Abstract
Chicken ovalbumin upstream promoter transcription factor-interacting proteins 1 and 2 (CTIP1 and CTIP2) are related transcriptional regulatory proteins. While overexpression of both of these proteins has been linked to the development of several lymphoid malignancies, lack of CTIP1 and CTIP2 expression results in defective lymphopoiesis and abnormal thymocyte development, respectively. Here, we describe the expression patterns of CTIP1 and CTIP2 during mouse embryogenesis and in the post-natal brain. Both CTIP1 and CTIP2 were expressed diffusely in the embryo at 10.5 days post-coitum (d.p.c.). However, the expression of both genes became increasingly restricted to the central nervous system (CNS) during the course of fetal development, culminating with high, but differential, expression levels throughout the hippocampal subregions, olfactory bulb and cortex, limbic system, basal ganglia and frontal cortex of the developing brain, and in dorsal cells of the spinal cord. The brain expression domains of CTIP1 and CTIP2 were maintained into adulthood. Outside the CNS, both genes exhibited differential expression within the facial mesenchyme at 12.5 d.p.c., and CTIP2 was selectively expressed from day 12.5 onwards in the olfactory epithelium and developing thymus, and to a lesser extent in oral and gut epithelia. Strong CTIP2 expression was maintained in the thymus at 18.5 d.p.c. These results support the selective contributions of both CTIP1 and CTIP2 in the development and function of both the central nervous and immune systems and the importance of future investigations to define the function(s) of both proteins.
Keywords: CTIP1, CTIP2, BCL11A, BCL11B, Transcription factors, Central nervous system, Immune system, Differentiation, Fetal development
CTIP1 (Bcl11a, Evi9) and CTIP2 (Bcl11b, Rit-1β) are two related C2H2 zinc finger proteins that have been demonstrated to modulate transcription by at least two mechanisms. Both of these transcription factors were originally identified as proteins that interact directly with chicken ovalbumin upstream promoter transcription factor (COUP-TF) family members (COUP-TFI/NR2F1, COUP-TFII/ARP1/NR2F2 and COUP-TFIII/Ear2/NR2F6) (Avram et al., 2000). Recruitment of either CTIP1 (Avram et al., 2000) or CTIP2 (Avram, unpublished studies) to the template by a COUP-TF family member was found to result in transcriptional repression of a reporter gene harboring a COUP-TF binding site. Additionally, CTIP1 and CTIP2 are sequence-specific DNA binding proteins that repress transcription via direct, COUP-TF-independent binding to a motif that is related to the canonical GC box (Avram et al., 2002). Thus, CTIPs may either be recruited to the template by a COUP-TF family member or bind directly to the template in a sequence-specific manner. In both cases, CTIP1 and CTIP2 appear to repress transcription in a manner that does not involve trichostatin A-sensitive histone deacetylation (Avram et al., 2000; Senawong et al., 2003). Although, the mechanistic basis of CTIP-mediated transcriptional repression has not been defined, overexpressed CTIP2 (Rohr et al., 2003) and CTIP1 (Avram, unpublished results) have been shown to interact with members of the heterochromatin-associated protein (HP1) family in mammalian cells, thus, implicating HP1 proteins and possibly a heterochromatin-based mechanism in the transcriptional repression activities of the CTIPs. In addition, recent studies have demonstrated that both CTIP2- (Senawong et al., 2003) and CTIP1 (Senawong et al., submitted)-mediated transcriptional repression may involve the action of the NAD+-dependent, TSA-insensitive histone deacetylase known as sirtuin 1 (SIRT1).
Expression of both CTIP1 and CTIP2 has recently been linked to the etiology of disease. Overexpression of CTIP1 following proviral integration led to the transformation of NIH3T3 cells and the development of myeloid leukemia in mice (Nakamura et al., 2000). This transformation event may be facilitated by a physical interaction of CTIP1 with BCL6, a known human B cell proto-oncogene product. Recently, an association between p53 and CTIP2 in mice has been implicated in the development of thymic lymphomas (Wakabayashi et al., 2003a). In addition, translocation of CTIP1 is involved in lymphoid malignancies, and the amplification and/or translocation of CTIP2 is thought to play a role in human leukemogenesis (Satterwhite et al., 2001). Finally, CTIP2 was recently demonstrated to repress Tat-mediated transcriptional activation and inhibit HIV-1 replication in human microglial cells (Rohr et al., 2003). Taken together, these reports underscore the potential importance of the CTIPs in human health and highlight the need for additional information regarding their function and expression patterns during development and in the adult organism.
