Abstract
Background
Mpdz gene variations are known contributors of acute alcohol withdrawal severity and seizures in mice.
Methods
To investigate the relevance of these findings for human alcoholism, we resequenced 46 exons, exon–intron boundaries, and 2 kilobases in the 5′ region of the human MPDZ gene in 61 subjects with a history of alcohol withdrawal seizures (AWS), 59 subjects with a history of alcohol withdrawal without AWS, and 64 Coriell samples from self-reported nonalcoholic subjects [all European American (EA) ancestry] and compared with the Mpdz sequences of 3 mouse strains with different propensity to AWS. To explore potential associations of the human MPDZ gene with alcoholism and AWS, single SNP and haplotype analyses were performed using 13 common variants.
Results
Sixty-seven new, mostly rare variants were discovered in the human MPDZ gene. Sequence comparison revealed that the human gene does not have variations identical to those comprising Mpdz gene haplotype associated with AWS in mice. We also found no significant association between MPDZ haplotypes and AWS in humans. However, a global test of haplotype association revealed a significant difference in haplotype frequencies between alcohol-dependent subjects without AWS and Coriell controls (p = 0.015), suggesting a potential role of MPDZ in alcoholism and or related phenotypes other than AWS. Haplotype-specific tests for the most common haplotypes (frequency > 0.05), revealed a specific high-risk haplotype (p = 0.006, maximum statistic p = 0.051), containing rs13297480G allele also found to be significantly more prevalent in alcoholics without AWS compared with nonalcoholic Coriell subjects (p = 0.019).
Conclusions
Sequencing of MPDZ gene in individuals with EA ancestry revealed no variations in the sites identical to those associated with AWS in mice. Exploratory haplotype and single SNP association analyses suggest a possible association between the MPDZ gene and alcohol dependence but not AWS. Further functional genomic analysis of MPDZ variants and investigation of their association with a broader array of alcoholism-related phenotypes could reveal additional genetic markers of alcoholism.
Keywords: Genomics, Alcoholism, MPDZ, Withdrawal, Seizures
Twin And Linkage studies have revealed that alcohol dependence is highly heritable and allelic variants of several candidate genes have been found to be associated with altered risks for alcohol dependence and withdrawal (Higuchi et al., 2006; Liu et al., 2004; Prescott and Kendler, 1999; Schuckit et al., 1972; Tyndale, 2003). Using a combination of interval-specific congenic strains and recombinant progeny tests in mice (Buck et al., 1997; Fehr et al., 2002; Shirley et al., 2004), it was shown that the Mpdz gene and its variants could affect alcohol and barbiturate withdrawal liability measured by presence and severity of seizures. The human analog, MPDZ, is located on chromosome 9p24-p22 and encodes for the Multiple-PDZ-Domain protein, which is also known as MUPP1 and contains 13 PDZ domains (Ullmer et al., 1998). PDZ domains are protein-interaction domains and are often part of the multi-domain scaffolding proteins that assemble large molecular complexes, control synaptic protein composition and structure, and allow the dynamic trafficking of synaptic proteins in specific cell locations (Kim and Sheng, 2004). The MPDZ protein is known to be involved in learning and memory-related synaptic plasticity as well as seizures and epilepsy through its interaction with the serotonin 5-HT2C receptor, glutamate NMDA receptors, GABAB receptor, and dopamine D2, D3, and D4 receptors (Bettler et al., 2004; Griffon et al., 2003; Krapivinsky et al., 2004; Parker et al., 2003).
Specific haplotypes associated with the production of the Mpdz protein variants II and III (NCBI# AAL37379.1 and AAL37390.1, respectively), differentiate strains of mice with severe withdrawal (including C57LJ and DBA/2J) from strains that have less severe withdrawal (including wild type C57BL/6J) and possess a haplotype that produces protein variant I (NCBI# AAL37377.2) (Fehr et al., 2002). Therefore, we hypothesized that genetic variability of the human MPDZ gene may also be associated with the risk of alcohol withdrawal seizures (AWS). This association may be due to variations in the coding sequence of the human MPDZ gene that are analogous to those described in mice (Fehr et al., 2002). Alternatively, variations in the regulatory regions could potentially affect protein production and contribute to the risk of alcohol withdrawal and AWS (Shirley et al., 2004).
To address these hypotheses, we performed direct comparisons between the human MPDZ gene and its mouse homolog Mpdz to identify potential variability in the human DNA sites that are homologous to the variability sites described as being part of the withdrawal seizure-related haplotype in mice (Fehr et al., 2002). The current version of the dbSNP (build 129) lists a total of 589 single nucleotide polymorphisms (SNPs) in the human MPDZ gene including 23 SNPs (12 nonsynonymous, 10 synonymous, and 1 frame shift) in coding regions (NCBI). In order to ensure the inclusion of all relevant variations, we performed resequencing of the MPDZ gene in a sample of alcohol-dependent subjects with European Ancestry and a sample of DNA from self reported nonalcoholic European Americans (EA) obtained from the Coriell Institute.
The results of this sequence comparison between mice and humans are reported as well as comparisons of sequence variation frequencies in groups of alcohol-dependent subjects with and without AWS and in the Coriell sample.
