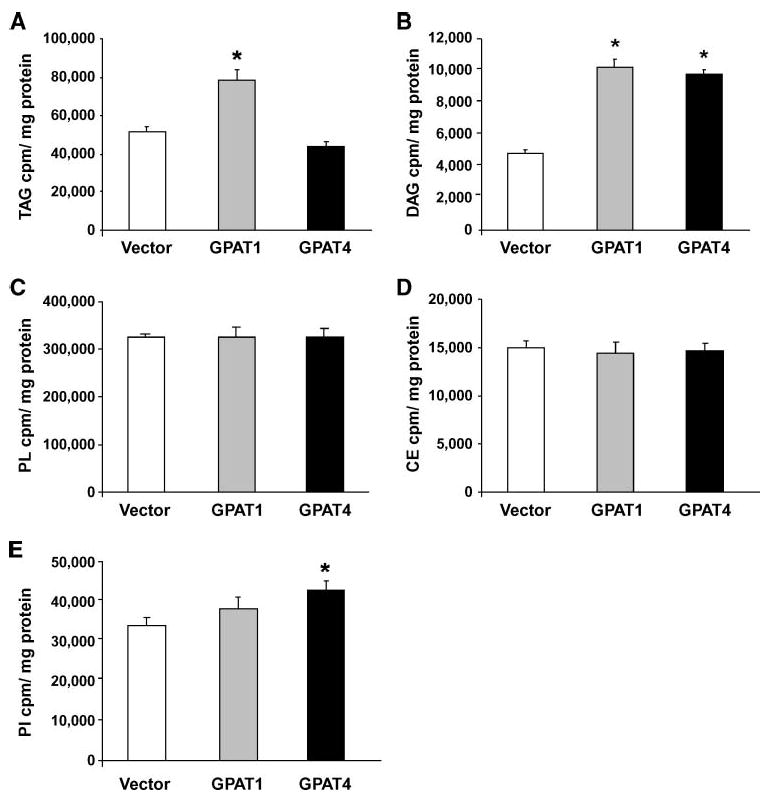
GPAT4 increases [14C]oleate incorporation into DAG in Cos-7 cells. Cos-7 cells were transfected with pcDNA3.1, Gpat1, or Gpat4 for 17 h and incubated with trace [14C]oleate for 3 h (n = 3). A: Label incorporation into cellular triacylglycerol (TAG). * P < 0.05 compared with vector. B: Label incorporation into cellular DAG. * P < 0.001 compared with vector. C: Label incorporation into total phospholipid (PL). D: Label incorporation into cholesteryl ester (CE). E: Label incorporation into phosphatidylinositol (PI). * P < 0.05 compared with vector. Data are shown as means ± SEM.
Discussion
Agpat6−/− mice have a subdermal lipodystrophy and reduced TAG content in BAT, WAT, liver, and mammary epithelium, suggesting that AGPAT6 plays an important role in TAG synthesis in these tissues (10, 15). Because AGPAT catalyzes the second step in de novo TAG synthesis, the phenotype of the Agpat6−/− mice was consistent with the idea that AGPAT6 catalyzes that second step. Based on protein size and amino acid sequence homology to AGPATs 1 and 2, mouse AGPAT6 had appeared to be an AGPAT, but as shown here, it is actually a GPAT. When expressed in Cos-7 cells, we observed an 80% increase in NEM-sensitive GPAT activity but no increase in AGPAT activity. It is unclear why previous expression constructs for this enzyme, which contained C-terminal V5 or V5-His epitope tags, failed to exhibit GPAT or AGPAT activity when expressed in insect or Cos-7 cells (15); it may be that these epitope tags interfered with enzyme action, whereas the FLAG epitope used here did not. Consistent with the true identity of AGPAT6 as a GPAT, the previously characterized Agpat6−/− mice have 65% lower NEM-sensitive GPAT specific activity in liver and BAT but no change in AGPAT specific activity. Thus, Agpat6 was misidentified and actually encodes an NEM-sensitive GPAT activity. As the fourth described GPAT, we have renamed it GPAT4. Based on previous localization studies of AGPAT6 (15), GPAT4 appears to represent a second endoplasmic reticulum resident GPAT (Fig. 6). Furthermore, because the Agpat6−/− mice are deficient in GPAT and not in AGPAT activity, they should be called Gpat4−/− mice.
Fig. 6.