Although CTIP1 and CTIP2 can contribute to immune system malignancies, both proteins are also essential for normal lymphocyte development. Using CTIP1−/− mice, Liu et al. (2003) demonstrated that this transcription factor was essential for post-natal development and normal lymphopoiesis. CTIP1-deficient mice lack B cells and have alterations in several types of T cells. Furthermore, mice transplanted with CTIP1-deficient cells died from T cell leukemia derived from the host suggesting that CTIP1/Bcl11a may function as a non-autonomous T cell suppressor gene. In a separate report, Kominami and colleagues evaluated the role of CTIP2 in the development of immune cells (Wakabayashi et al., 2003b). In CTIP2-null mice, impairment of B cells or γδ T cell lineages was not observed, however, the generation of αβ T lymphocytes was blocked at the CD4− CD8− double-negative stage of thymocyte development. Although these mutant animals were not reported to develop subsequent malignancies, these results clearly define the importance of CTIP2 in the differentiation and survival of thymocytes.
Substantial information has been published regarding the expression patterns of COUP-TF family members (Jonk et al., 1994; Liu et al., 2000; Qiu et al., 1994; Zhou et al., 1999). However, contrary to the conclusions of a previous paper (Gunnersen et al., 2002), little information exists concerning the patterns of CTIP1 and CTIP2 expression during central nervous system (CNS) development and that which has been published was primarily derived from RT-PCR studies and/or northern analyses. For example, expression of CTIP1 in mice has been detected as early as day 10–12.5 post-coitum and persists until at least day 17 (Avram et al., 2000; Nakamura et al., 2000). CTIP1 is highly expressed in both mouse and rat cortex during embryogenesis and may thus be required for neuronal development and differentiation (Gunnersen et al., 2002). In contrast, CTIP2 transcripts have been detected in the embryo at 10–12.5 (Avram et al., 2000), however, no additional information exists on its expression during fetal development. In adult animals, CTIP1 is highly expressed in the brain and spleen, and found at lower levels in the heart, liver, testis and lung (Avram et al., 2000; Nakamura et al., 2000). In addition, in hematopoietically derived human cells, CTIP1 mRNA was detected in CD34 + myeloid precursor cells, B cells, monocytes, and megakaryocytes (Saiki et al., 2000).
In this report, we describe the expression of both CTIP1 and CTIP2 during mouse fetal development as revealed by in situ hybridization (ISH). Our results highlight the expression of both CTIP1 and CTIP2 during fetal development in the brain and early immune tissues such as the embryonic thymus.
1. Results
Developmental expression of CTIPs has been suggested by previous findings demonstrating that CTIP1 and CTIP2 are expressed in both P19 and F9 embryonal carcinoma cell lines (data not shown) as well in 10–12.5 days post-coitum (d.p.c.) embryos (Avram et al., 2000). Thus, ISH studies were conducted to define the spatial and temporal patterns of expression of CTIP1 and CTIP2 during mouse embryogenesis.