MATERIALS AND METHODS
Study Subjects
This study was conducted in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and the standards established by the Institutional Review Board of the Mayo Clinic Rochester. Participants were recruited from subjects treated in the outpatient and residential addictions treatment programs or on the medical and surgical floors of the hospitals affiliated with the Mayo Clinic in Rochester, MN. All subjects provided informed consent. DNA samples were collected from 120 male and female EA subjects, 18 years or older, who met DSM-IVTR criteria for diagnosis of alcohol dependence (APA, 2000). Of these, 61 subjects had a history of alcohol withdrawal with seizures (AWS), including 15 subjects who also had a history of delirium tremens (DT). The remaining 59 subjects reported a history of alcohol withdrawal without seizures or DT. Potential study subjects were excluded if they had a history of: psychotic or bipolar I disorders; use of substances known to potentially cause seizures or delirium (e.g., barbiturates, benzodiazepines, opiates, hallucinogens, stimulants, cocaine, hallucinogens, PCP, anticholinergics) at the time of the first AWS; neurologic conditions with seizures and or EEG or imaging data indicating presence of the potentially epileptogenic focus prior to the first AWS; presence of somatic or metabolic conditions capable of provoking seizures and or delirium at the time of the first AWS (e.g., fever > 40°C, sodium < 120 or >160 mEq/l, glucose < 60 mg%, magnesium < 1.0 mEq/l, calcium < 6.0 mEq/l, ammonia > 50 μgN/dl, BUN > 100 mg%, creatinine > 5.0 mg%, or osmolality 350 mOsm/l). A review of the medical records and diagnostic evaluation of each study participant by a Board Certified psychiatrist (VMK) was performed to ensure that DSM IV-TR criteria for alcohol dependence are met and none of the exclusion criteria are present. Genomic DNA from 64 self-reported EA nonalcoholic subjects obtained from the Coriell Institute was used as nonalcoholic control.
DNA Resequencing
Genomic DNA was extracted from peripheral blood lymphocytes using AutoPure LS (Gentra, Minneapolis, MN), according to the manufacturer’s protocol. New pairs of primers were generated for all exons of the human MPDZ gene, including the exon–intron boundaries and 2 kilobases in the 5′ region based on the chromosome 9p24-p22 genomic sequence (NT_008413). The coding sequence of the human MPDZ gene was compared with the Ensembl transcript OTTHUMT00000055488. All exons and introns, including 75 to 150 bp of the boundary regions of the respective exons, were amplified using polymerase chain reaction and sequenced using an ABI PRISM Big Dye Terminator, Cycle sequencing assay (Applied Biosystems, Foster City, CA), and an ABI 3730xl automated sequencer (Applied Biosystems; Genomics Technology Center, Mayo Clinic, Rochester, MN). After the sequencing, sequence variants were screened using the Polyphred program.
Data Analysis and Statistics
The ALIGN software (Pearson et al., 1997) and ClustalW program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) were used to compare coding DNA (cDNA) sequence of the human MPDZ gene and protein with the sequences of Mpdz gene and proteins of 3 mouse strains. The Branchpoint (BP) Analysis program of EMBL-EBI web site (http://www.ebi.ac.uk/asd-srv/wb.cgi?method=2) was used to determine if variations were located in consensus areas potentially important for alternative splicing process. The SIGSCAN (http://bimas.dcrt.nih.gov/molbio/signal/) program was used to identify potential transcription enhancer/repressor consensus sequences in the putative promotor areas.
Haploview v3.2 software (Barrett et al., 2005) was used to determine linkage disequilibrium (LD) of MPDZ gene variants. Allele frequencies of all variants with a minor allele frequency (MAF) ≥0.05 were compared in the 3 groups of subjects using a Fisher’s exact test. Associations of these variants with AWS were explored by comparing allele frequencies between alcohol-dependent subjects with a history of AWS and alcohol-dependent subjects without AWS history. Associations with alcoholism were explored by comparing allele frequencies of alcohol-dependent subjects without AWS history and frequencies in the nonalcoholic Coriell subjects.
Haplotype association tests were performed on haplotypes composed of all SNPs with a MAF ≥ 0.05, as well as tag-SNPs representative of these SNPs, using the HaploStats package (Schaid et al., 2002). First, a global test of association was performed. This test simultaneously considers all haplotypes (allelic combinations) of a specific set of tightly linked polymorphisms, uses a single statistic to test whether frequencies of any of these haplotypes differ between cases and controls and, thus, eliminates the need for correction for multiple testing. Subsequently, haplotype-specific tests for the most common haplotypes (frequency > 0.05) were performed. Each haplotype-specific test assesses whether the frequency of a particular haplotype differs between cases and controls. For the haplotype with the largest score statistic, demonstrating the strongest evidence of association with the phenotype of interest, a simulation-based p-value for the maximum score statistic was calculated. The maximum statistic simulated p-value is used to control for multiple testing in this case. It is expected that the global test may be more powerful when several haplotypes are associated with the trait, while the maximum score statistic is expected to be more powerful than the global test of association when the effect of a single haplotype is much larger than that of all the other haplotypes (see Schaid et al., 2002, for details).
RESULTS
Study Subjects
The group of alcohol-dependent subjects who had a history of AWS included 44 males (72%) and 17 females (28%) with a mean age of 50 ± 10. The group of alcohol-dependent subjects without an AWS history included 45 males (76%) and 14 females (24%) who had a mean age of 52 ± 8. The average age of the beginning of regular use of alcohol was 18 ± 4 in the AWS group and 18 ± 3 in the group of subjects without AWS history. Maximum drinking tolerance (defined as the maximum number of drinks ever consumed per 24 hour) was 29 ± 18 in the AWS group and 23 ± 10 in the group of alcohol-dependent subjects without AWS. This difference in maximum drinking tolerance was not statistically significant (Kruskal–Wallis test p > 0.05).
Resequencing of the Human MPDZ Gene
The 46 exons, exon–intron boundaries, and proximal promoter region of the MPDZ gene were resequenced and 92 variants were identified including 67 new variants. A detailed description of the identified variations is presented in Table 1 and their locations are illustrated in Fig. 1. Primer sequences are available in the Table S1. Of the 92 variants, 13 had MAF ≥ 0.05 in the combined set of all subjects (Table 1, Fig. 1). Figure 2 shows the LD pattern of these 13 SNPs with a MAF ≥ 0.05.
Table 1.