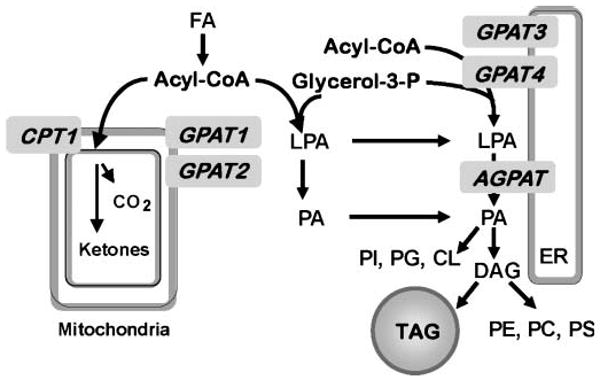
Relationship of GPAT4 to other enzymes in the pathway of de novo glycerolipid synthesis. GPAT isoforms located in mitochondria (GPAT1 and GPAT2) and endoplasmic reticulum (GPAT3 and GPAT4) catalyze the initial step in the synthesis of glycerophospholipids and TAG. AGPAT isoforms catalyze the second step in this pathway by acylating LPA to form PA. Acyl-CoA used for β-oxidation or TAG synthesis is regulated reciprocally by carnitine palmitoyl transferase-1 (CPT1) and GPAT1 (26). CL, cardiolipin; ER, endoplasmic reticulum; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PS, phosphatidylserine.
Gpat4 mRNA is highly expressed in liver (15) and accounts for almost 50% of the total GPAT activity in wild-type mouse liver, as demonstrated by the 49% reduction observed in Gpat4−/− mice. This loss in hepatic GPAT activity may be responsible for the 40–50% decrease in liver TAG reported for Gpat4−/− mice (10). GPAT4 represented 65% of liver NEM-sensitive GPAT specific activity, suggesting that GPAT4 is the major NEM-sensitive isoform in liver but that one or more additional NEM-sensitive GPAT isoforms is also present. To determine whether the mRNA expression of other GPAT isoforms increased in Gpat4−/− mouse liver to compensate for the loss of GPAT4, we measured the expression of Gpat1, -2, and -3. Neither Gpat2 nor Gpat3 mRNA expression was increased in liver from the Gpat4−/− mice, but Gpat1 mRNA was increased by 2-fold, although this increase did not correspond to an increase in NEM-resistant (GPAT1) GPAT specific activity. Gene expression does not invariably correspond to protein expression, and GPAT1 enzyme activity may be regulated by posttranslational mechanisms (22). We previously reported that the mRNAs for Agpat1 to -5, -7, and -8 did not change in BAT from the Gpat4−/− mice (10).
GPAT4 also appears to be the primary NEM-sensitive GPAT in BAT, as 65% of the NEM-sensitive GPAT activity was lost in BAT from male Gpat4−/− mice. NEM-resistant activity in Gpat4−/− BAT was unaltered, and none of the known GPAT isoforms was upregulated at the transcriptional level to compensate for the loss of GPAT4. Interestingly, total and NEM-sensitive GPAT activities were not reduced in gonadal WAT from Gpat4−/− mice, despite the previous demonstration that the TAG content of WAT was reduced in chow-fed male Gpat4−/− mice (10). It may be that a compensatory increase in the activity of known GPAT isoforms occurs at a posttranscriptional level. It is also possible that the reduced TAG content in WAT of Gpat4−/− mice is a secondary effect, as these animals also exhibit increased energy expenditure (10).
Analysis of GPAT assay products from transfected Cos-7 cells showed that LPA was increased by 2-fold as a result of GPAT4 overexpression, consistent with a GPAT activity. Labeled PA was also increased by 70% in samples from GPAT4-transfected cells, indicating that the LPA produced by GPAT4 can be used as a substrate by AGPAT enzymes present in the membrane preparation. Because GPAT activity is rate-limiting, it is not surprising that most of the product is normally captured as PA in enzyme assays (23). Subsequent formation of DAG did not occur because the NaF in the assay mixture inhibits phosphatases and because phosphatidic acid phosphohydrolase (lipin) is primarily resident in the cytosol and therefore would be largely absent in the membrane preparation (24).