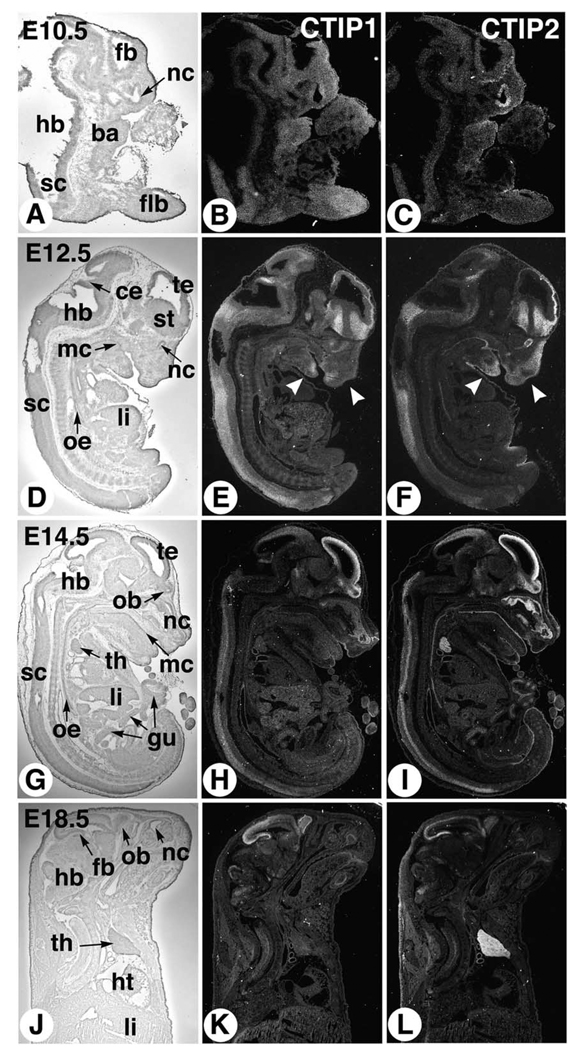
Expression of both CTIP1 and CTIP2 was observed at the earliest time point examined, 10.5 d.p.c., at which the expression pattern of the two genes partially overlapped (Fig. 1). CTIP2 transcripts were detected primarily in the nasal cavity epithelium and in the outer layers of the developing cerebral cortex (Fig. 1C). Diffuse, low-level CTIP2 signal was observed in the rest of the 10.5 d.p.c. embryo including the branchial arches and forelimb bud (Fig. 1C). CTIP1 expression appeared quite ubiquitous at 10.5 d.p.c. as diffuse levels of transcripts were observed in the forelimb bud, branchial arches, and throughout the CNS (Fig. 1B).
Fig. 1.
Expression of CTIP2 and CTIP2 during mouse development. Sagittal sections of mouse embryos or fetuses at 10.5 (A–C), 12.5 (D–F), 14.5 (G–I), and 18.5 d.p.c. (J–L), hybridized with antisense [35S]-labeled cRNAs corresponding to CTIP1 (middle column) and CTIP2 (right column), were examined by dark-field microscopy for signal detection (signal grain appears in white). A bright-field view of one of the corresponding sections is shown in the left column for histological orientation. ba, branchial arches; ce, cerebellum; fb, forebrain; flb, forelimb bud; gu, gut; hb, hindbrain; ht, heart; li, liver; mc, mouth cavity; nc, nasal cavity; ob, olfactory bulb; oe, esophagus; sc, spinal cord; st, striatum; te, telencephalic vesicle; th, thymus. Arrowheads in (E,F) point to differential spatial expression domains in the developing mandibular and frontonasal region.
The expression of both CTIP1 and CTIP2 became more restricted at 12.5 (Fig. 1D–F) and 14.5 d.p.c. (Fig. 1G–I), and a differential spatial pattern of expression of the two genes clearly emerged. Highest CTIP1 expression was observed in the developing forebrain within the striatum, telencephalic vesicles and olfactory bulb, in part of the cerebellum anlage, and in dorsal cells of the spinal cord (Fig. 1E,H). High expression of CTIP2 was detected in the same regions of the CNS, except in the cerebellar domain (Fig. 1F,I), and CTIP2 transcripts appeared to be expressed at higher levels in the intermediate region of the spinal cord at E14.5 (Fig. 1I).
Outside the CNS, CTIP1 remained expressed at low and diffuse levels at 12.5 (Fig. 1H) and 14.5 d.p.c. (Fig. 1I). Areas of higher expression were seen, however, at 12.5 d.p.c. in the mandibular and nasal mesenchymes (Fig. 1E, arrowheads). The liver also exhibited slightly elevated CTIP1 expression levels at 12.5 and 14.5 d.p.c. (Fig. 1E,H). CTIP2 was expressed in mandibular and nasal mesenchymes at 12.5 d.p.c., albeit in a more extended domain than CTIP1 (Fig. 1F, arrowheads). High CTIP2 expression persisted in the olfactory epithelium at 12.5 and 14.5 d.p.c., whereas lower transcript levels were detected in the mouth cavity, esophagus and gut epithelia (Fig. 1F,I). In addition, relatively high levels of CTIP2, but not CTIP1, transcripts were detected in the thymus at 14.5 d.p.c., a time at which most of the thymic cells are double negative (CD4− CD8−), T cell precursors.
The CNS expression patterns of both CTIP genes remained strong and restricted at 18.5 d.p.c. (Fig. 1K,L, and see below). Lower levels of CTIP1 transcripts were diffusely distributed throughout the 18.5 d.p.c. fetus (Fig. 1K). In contrast, the dominant tissue expressing CTIP2 transcripts at this developmental stage was clearly the thymus (Fig. 1L). Together with the 14.5 d.p.c. data, these results suggest that CTIP2 is expressed in both CD4− CD8− T cell precursors (14.5 d.p.c.) and in more mature T cells in the thymus (detailed analysis of CTIP expression features in cells of the lymphoid system will be reported elsewhere; Shepherd et al., in preparation).