Polymorphisms of MPDZ Gene of Alcoholic Subjects and Coriell Controls
| Minor allele frequency |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr. position | Gene position | dbSNP ID | Variation | Amino acid change | Location | Position | Coriell n = 64 | No AWS n = 60 | AWS n = 60 | |
| 1 | 13242041 | −1727 | G > C | Upstream-1727 | Upstream-1727 | 0.0000 | 0.0085 | 0.0000 | ||
| 2 | 13241615 | −1301 | C > G | Upstream-1301 | Upstream-1301 | 0.0078 | 0.0000 | 0.0000 | ||
| 3 | 13240778 | −464 | G > A | Upstream-464 | Upstream-464 | 0.0000 | 0.0085 | 0.0000 | ||
| 4 | 13240750 | −436 | insT | Upstream-436 | Upstream-436 | 0.2344 | 0.2458 | 0.1967 | ||
| 5 | 13240302 | 13 | A > G | Ile5Val | Exon 1 | c13 | 0.0000 | 0.0085 | 0.0082 | |
| 6 | 13240274 | 41 | C > T | Intron 1 | IVS1 + 25 | 0.0078 | 0.0085 | 0.0164 | ||
| 7 | 13237636 | 2679 | C > G | Gln61Glu | Exon 2 | c181 | 0.0000 | 0.0085 | 0.0000 | |
| 8 | 13237519 | 2796 | A > G | Intron 2 | IVS2 + 115 | 0.0000 | 0.0085 | 0.0082 | ||
| 9 | 13214491 | 25824 | rs17273542 | C > T | Ser92Leu | Exon 3 | c275 | 0.0078 | 0.0169 | 0.0000 |
| 10 | 13214441 | 25874 | G > T | Gly109Cys | Exon 3 | c325 | 0.0000 | 0.0000 | 0.0082 | |
| 11 | 13214410 | 25905 | G > C | Cys119Ser | Exon 3 | c356 | 0.0000 | 0.0000 | 0.0082 | |
| 12 | 13214354 | 25961 | A > C | Intron 3 | IVS3 + 31 | 0.0156 | 0.0000 | 0.0000 | ||
| 13 | 13213592 | 26723 | C > G | Gln171Glu | Exon 4 | c511 | 0.0000 | 0.0000 | 0.0082 | |
| 14 | 13213515 | 26800 | G > A | Intron 4 | IVS4 + 55 | 0.0000 | 0.0000 | 0.0082 | ||
| 15 | 13212200 | 28115 | A > C | Intron 5 | IVS5 + 32 | 0.0156 | 0.0085 | 0.0082 | ||
| 16 | 13211401 | 28914 | A > G | Lys282Lys | Exon 6 | c846 | 0.0078 | 0.0000 | 0.0000 | |
| 17 | 13209839 | 30476 | C > T | Intron 6 | IVS6 + 1532 | 0.0078 | 0.0000 | 0.0000 | ||
| 18 | 13209797 | 30517–30518 | delTA | Intron 6 | IVS6 + 1573–74 | 0.0156 | 0.0000 | 0.0000 | ||
| 19 | 13209603 | 30712 | rs34911705 | G > C | Leu347Phe | Exon 7 | c1041 | 0.0078 | 0.0000 | 0.0000 |
| 20 | 13209570 | 30745 | rs13297480 | A > G | Thr358Thr | Exon 7 | c1074 | 0.0938 | 0.2034 | 0.0902 |
| 21 | 13209459 | 30856 | A > G | Intron 7 | IVS7 + 99 | 0.0000 | 0.0085 | 0.0000 | ||
| 22 | 13195789 | 44526 | A > T | Intron 10 | IVS10 + 126 | 0.0313 | 0.0000 | 0.0164 | ||
| 23 | 13195264 | 45051 | rs2039333 | T > C | Intron 10 | IVS10 + 651 | 0.0313 | 0.0000 | 0.0164 | |
| 24 | 13195020 | 45295 | C > T | Intron 11 | IVS11 + 15 | 0.0000 | 0.0085 | 0.0000 | ||
| 25 | 13186032 | 54283 | T > C | Intron 12 | IVS12 + 88 | 0.0078 | 0.0000 | 0.0082 | ||
| 26 | 13182360 | 57955 | insA | Intron 13 | IVS13 + 806 | 0.0156 | 0.0169 | 0.0164 | ||
| 27 | 13182320 | 57995 | rs1331676 | G > C | Intron 13 | IVS13 + 846 | 0.0000 | 0.0000 | 0.0082 | |
| 28 | 13182201 | 58114 | C > T | Arg633STOP | Exon 14 | c1897 | 0.0000 | 0.0085 | 0.0000 | |
| 29 | 13182048 | 58267 | rs13291548 | A > G | Intron 14 | IVS14 + 82 | 0.0391 | 0.0169 | 0.0000 | |
| 30 | 13182024 | 58291 | rs16930194 | C > T | Intron 14 | IVS14 + 106 | 0.0234 | 0.0339 | 0.0164 | |
| 31 | 13180292 | 60023 | G > A | Val659Ile | Exon 15 | c1975 | 0.0078 | 0.0085 | 0.0082 | |
| 32 | 13180272 | 60043 | C > T | Ile665Ile | Exon 15 | c1995 | 0.0078 | 0.0000 | 0.0000 | |
| 33 | 13180235 | 60080 | A > T | Thr678Ser | Exon 15 | c2032 | 0.0078 | 0.0000 | 0.0082 | |
| 34 | 13180163 | 60152 | rs4741289 | G > A | Glu702Lys | Exon 15 | c2104 | 0.0078 | 0.0000 | 0.0164 |