Because hepatic and adipose TAG content is reduced in Gpat4−/− mice, we expected that overexpression of GPAT4 in Cos-7 cells would increase cellular TAG content (10). Instead, we found that overexpression of GPAT4 in Cos- 7 cells did not affect total cellular TAG content or [14C]oleate incorporation into TAG. Interestingly, however, overexpression of both GPAT4 and GPAT1 in Cos-7 cells increased [14C]oleate incorporation into glycerolipid. With GPAT1, overexpression doubled the incorporation of [14C]oleate into DAG and TAG, whereas overexpression of GPAT4 increased label incorporation only into DAG and phosphatidylinositol. Together, these data suggest that available DAG substrate was not lacking but that the LPA, PA, and DAG products of GPAT1 and GPAT4 form different intracellular pools. Perhaps GPAT1 overexpression increases the synthesis of a DAG pool that is accessible to diacylglycerol acyltransferase, whereas the DAG formed by GPAT4 is not accessible. Because the labeling study in Cos-7 cells (African green monkey kidney fibroblasts) did not explain why hepatic and adipose TAG content is markedly reduced in Gpat4−/− mice, it is possible that the role of GPAT4 in initiating TAG synthesis varies by cell type. A 20% increase in GPAT4 activity in HepG2 cells was associated with a 20% increase in cellular TAG, suggesting that GPAT4 may play an important role in TAG synthesis and that overexpression of GPAT4 in primary liver cells could, indeed, result in enhanced label incorporation into TAG. Further studies will be required to determine whether GPAT1 and GPAT4 play different regulatory roles in glycerolipid synthesis in hepatocytes and adipocytes.
The recent identification of two novel NEM-sensitive GPAT isoforms, GPAT3 (7) and GPAT4 (this paper), with different tissue-specific expression patterns has revealed the complexity of de novo glycerolipid synthesis and regulation. More studies will be required to fully characterize the roles of GPAT3 and GPAT4 in the synthesis of TAG and phospholipids in liver and adipose tissue, the mechanisms by which these isoforms are regulated during different physiologic states, and the effects of their glycerolipid synthetic products on downstream signaling pathways (25). Importantly, it is unknown whether additional GPAT isoforms remain to be identified. Additional putative AGPAT enzymes for which AGPAT activity has not been convincingly demonstrated (AGPATs 3, 4, 5, and 7) might also actually be GPAT isoforms. Further studies are needed to confirm the specific acyltransferase activities of these enzymes.
Acknowledgments
This work was supported by grants DK-56598 (R.A.C.), HL-66621 (K.R.), and DK-56350 to the University of North Carolina Clinical Nutrition Research Unit from the National Institutes of Health and by a grant from the American Heart Association Mid-Atlantic Division (C.A.N.).
Abbreviations
- AGPAT
sn-1-acylglycerol-3-phosphate O-acyltransferase
- BAT
brown adipose tissue
- DAG
diacylglycerol
- GPAT
sn-glycerol-3-phosphate acyltransferase
- LPA
lysophosphatidic acid
- NEM
N-euiylmaleimide
- PA
phosphatidic acid
- TAG
triacylglycerol
- WAT
white adipose tissue
References
- 1.Unger RH, Orci L. Diseases of liporegulation: new perspective on obesity and related disorders. FASEB J. 2001;15:312–321. doi: 10.1096/fj.00-0590. [DOI] [PubMed] [Google Scholar]
- 2.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 3.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- 5.Harada N, Hara S, Yoshida M, Zenitani T, Mawatari K, Nakano M, Takahashi A, Hosaka T, Yoshimoto K, Nakaya Y. Molecular cloning of a murine glycerol-3-phosphate acyltransferase-like protein 1 (xGPAT1) Mol Cell Biochem. 2007;297:41–51. doi: 10.1007/s11010-006-9321-5. [DOI] [PubMed] [Google Scholar]
- 6.Shin DH, Paulauskis JD, Moustaid N, Sul HS. Transcriptional regulation of p90 with sequence homology to Escherichia coli glycerol-3-phosphate acyltransferase. J Biol Chem. 1991;266:23834–23839. [PubMed] [Google Scholar]