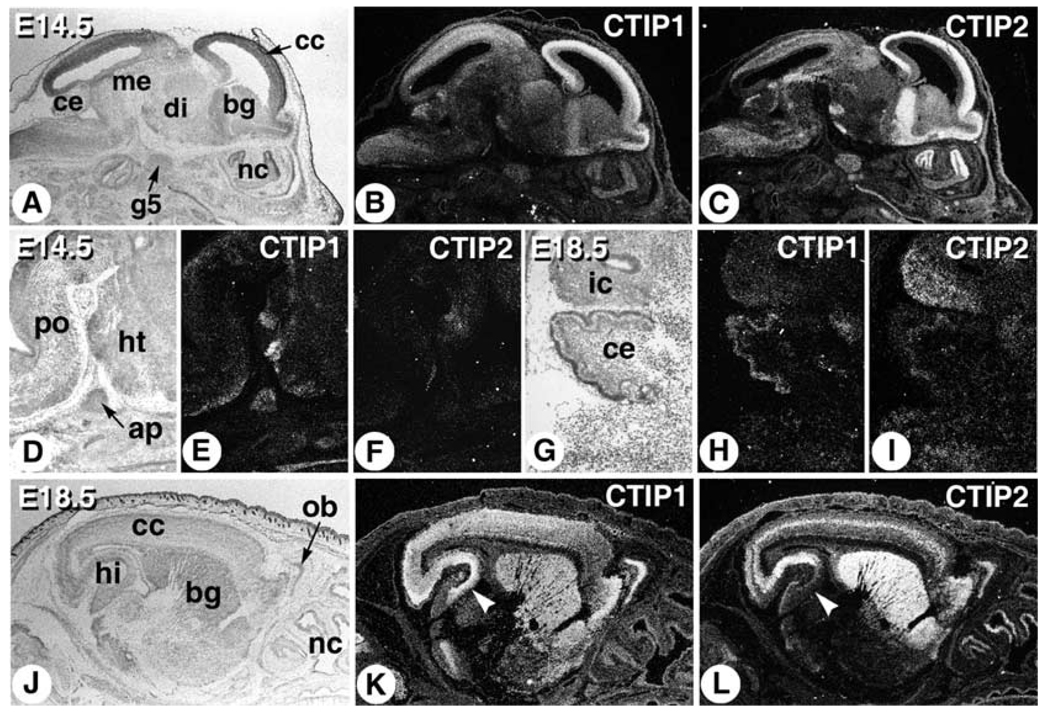
Further investigation of the brain expression patterns of CTIP1 and CTIP2 was conducted at 14.5 and 18.5 d.p.c. to define their expression patterns more precisely. The CTIP1 probe strongly labeled the cerebral cortex, olfactory bulb and midbrain, and to a lesser extent, the cerebellum, hindbrain, basal ganglia, and nasal epithelium (Fig. 2A,B,H). Additional CTIP1 labeling was observed in the hypothalamus, anterior pituitary, and pontine nucleus (Fig. 2D,E). CTIP2 labeling was most prominent in the cerebral cortex, basal ganglia, fifth cranial (trigeminal) ganglion and olfactory region of the nasal epithelium, while lower level expression of CTIP2 was observed in the mesencephalon, cerebellum, and inferior colliculus (Fig. 2A,C,I).
Fig. 2.
Expression of CTIP1 and CTIP2 in the developing mouse brain. Embryos were sagittally sectioned at 14.5 (A–F) and 18.5 d.p.c. (G–L), and hybridized with [35S]-labeled probes as indicated. ap, anterior pituitary; bg, basal ganglia; cc, cerebral cortex; ce, cerebellum; di, diencephalon; g5, 5th cranial nerve ganglion (trigeminal ganglion); hi, hippocampus; ht, hypothalamus; ic, inferior colliculus; me, mesencephalon; nc, nasal cavity; ob, olfactory bulb; po, pontine nucleus.