| 35 | 13180162 | 60153 | rs4740548 | A > T | Glu702Val | Exon 15 | c2105 | 0.0078 | 0.0000 | 0.0164 |
| 36 | 13179093 | 61222 | C > T | Intron 15 | IVS15 + 1020 | 0.0000 | 0.0000 | 0.0082 | ||
| 37 | 13179017 | 61298 | rs41265290 | A > G | Intron 15 | IVS15 + 1096 | 0.0313 | 0.0000 | 0.0082 | |
| 38 | 13178754 | 61561 | T > G | Intron 16 | IVS16 + 29 | 0.0078 | 0.0000 | 0.0000 | ||
| 39 | 13178719 | 61596 | T > C | Intron 16 | IVS16 + 64 | 0.0000 | 0.0085 | 0.0000 | ||
| 40 | 13176355 | 63960 | A > G | Lys799Glu | Exon 17 | c2395 | 0.0078 | 0.0000 | 0.0082 | |
| 41 | 13173701 | 66614 | A > G | Intron 17 | IVS17 + 2568 | 0.0000 | 0.0085 | 0.0000 | ||
| 42 | 13173486 | 66829 | rs34704118 | A > G | Leu860Leu | Exon 18 | c2580 | 0.0156 | 0.0000 | 0.0082 |
| 43 | 13166312 | 74003 | rs2274856 | G > A | Ser918Ser | Exon 19 | c2754 | 0.2734 | 0.3051 | 0.2049 |
| 44 | 13165987 | 74328 | rs1331674 | G > A | Intron 19 | IVS19 + 148 | 0.2734 | 0.2966 | 0.2131 | |
| 45 | 13165908 | 74407 | C > T | Intron 19 | IVS19 + 227 | 0.0391 | 0.0339 | 0.0164 | ||
| 46 | 13165719 | 74594–74596 | rs3831219 | delCTC | Intron 20 | IVS20 + 30–32 | 0.3359 | 0.3390 | 0.2541 | |
| 47 | 13165639 | 74676 | G > T | Intron 20 | IVS20 + 112 | 0.0000 | 0.0085 | 0.0000 | ||
| 48 | 13158588 | 81727 | T > C | Intron 20 | IVS20 + 7163 | 0.0000 | 0.0000 | 0.0082 | ||
| 49 | 13158251 | 82064 | insT | Intron 21 | IVS21 + 114 | 0.0000 | 0.0085 | 0.0000 | ||
| 50 | 13153010 | 87305 | rs1331672 | C > G | Intron 21 | IVS21 + 5355 | 0.3359 | 0.3220 | 0.2295 | |
| 51 | 13148261 | 92054 | delC | Intron 22 | IVS22 + 4429 | 0.0156 | 0.0085 | 0.0164 | ||
| 52 | 13148062 | 92253 | rs41265286 | G > A | Ser1136Asn | Exon 23 | c3407 | 0.0078 | 0.0085 | 0.0000 |
| 53 | 13140714 | 99601 | G > A | Intron 23 | IVS23 + 7303 | 0.0313 | 0.0000 | 0.0082 | ||
| 54 | 13140558 | 99757 | T > G | Ser1194Arg | Exon 24 | c3582 | 0.0078 | 0.0085 | 0.0000 | |
| 55 | 13140531 | 99784 | rs10756457 | A > G | Lys1203Lys | Exon 24 | c3609 | 0.3359 | 0.3475 | 0.2377 |
| 56 | 13140506 | 99809 | C > T | Intron 24 | IVS24 + 4 | 0.0000 | 0.0000 | 0.0082 | ||
| 57 | 13137723 | 102592 | rs7024892 | A > G | Intron 24 | IVS24 + 2787 | 0.3281 | 0.3644 | 0.2377 | |
| 58 | 13137602 | 102713 | G > A | Arg1229Gln | Exon 25 | c3686 | 0.0000 | 0.0085 | 0.0000 | |
| 59 | 13137569 | 102746 | A > T | Gln1240Leu | Exon 25 | c3719 | 0.0000 | 0.0000 | 0.0082 | |
| 60 | 13133591 | 106724 | G > A | Intron 25 | IVS25 + 3956 | 0.0000 | 0.0000 | 0.0082 | ||
| 61 | 13133470 | 106845 | G > T | Asp1279Tyr | Exon 26 | c3835 | 0.0000 | 0.0085 | 0.0000 | |
| 62 | 13133254 | 107061 | T > C | Intron 26 | IVS26 + 211 | 0.0078 | 0.0000 | 0.0000 | ||
| 63 | 13130070 | 110245 | G > A | Glu1307Lys | Exon 27 | c3919 | 0.0000 | 0.0085 | 0.0000 | |
| 64 | 13127982 | 112333 | delT | Leu1392a | Exon 28 | c4174 | 0.0078 | 0.0000 | 0.0000 | |
| 65 | 13127889 | 112426 | A > G | Intron 28 | IVS28 + 67 | 0.0000 | 0.0085 | 0.0082 | ||
| 66 | 13126160 | 114155 | G > C | Gln1438His | Exon 30 | c4314 | 0.0000 | 0.0000 | 0.0082 | |
| 67 | 13123801 | 116514 | A > G | Intron 31 | IVS31 + 22 | 0.0000 | 0.0000 | 0.0082 | ||
| 68 | 13116836 | 123479 | G > T | Intron 31 | IVS31 + 6987 | 0.0078 | 0.0085 | 0.0000 | ||
| 69 | 13115415 | 124900 | T > C | Intron 33 | IVS33 + 1100 | 0.0078 | 0.0000 | 0.0000 | ||
| 70 | 13115201 | 125114 | rs2297003 | A > G | Intron 34 | IVS34 + 14 | 0.3359 | 0.3390 | 0.2459 | |
| 71 | 13113330 | 126985 | T > C | Intron 34 | IVS34 + 1885 | 0.0000 | 0.0000 | 0.0082 | ||