- 7.Cao J, Li JL, Li D, Tobin JF, Gimeno RE. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc Natl Acad Sci USA. 2006;103:19695–19700. doi: 10.1073/pnas.0609140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewin TM, Schwerbrock NMJ, Lee DP, Coleman RA. Identification of a new glycerol-3-phosphate acyltransferase isoenzyme, mtGPAT2, in mitochondria. J Biol Chem. 2004;279:13488–13495. doi: 10.1074/jbc.M314032200. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Lee DP, Gong N, Schwerbrock NMJ, Mashek DG, Gonzalez-Baró MR, Stapleton CM, Li LO, Lewin TM, Coleman RA. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2) Arch Biochem Biophys. 2007;465:347–358. doi: 10.1016/j.abb.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergnes L, Beigneux AP, Davis RG, Watkins SM, Young SG, Reue K. Agpat6 deficiency causes subdermal lipodystrophy and resistance to obesity. J Lipid Res. 2006;47:745–754. doi: 10.1194/jlr.M500553-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguado B, Campbell RD. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatability complex. J Biol Chem. 1998;273:4096–4105. doi: 10.1074/jbc.273.7.4096. [DOI] [PubMed] [Google Scholar]
- 12.Eberhardt C, Gray PW, Tjoelker LW. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J Biol Chem. 1997;272:20299–20305. doi: 10.1074/jbc.272.32.20299. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal AK, Arioglu E, De Almeida S, Akkoc N, Taylor SI, Bowcock AM, Barnes RI, Garg A. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat Genet. 2002;31:21–23. doi: 10.1038/ng880. [DOI] [PubMed] [Google Scholar]
- 14.Lu B, Jiang YJ, Zhou YT, Xu FY, Hatch GM, Choy PC. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem J. 2005;385:469–477. doi: 10.1042/BJ20041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beigneux AP, Vergnes L, Qiao X, Quatela S, Davis RG, Watkins SM, Coleman RA, Walzem RL, Philips M, Reue K, et al. Agpat6—a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J Lipid Res. 2006;47:734–744. doi: 10.1194/jlr.M500556-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewin TM, Wang P, Coleman RA. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 1999;38:5764–5771. doi: 10.1021/bi982805d. [DOI] [PubMed] [Google Scholar]
- 17.Coleman RA, Haynes EB. Selective changes in microsomal enzymes of triacylglycerol and phosphatidylcholine synthesis in fetal and postnatal rat liver: induction of microsomal sn-glycerol 3-P and dihydroxyacetone-P acyltransferase activities. J Biol Chem. 1983;258:450–465. [PubMed] [Google Scholar]
- 18.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 19.Phan J, Peterfy M, Reue K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J Biol Chem. 2004;279:29558–29564. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- 20.Igal RA, Wang S, Gonzalez-Baró M, Coleman RA. Mitochondrial glycerol phosphate acyltransferase directs incorporation of exogenous fatty acids into triacylglycerol. J Biol Chem. 2001;276:42205–42212. doi: 10.1074/jbc.M103386200. [DOI] [PubMed] [Google Scholar]
- 21.Nagle CA, An J, Shiota M, Torres TP, Cline GW, Liu ZX, Wang S, Catlin RL, Shulman GI, Newgard CB, et al. Hepatic overexpression of glycerol-sn-3-phosphate acyltransferase 1 in rats causes insulin resistance. J Biol Chem. 2007;282:14807–14815. doi: 10.1074/jbc.M611550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin TM, Granger DA, Kim JH, Coleman RA. Regulation of mitochondrial sn-glycerol-3-phophate acyltransferase activity: response to feeding status is unique in various rat tissues and is discordant with protein expression. Arch Biochem Biophys. 2001;396:119–127. doi: 10.1006/abbi.2001.2604. [DOI] [PubMed] [Google Scholar]
- 23.Schlossman DM, Bell RM. Microsomal sn-glycerol 3-phosphate and dihydroxyacetone phosphate acyltransferase activities from liver and other tissues. Arch Biochem Biophys. 1977;182:732–742. doi: 10.1016/0003-9861(77)90555-0. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Munoz A, Hatch GM, Martin A, Jamal Z, Vance DE, Brindley DN. Effects of okadaic acid on the activities of two distinct phosphatidate phosphohydrolases in rat hepatocytes. FEBS Lett. 1992;301:103–106. doi: 10.1016/0014-5793(92)80219-7. [DOI] [PubMed] [Google Scholar]
- 25.Coleman RA. How do I fatten thee: let me count the ways…. Cell Metab. 2007;5:87–89. doi: 10.1016/j.cmet.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammond LE, Neschen S, Romanelli AJ, Cline GW, Ilkayeva OR, Shulman GI, Muoio DM, Coleman RA. Mitochondrial glycerol-3-phosphate acyltransferase-1 is essential in liver for the metabolism of excess acyl-CoAs. J Biol Chem. 2005;280:25629–25636. doi: 10.1074/jbc.M503181200. [DOI] [PubMed] [Google Scholar]