Strong CTIP1 labeling was observed in the olfactory bulb and throughout the cerebral cortex, hippocampus, and basal ganglia of the 18.5 d.p.c. fetus (Fig. 2J,K). CTIP2 transcripts were also detected in the olfactory bulb and cortex, as well as in the basal ganglia and nasal epithelium (Fig. 2J,L). However, the CNS expression patterns of CTIP1 and CTIP2 differed in two important aspects in the 18.5 d.p.c. fetus. First, the CTIP2 probe preferentially labeled a relatively narrow band of cells in the cerebral cortex, which appeared to correspond to the intermediate zone, whereas CTIP1 transcripts were diffusely distributed throughout the developing cerebral cortex (Fig. 2L,K, respectively). Second, CTIP1 transcripts were observed in all subregions (Fig. 2K), whereas CTIP2 mRNA was observed primarily in the more rostral portions (Fig. 2L), of the developing hippocampus.
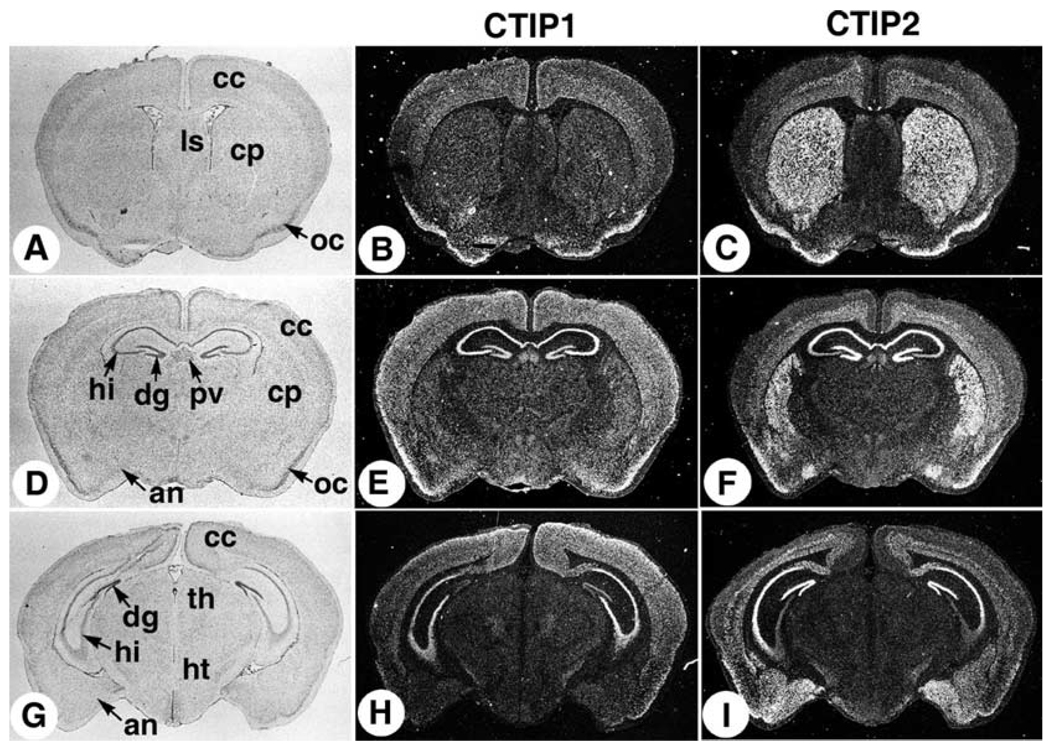
The differential distribution of CTIP1 and CTIP2 transcripts in the brain was further investigated in coronal sections of young animals (3 weeks of age) and adult mice. Again, the CTIP1 probe diffusely stained all layers of the cerebral cortex as well as the olfactory cortex, paraventricular thalamic nucleus, and hippocampus in 3-week-old animals (Fig. 3A,E,H). CTIP1 transcripts were observed in all subregions of the hippocampus including CA1, CA2, CA3, and CA4, as well as the dentate gyrus (Fig. 3E,H; see also Fig. 4). CTIP2 expression in the 3-week-old animals was observed in the middle to deep layers of the cerebral cortex (Fig. 3C,F,I), which most likely correspond to layers VI (multiform layer) and IV (internal granule cell layer). Intense CTIP2 staining was observed in the young animals within the caudate nucleus and putamen regions of the striatum (Fig. 3C,F), one of the four principal nuclei of the basal ganglia. CTIP2 transcripts were also detected in the cell bodies of hippocampal neurons located primarily in the CA1 and CA2 subregions, and the dentate gyrus (Fig. 3F,I), a pattern consistent with that observed in 18.5 d.p.c. (see Fig. 2L). While the CTIP2 probe also hybridized to cell bodies in the CA3 and CA4 subregions of the hippocampus in young animals (Fig. 3F,I; see also Fig. 4C), this staining was clearly less intense than that observed in either the CA1 and CA2 regions or the dentate gyrus. Relatively high levels of CTIP2 transcripts were also detected in the olfactory cortex and amygdala including the posteromedial cortical amygdaloid nucleus and the posteromedial part of the amygdalohippocampal area (Fig. 3F,I).