| 72 | 13109766 | 130549 | A > G | Intron 37 | IVS37 + 1972 | 0.0000 | 0.0085 | 0.0082 | ||
| 73 | 13105422 | 134893 | rs2274647 | A > C | Intron 38 | IVS38 + 4079 | 0.3125 | 0.3644 | 0.2705 | |
| 74 | 13105350 | 134965 | insC | Intron 38 | IVS38 + 4151 | 0.0078 | 0.0085 | 0.0082 | ||
| 75 | 13105349 | 134966 | rs41265282 | C > G | Intron 38 | IVS38 + 4152 | 0.0313 | 0.0000 | 0.0082 | |
| 76 | 13103159 | 137156 | delA | Intron 40 | IVS40 + 771 | 0.0313 | 0.0000 | 0.0082 | ||
| 77 | 13103006 | 137309 | A > G | Intron 41 | IVS41 + 4 | 0.0000 | 0.0085 | 0.0000 | ||
| 78 | 13102228 | 138087 | C > G | Intron 41 | IVS41 + 782 | 0.0078 | 0.0000 | 0.0000 | ||
| 79 | 13102024 | 138291 | rs34605667 | G > A | Arg1880Lys | Exon 42 | c5639 | 0.0547 | 0.0339 | 0.0492 |
| 80 | 13102015 | 138300 | G > A | Intron 42 | IVS42 + 8 | 0.0000 | 0.0000 | 0.0082 | ||
| 81 | 13100138 | 140177 | T > A | Intron 43 | IVS43 + 497 | 0.0078 | 0.0000 | 0.0000 | ||
| 82 | 13100068 | 140247 | C > T | Intron 43 | IVS43 + 567 | 0.0078 | 0.0000 | 0.0000 | ||
| 83 | 13099969 | 140346 | T > C | Ile1947Thr | Exon 44 | c5840 | 0.0000 | 0.0085 | 0.0000 | |
| 84 | 13099015 | 141300 | G > T | Gly1968Cys | Exon 45 | c5902 | 0.0000 | 0.0000 | 0.0082 | |
| 85 | 13098915 | 141400 | C > A | Intron 45 | IVS45 + 20 | 0.0000 | 0.0000 | 0.0082 | ||
| 86 | 13098824 | 141491 | G > A | Intron 45 | IVS45 + 111 | 0.0078 | 0.0000 | 0.0000 | ||
| 87 | 13098794 | 141521 | rs3765550 | A > G | Intron 45 | IVS45 + 141 | 0.4063 | 0.3729 | 0.4426 | |
| 88 | 13097222 | 143093 | C > T | Intron 45 | IVS45 + 1713 | 0.0000 | 0.0000 | 0.0082 | ||
| 89 | 13097121 | 143194 | A > G | Intron 45 | IVS45 + 1814 | 0.0078 | 0.0085 | 0.0000 | ||
| 90 | 13096598 | 143717 | rs3264 | A > G | Exon 46 | 3′-UTR + 366 | 0.3359 | 0.4237 | 0.3689 | |
| 91 | 13096469 | 143846 | A > G | Intron 46 | Intergenic + 109 | 0.0000 | 0.0000 | 0.0082 | ||
| 92 | 13096335 | 143980 | rs722651 | C > T | Intron 46 | Intergenic + 243 | 0.3828 | 0.2627 | 0.3607 | |
AWS, alcohol withdrawal seizure.
Variations included in the haplotype analysis are highlighted. AWS group included alcohol-dependent subjects with a history of withdrawal seizures. No AWS group included alcohol-dependent subjects without history of withdrawal seizures.
A frameshift.
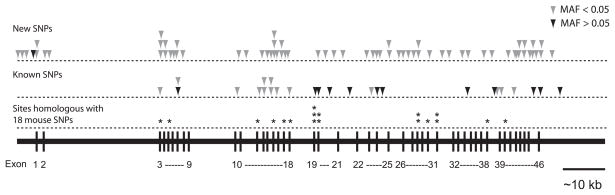
Fig. 1.
Schematic representation of the known and newly discovered sequence variations in the human MPDZ gene. The arrows indicate approximate locations of the variant alleles. Pale arrows indicate variants with minor allele frequency (MAF) < 0.05 and dark arrows indicate variants with MAF > 0.05. The numbering of cSNPs corresponds to the start codon, ATG. Detailed information about each variant is presented in Table 1. Positions in the human MPDZ sequence homologous to 18 sites in mouse Mpdz sequence associated with variable propensity to alcohol-withdrawal seizures are indicated by *. Detailed information about each of these positions in the cDNA sequence is presented in Table 2.
Fig. 2.
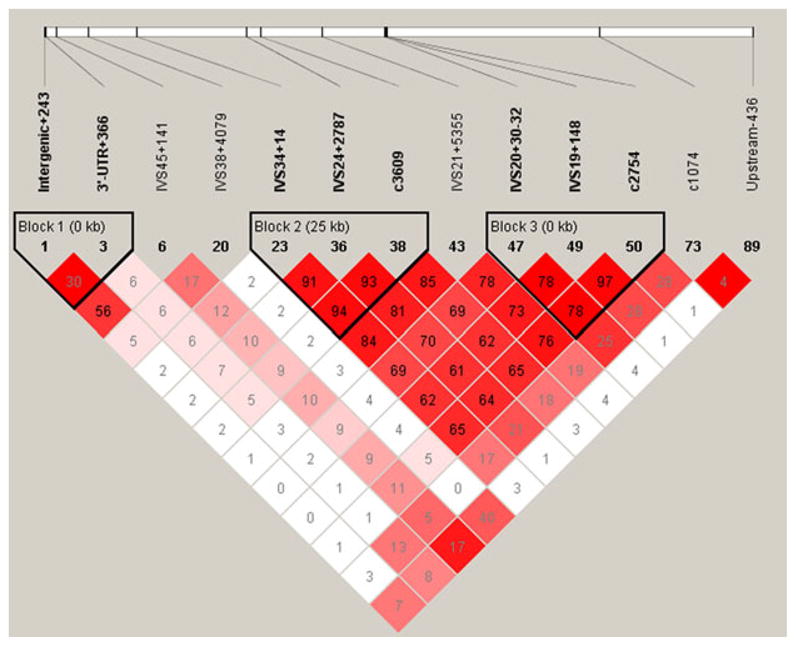
LD and haplotype block analysis of 13 MPDZ variations with MAF ≥ 0.05. LD plot prepared using Haploview (Barrett et al., 2005). Colors are used to display pairwise LD as follows: bright red, LOD ≥ 2 and D′ = 1; shades of pink/red, LOD ≥ 2 and D′ < 1; white, LOD < 2 and D′ < 1. The numbers indicate pairwise r2 values shown as a percentage. Blocks are defined based on the confidence interval method as described by Gabriel and colleagues (2002).