Fig. 3.
Mouse brain expression of CTIP1 and CTIP2 at post-natal day 21. Mouse brain coronal sections from post-natal day 21 animals were probed with [35S]-labeled riboprobes as indicated. A bright-field image is shown in the left column for histological orientation of the in situ hybridization panels (dark-field images in the middle and right columns). The sections shown are progressively more caudal from the top to the bottom rows. an, amydaloid nucleus; cc, cerebral cortex; cp, caudate-putamen; dg, dentate gyrus; hi, hippocampus; ht, hypothalamus, lateral septal nucleus; oc, olfactory cortex; pv, paraventricular thalamic nucleus; th, thalamus.
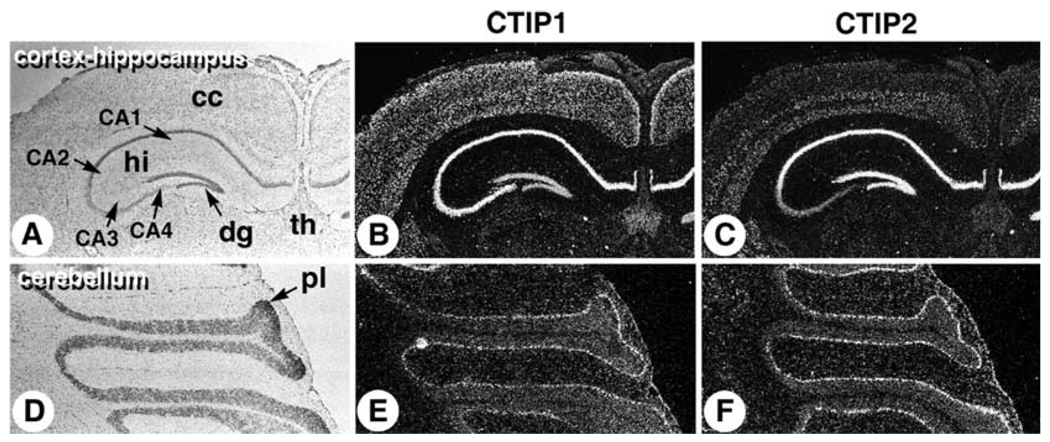
Fig. 4.
Expression of CTIP1 and CTIP2 in adult cerebral cortex, hippocampus, and cerebellum. Coronal cryosections of adult mouse brain were probed with [35S]-labeled CTIP1 (B) and CTIP2 (C) riboprobes. (A) Bright-field image for panels B and C. (D–F) Sagittal sections of adult mouse cerebellum hybridized with CTIP1 (E) and CTIP2 (F) probes and the corresponding bright-field image (D). CA1–CA4, hippocampal subregions; cc, cerebral cortex; hi, hippocampus; dg, dentate gyrus; th, thalamus; pl, Purkinje cell layer.
The differential patterns of CTIP1 and CTIP2 expression observed in the 18.5 d.p.c. and 3-week-old animals were maintained in adult brain at the level of the hippocampal subregions and the cerebral cortex (Fig. 4B,C). However, the expression patterns of CTIP1 and CTIP2 were completely overlapping in the cerebellum of adult animals as both probes appeared to label the Purkinje cell layer (Fig. 4E,F). Purkinje cells, which are large GABAergic neurons that extend dendritic projections upwards into the molecular layer and axonal projections downward to deep cerebellar nuclei, are considered to be the primary output of the cerebellum controlling multiple components of descending motor pathways (Ghez and Thack, 2000).
In summary, the results of these ISH studies demonstrate that CTIP1 and CTIP2 exhibit spatially and temporally restricted patterns of expression during mouse embryogenesis. The two genes appeared to be differentially expressed in the brain of both neonatal and adult animals within several regions including, most prominently, the hippocampus, cerebral cortex, basal ganglia, and limbic system as well as in the spinal cord. CTIP1 and CTIP2 were co-expressed in many tissues such as the olfactory cortex, CA1 and CA2 hippocampal subregions and in cerebellar Purkinje cells. Considered together, these findings suggest that CTIP1 and CTIP2 may play non-redundant roles during CNS development and in the adult brain.
2. Discussion
CTIP1 and CTIP2 were found to exhibit specific spatial and temporal patterns of expression during mouse embryogenesis that became increasingly restricted to and within the CNS as development progressed towards birth. The primary exceptions to this observation were the cells and tissues of the immune system in which CTIP1 and CTIP2 were highly but differentially expressed (see below), and the olfactory epithelium, which stained intensely with the CTIP2 probe from 10.5 to 14.5 d.p.c.
Both CTIP1 and CTIP2 were originally identified as proteins that interacted with and stimulated transcriptional repression activity of the COUP-TF family of orphan nuclear receptors (Avram et al., 2000). Correspondingly, the expression patterns of CTIPs in the developing CNS described herein partially, but not entirely, overlap with those of both COUP-TFI and -TFII (Jonk et al., 1994; Qiu et al., 1994) during the early to mid-stages of neurodevelopment. For example, COUP-TFI and -TFII are expressed in the developing telencephalon from 10.5 to 14.5 d.p.c. (Jonk et al., 1994; Qiu et al., 1994), and we observed a high level of expression of CTIP1 and CTIP2 in this brain region at 14.5 d.p.c. (forebrain, Fig. 1). Similarly, COUP-TFII expression was previously observed in the hypothalamus at 14.5 d.p.c. (Jonk et al., 1994), a brain region found to express predominantly CTIP1 transcripts at this developmental stage (Fig. 2E,F). Other developing brain regions that would appear to co-express transcripts corresponding to both COUP-TF family members and CTIPs include the midbrain and olfactory bulb (Jonk et al., 1994; Qiu et al., 1994; Fig. 1, Fig 2 of this paper). Outside the developing CNS, CTIP2 expression was primarily detected in the olfactory epithelium and trigeminal ganglion at 14.5 d.p.c. (Fig. 1, Fig 2C), tissues that have been previously shown to express COUP-TFI and COUP-TFII transcripts (Jonk et al., 1994).
The primary regions expressing both COUP-TF (Lopes da Silva et al., 1995) and CTIP transcripts in the adult brain are: olfactory bulb (COUP-TFI, CTIP1, CTIP2), amygdaloid nucleus (COUP-TFII, CTIP2), and the CA1–CA4 regions of the hippocampus including the dentate gyrus (COUP-TFI, CTIP1, CTIP2). Although COUP-TFI and CTIP1/2 are expressed in the cerebellum, the cellular distribution of transcripts differs as COUP-TFI is expressed in cerebellar granule cells (Lopes da Silva et al., 1995), whereas the expression of both CTIP1 and CTIP2 is restricted to the Purkinje cell layer (Fig. 4E,F).
It is clear from the results of the present and previous (Jonk et al., 1994; Lopes da Silva et al., 1995; Qiu et al., 1994) studies that the expression patterns of CTIPs and COUP-TF family members partially overlap in the developing and adult mouse brain in both spatial and temporal contexts. This finding would be consistent with the hypothesis that the two classes of proteins may function together to regulate the expression of target genes in vivo. However, both CTIP1 and CTIP2 are also sequence-specific DNA binding proteins that regulate transcription in a manner that is independent of COUP-TF family members (Avram et al., 2002). Correspondingly, co-expression with a COUP-TF family member may not be a pre-requisite for the manifestation of transcriptional regulatory activity of either CTIP1 or CTIP2. Indeed, some neuronal cells and tissues that do not express either COUP-TFI or -TFII were found to express CTIP1 and/or CTIP2; most notably, the Purkinje cell layer of the cerebellum (CTIP1 and CTIP2) and the basal ganglia (CTIP2). However, the cells of the immune system were clearly the most striking example of the divergent expression of COUP-TF and CTIP family members as no COUP-TF mRNA was detected in any of the immune tissues examined whereas CTIP expression was abundant (Shepherd et al., in preparation). These data reinforce the premise that CTIPs can function as COUP-TF-independent transcription factors.