Comparison of the Human MPDZ Gene With the Mouse Homolog
As allelic variation in specific gene locations was associated with differences in severity of alcohol withdrawal as defined by presence of seizures in inbred strains of mice (Fehr et al., 2002), sequence similarity between human and mouse variants was explored. The human gene sequence (Ensembl transcript OTTHUMT00000055488 updated with our resequencing data) was compared with the gene sequence of 3 strains of mice: the wild type C57BL/6J (NCBI# AF326531), the C57L/J (NCBI# AF326533), and the DBA/2J (NCBI# AF326544). The alignment showed 76% global sequence identity between human MPDZ gene and its mouse homolog Mpdz. As shown in Table 2, at 12 of 18 cDNA positions forming withdrawal seizures-related haplotype in mice, the nucleotides in human sequence were identical with C57L/J strain and 10 of 18 of these haplotype-forming sites were the same in human and the DBA/2J strain. Both of these strains are prone to severe withdrawal and seizures. In contrast, only in 5 of 18 haplotype-forming sites nucleotides were identical in humans and the withdrawal seizure-resistant C57BL/6J strain. However, no genetic variability of these sites in human MPDZ gene was found in our sample of 240 chromosomes from alcoholics and 128 chromosomes from Coriell subjects (all with EA ancestry). This analysis had sufficient power (97.5%) to reveal rare polymorphisms and mutations (MAF = 0.01).
Table 2.
Comparison Between cDNA Sequence Positions of the Nucleotides FormingWithdrawal Seizures-Related Haplotype in Mouse Mpdz Gene and Nucleotides in the Homologues Positions in the Human MPDZ Gene
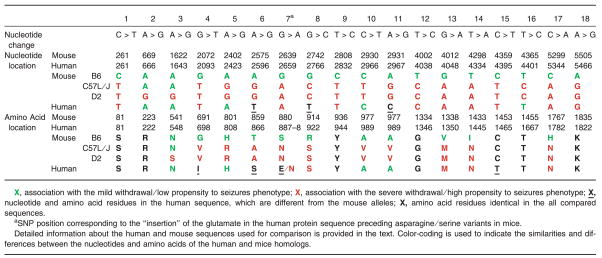 |
The amino acid sequence of the human MPDZ protein (NP_003820) was compared to those of the inbred lines, C57BL/6J (AAL37377), C57L/J (AAL37379), and DBA/2J (AAL37390). Alignment showed 83% global sequence identity between the human MPDZ protein and its mouse homolog. As shown in Table 2, of the 18 amino acids corresponding to the SNPs described as being part of the seizure-prone haplotype in mice, 6 were completely conserved between human and mice. Of the remaining 12 amino acids, 5 amino acids in C57L/J strain, 4 amino acids in DBA/2J strain, and 4 in C57BL/6J strain were identical to the human sequence (Table 2).
Comparison of the MPDZ Variations Frequency in the Alcoholic and Coriell Samples
To explore potential associations of the human MPDZ gene variations with AWS and with alcoholism, we studied 13 variants (Table 3) with MAF ≥ 0.05. The synonymous rs13297480 G variant, in exon 7, was found to be significantly more prevalent in alcoholics without history of AWS compared to nonalcoholic Coriell subjects (p = 0.019), or to alcoholics with history of AWS (p = 0.017). Also, the intron 24 rs7024892 G variant was found to be less prevalent in AWS group compared to the group without AWS history. Given that these results are not corrected for the multiple comparisons performed, replication is necessary to confirm these findings.
Table 3.
Polymorphic Variants of the MPDZ Gene Selected for LD and Haplotype Analysis
| Minor allele frequency |
No AWSa versus Coriellb |
AWS versus Alcoholicb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP ID | Variant | Location | AWS | No AWSa | Coriell | p value | OR | 95% CI | p value | OR | 95% CI |
| N/A | insT | −436 upstream | 0.197 | 0.246 | 0.234 | 0.882 | 1.064 | 0.567–1.994 | 0.437 | 0.752 | 0.388–1.450 |
| rs13297480 | A > G | Exon 7 | 0.090 | 0.203 | 0.094 | 0.019 | 2.459 | 1.113–5.701 | 0.017 | 0.390 | 0.163–0.878 |
| rs2274856 | G > A | Exon 19 | 0.205 | 0.305 | 0.273 | 0.673 | 0.858 | 0.475–1.546 | 0.078 | 1.700 | 0.909–3.218 |
| rs1331674 | G > A | Intron 19 | 0.213 | 0.297 | 0.273 | 0.777 | 0.893 | 0.494–1.614 | 0.142 | 1.554 | 0.832–2.930 |
| rs3831219 | delCTC | Intron 20 | 0.254 | 0.339 | 0.336 | 1.000 | 1.014 | 0.577–1.780 | 0.160 | 0.665 | 0.365–1.204 |
| rs1331672 | C > G | Intron 21 | 0.230 | 0.322 | 0.336 | 0.892 | 0.939 | 0.532–1.655 | 0.114 | 0.628 | 0.339–1.154 |
| rs10756457 | A > G | Exon 24 | 0.238 | 0.347 | 0.336 | 0.893 | 1.052 | 0.600–1.845 | 0.066 | 0.587 | 0.320–1.067 |
| rs7024892 | A > G | Intron 24 | 0.238 | 0.364 | 0.328 | 0.593 | 1.173 | 0.670–2.055 | 0.035 | 0.545 | 0.298–0.988 |
| rs2297003 | A > G | Intron 34 | 0.246 | 0.339 | 0.336 | 1.000 | 0.987 | 0.562–1.734 | 0.120 | 1.570 | 0.864–2.873 |
| rs2274647 | A > C | Intron 38 | 0.270 | 0.364 | 0.313 | 0.420 | 1.260 | 0.718–2.217 | 0.129 | 0.648 | 0.360–1.159 |
| rs3765550 | A > G | Intron 45 | 0.443 | 0.373 | 0.406 | 0.603 | 0.870 | 0.503–1.499 | 0.295 | 1.334 | 0.772–2.315 |
| rs3264 | A > G | Exon 46 | 0.369 | 0.424 | 0.336 | 0.188 | 1.451 | 0.838–2.522 | 0.429 | 0.796 | 0.458–1.379 |
| rs722651 | C > T | Intron 46 | 0.361 | 0.263 | 0.383 | 0.056 | 0.576 | 0.321–1.023 | 0.125 | 1.580 | 0.880–2.862 |
AWS, group of alcoholic subjects with a history of withdrawal seizure; OR, odd ratio; CI, confidence intervals.
No AWS group included alcohol-dependent subjects without history of withdrawal seizure and/or delirium tremens.
Statistical analyses were performed using Fisher’s exact test.
p values below 0.05 are presented in bold.
The association between MPDZ haplotypes and alcoholism with and without AWS was further investigated by considering the haplotype that comprised all 13 SNPs with MAF ≥ 0.05. The comparisons were made using the approach proposed by Schaid and colleagues (2002), as described in the Methods section. The seven 13-SNP haplotypes with frequencies > 0.05 used for these comparisons are presented in Table 4.
Table 4.
Association of MPDZ Haplotypes With AlcoholismWithout AWS
| SNP |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | −436 Upstream* | rs13297480* | rs2274856* | rs1331674 | rs3831219* | rs1331672 | rs10756457* | rs7024892 | rs2297003 | rs2274647* | rs3765550* | rs3264* | rs722651* | Haplotype frequency | Simulated p-value | Max statistic simulated p-value |
| 1 | A | G | G | C | A | A | G | A | A | G | C | 0.0621 | 0.087 | |||
| 2 | A | G | G | C | A | A | G | A | A | A | C | 0.0725 | 0.164 | |||
| 3 | A | G | G | C | A | A | G | A | G | A | T | 0.2421 | 0.162 | |||
| 4 | insT | A | A | A | delCTC | G | G | G | A | C | A | A | C | 0.0520 | 0.916 | |
| 5 | insT | A | G | G | C | A | A | G | C | A | A | C | 0.0980 | 0.179 | ||
| 6 | A | G | G | C | A | A | G | A | G | G | C | 0.0774 | 0.063 | |||
| 7 | G | A | A | delCTC | G | G | G | A | A | A | G | C | 0.0746 | 0.006 | 0.051 | |
AWS, alcohol withdrawal seizure.
Global score test statistic = 16.90, df = 7, Global simulation p-value = 0.015.
Tag-SNP.
p values below 0.05 are presented in bold.
First, to test for association between the MPDZ gene and AWS, the frequencies of the 13-SNP haplotypes were compared between subjects with AWS history and alcohol-dependent subjects without AWS. No significant haplotype associations were identified (data not shown).
To test for association between MPDZ gene and alcoholism without AWS, we compared frequencies of the same 13-SNP haplotypes between the alcohol-dependent subjects without AWS and Coriell samples. The results of this comparison, including frequencies of the 7 most common haplotypes (frequency > 0.05) and score test simulation p-values are shown in Table 4. The global score test (Schaid et al., 2002) led to a simulation-based p-value of 0.015. As explained in the Methods section, only one global haplotype test was performed for this comparison, and the p-value does not require correction for multiple testing. Tests of individual haplotype effects shown in the last column of Table 4 indicate that haplotype #7 had the strongest association with alcoholism (uncorrected p = 0.006). The corresponding maximum statistic simulation p-value, which corrects for the multiple tests performed on all individual haplotypes shown in Table 4, was 0.051 suggesting that haplotype #7 confers a higher risk of alcoholism than the other haplotypes. This haplotype is one of only 2 common haplotypes that carries the high risk rs13297480 G variant in the exon 7, indicating its potentially important role.
We also investigated association of the studied phenotypes with haplotypes composed of 9 tag-SNPs selected from the 13 common SNPs. The tag-SNPs are indicated in Table 4 with an asterisk (*) following the SNP rs number. Results were very similar to those described above for the 13-SNP haplotype: There was no association between the tag-SNP haplotypes and history of AWS; however, there was evidence of association of the tag-SNP haplotypes with alcoholism without AWS (global test simulation p = 0.031; uncorrected p-value for most strongly associated haplotype = 0.007; maximum statistic simulation p-value = 0.045).
DISCUSSION
Animal studies have demonstrated that genetic variations in coding sequence of Mpdz gene and changes in the expression of this gene might be associated with the severity of acute alcohol withdrawal and seizures (Fehr et al., 2002; Shirley et al., 2004). To investigate the signficance of this finding in human subjects with alcoholism, sequence variations of the human MPDZ gene were examined in alcohol-dependent subjects (EA ancestry) with and without history of AWS and in the DNA samples from EA subjects available at the Coriell Institue (as representatives of the EA population). Sequencing of 240 chromosomes from the alcohol-dependent subjects and 128 chromosomes from the Coriell subjects provided sufficient power to reveal rare polymorphisms and mutations, with 97.5% power to detect variations with a MAF of 0.01 within specific ethnic group. We identified 67 new sequence variants but found no genetic variability in the human MPDZ gene sites homologous to the variability sites described as being part of a withdrawal seizure-related haplotype in mice. We also found no significant haplotype association of MPDZ gene with a history of AWS, although a larger study would be recommended to reliably rule out this possibility.