3. Experimental procedures
Mouse embryos and brains from 3-week-old and adult animals were cryosectioned and probed with [35S]-labeled cRNAs corresponding to CTIP1 and CTIP2 as previously described (Niederreither and Dolle, 1998). In all cases, sections were analyzed by light- and dark-field microscopy for histological purposes and to detect the ISH signal, respectively.
Acknowledgements
The studies were supported by grants from the National Institutes of Health (GM60852) to M.L. and by a NIEHS center grant (ES00210) to the Oregon State University Environmental Health Sciences Center.
References
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem. J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghez C, Thack WT. The cerebellum. In: Kandel ER, et al., editors. Principles of Neural Science. New York: McGraw-Hill; 2000. pp. 832–852. [Google Scholar]
- Gunnersen JM, Augustine C, Spirkoska V, Kim M, Brown M, Tan SS. Global analysis of gene expression patterns in developing mouse neocortex using serial analysis of gene expression. Mol. Cell. Neurosci. 2002;19:560–573. doi: 10.1006/mcne.2001.1098. [DOI] [PubMed] [Google Scholar]
- Jonk LJ, de Jonge ME, Pals CE, Wissink S, Vervaart JM, Schoorlemmer J, Kruijer W. Cloning and expression during development of three murine members of the COUP family of nuclear orphan receptors. Mech. Dev. 1994;47:81–97. doi: 10.1016/0925-4773(94)90098-1. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dwyer ND, O’Leary DD. Differential expression of COUP-TFI, CHL1, and two novel genes in developing neocortex identified by differential display PCR. J. Neurosci. 2000;20:7682–7690. doi: 10.1523/JNEUROSCI.20-20-07682.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, et al. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- Lopes da Silva S, Cox JJ, Jonk LJ, Kruijer W, Burbach JP. Localization of transcripts of the related nuclear orphan receptors COUP-TF I and ARP-1 in the adult mouse brain. Brain Res. Mol. Brain Res. 1995;30:131–136. doi: 10.1016/0169-328x(94)00289-q. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yamazaki Y, Saiki Y, Moriyama M, Largaespada DA, Jenkins NA, Copeland NG. Evi9 encodes a novel zinc finger protein that physically interacts with BCL6, a known human B-cell proto-oncogene product. Mol. Cell. Biol. 2000;20:3178–3186. doi: 10.1128/mcb.20.9.3178-3186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. In situ hybridization with 35S-labeled probes for retinoid receptors. Methods Mol. Biol. 1998;89:247–267. doi: 10.1385/0-89603-438-0:247. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Cooney AJ, Kuratani S, DeMayo FJ, Tsai SY, Tsai MJ. Spatiotemporal expression patterns of chicken ovalbumin upstream promoter-transcription factors in the developing mouse central nervous system: evidence for a role in segmental patterning of the diencephalon. Proc. Natl Acad. Sci. USA. 1994;91:4451–4455. doi: 10.1073/pnas.91.10.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr O, Lecestre D, Chasserot-Golaz S, Marban C, Avram D, Aunis D, et al. Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells. J. Virol. 2003;77:5415–5427. doi: 10.1128/JVI.77.9.5415-5427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki Y, Yamazaki Y, Yoshida M, Katoh O, Nakamura T. Human EVI9, a homologue of the mouse myeloid leukemia gene, is expressed in the hematopoietic progenitors and down-regulated during myeloid differentiation of HL60 cells. Genomics. 2000;70:387–391. doi: 10.1006/geno.2000.6385. [DOI] [PubMed] [Google Scholar]
- Satterwhite E, Sonoki T, Willis TG, Harder L, Nowak R, Arriola EL, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, Leid M. Involvement of the histone deacetylase SIRT1 in COUP-TF-interacting protein 2-mediated transcriptional repression. J. Biol. Chem. 2003;278:43041–43050. doi: 10.1074/jbc.M307477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Inoue J, Takahashi Y, Matsuki A, Kosugi-Okano H, Shinbo T, et al. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem. Biophys. Res. Commun. 2003a;301:598–603. doi: 10.1016/s0006-291x(02)03069-3. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat. Immunol. 2003b;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Zhou C, Qiu Y, Pereira FA, Crair MC, Tsai SY, Tsai MJ. The nuclear orphan receptor COUP-TFI is required for differentiation of subplate neurons and guidance of thalamocortical axons. Neuron. 1999;24:847–859. doi: 10.1016/s0896-6273(00)81032-6. [DOI] [PubMed] [Google Scholar]