At the same time, a statistically significant haplotype association of MPDZ gene with alcoholism was demonstrated when subjects without AWS history were compared to Coriell controls. This association disappeared when haplotype association was tested in all 120 alcoholics (with and without AWS) versus Coriell controls (data not shown). Several important issues need to be considered in order to interpret this finding. First, different alcoholism-related phenotypes may have shared as well as independent genetic risk factors. For example, it is well known that AWS-resistant C57BL/6J mice consume significantly more alcohol than the AWS-prone DBA/2J mice (Belknap et al., 1993; Metten and Crabbe, 2005). Two (or more) different genetic variations may be responsible for the increased propensity to AWS and decreased preference of alcohol. Alternatively, the pleiotropic effects of a single genetic variation may contribute to both phenotypes, leading to inverse correlation between propensity to AWS and alcohol preference phenotypes in mice. Both of these possibilities are potentially applicable to humans and need to be investigated. Second, all alcohol-dependent subjects in our study had a history of alcohol withdrawal and the number of subjects with the history of AWS was increased compared to their frequency in general population or among treatment seeking alcoholics (Caetano et al., 1998). Consequently, it is possible that increased presence of the AWS phenotype may mask the association between MPDZ gene and other alcohol-related phenotypes, including alcoholism in general and alcohol withdrawal symptoms other than seizures. Comparison of alcohol-dependent subjects with and without history of withdrawal is necessary in order to investigate this posibility. In fact, in the model studies in mice, only one alcoholism-related phenotype, defined as “acute alcohol withdrawal,” has been studied for association with Mpdz gene variations and mRNA expression (Fehr et al., 2002; Shirley et al., 2004). The results of this study indicate that the role of MPDZ gene variability in a broader array of alcohol-related phenotypes needs to be considered. As a scaffolding protein, MPDZ is known to intereact with serotonin, dopamine, glutamate, and GABA receptors in the brain (Bettler et al., 2004; Griffon et al., 2003; Krapivinsky et al., 2004; Parker et al., 2003). Therefore, if particular variations of the MPDZ gene alter its expression or protein structure, these variants could be associated with changes in addictive behaviors (e.g., impulsivity, craving, level of response and/or withdrawal), which have been associated with serotonin, dopamine, glutamate, and GABA neurotransmission and are implicated in alcoholism-related phenotypes in humans and in animal models (Dahchour and De Witte, 2003; De Witte et al., 2003; Heinz et al., 2004; Kalivas and Volkow, 2005; Krystal et al., 2003; Nakamura et al., 1999).
The discrepancy between allelic variations in the coding sequence of the mouse Mpdz gene associated with severity of alcohol withdrawal and corresponding human sequence might explain the lack of genetic association of human MPDZ haplotypes and the history of withdrawal seizures. However, this conclusion should not be extended to an association between the abundance of the Mpdz mRNA in the different strains of mice and severity of alcohol withdrawal (Shirley et al., 2004). It is possible that noncoding variations may be responsible for the difference of Mpdz mRNA expression in different strains of mice and that these variations may potentially be conserved between species. Our findings also suggest that species-specific genetic variations may contribute to the susceptibility to the disease while the role of the gene remains similar between species. Although the pattern of results emerging in psychiatric genetics is generally consistent with the findings of behavioral genetics in animal models, for some behaviors, the pathway from genes to behavior differs meaningfully between species (Kendler and Greenspan, 2006). Further research in mice and human studies are necessary to address this issue.
Our analyses indicated that a common rs7024892 variant in intron 24 was less frequent in the AWS group when compared to alcoholics without AWS while the synonymous rs13297480 (c1074A/G, Thr358Thr) substitution in exon 7 showed association with alcoholism (although not after correction for multiple testing). The minor 1074G allele was twice as frequent in the alcoholic group without AWS when compared to either subjects with AWS or Coriell groups. The same 1074G allele also differentiated haplotype #7, which showed strongest association with alcoholism. This warrants further investigation of its functional importance. Although synonymous single-nucleotide polymorphisms do not produce altered coding sequences, the presence of a rare codon, marked by a synonymous polymorphism, may affect the timing of cotranslational folding and alter protein translation kinetics (Kimchi-Sarfaty et al., 2007). Thus, studies investigating effects of the rs13297480 SNP on MPDZ protein translation kinetics in the context of alcohol effects could be of interest. In addition, it was demonstrated that the expression of the Thr (TGT) tRNA is higher compared to the Thr (CGT) tRNA in most body tissues (Dittmar et al., 2006). These tRNA anticodons correspond to the ACA1074ACG variation of the Exon 7 SNP in the MPDZ gene. Thus, differences in the MPDZ expression in individuals with the c1074A and c1074G alleles may be tissue specific (e.g., different in the brain vs. liver). In this case, study of the tissue-specific response to alcohol and its withdrawal in subjects with different c1074 alleles would be of interest.
Although association tests were not performed for rare SNPs, we did note that 2 rare intronic variants (rs41265290 in Intron 15 and 7303GA in Intron 23) were more common in Coriell sample (3%) than in subjects with AWS history (0.8% in AWS group and 0.0% in alcohol-dependent subjects without AWS) (Table 1). Our sample does not provide adequate power to test for statistical significance of these differences and association with the group status. However, the last 2 variants are located near splice junctions and may influence alternative splicing patterns. Genomic variants located in or near consensus splice junctions at the exon–intron boundaries are important for specifying the splice sites, and estimated to affect more than 15% of human genetic diseases (Faustino and Cooper, 2003; Garcia-Blanco et al., 2004; Pagani and Baralle, 2004). Several transcripts of the MPDZ gene were reported in humans. Thus, further investigation involving a sufficiently high number of study subjects should focus on these rare, but potentially important, variants and their contribution to the differential response to alcohol and its withdrawal in different subjects.
A limitation of this study is that only DNA from EA subjects was utilized, so it was not possible to assess variability in the human MPDZ gene in other ethnic groups. In addition, as the nonalcoholic control DNA was obtained from the Coriell Institute, rather than through random sampling from the same population as the alcoholic patients, potential effects of population stratification should be considered. As only EA subjects were enrolled in the study, we do not expect that population stratification was a serious problem. Another limitation is that there is no information about alcohol use by the subjects who provided DNA samples for the Coriell Institute. A consequence of using a control group for which detailed phenotyping is not available relates to the potential for mis-classification bias. Specifically, a proportion of the controls is likely to have the disease of interest or may develop it in the future. However, the resultant decrease in power should not be high. For example, if 5% of controls would meet the definition of cases, the loss of power is approximately the same as that due to a reduction of the sample size by 10% (Colhoun et al., 2003). As the prevalence of alcohol dependence is only 3.85% of white males and 2.37% in white females (Grant et al., 2004), the potential effect this has on the power of our study is modest. These considerations are in line with the approach used in several large case-control association studies [e.g., WTCCC Consortium (2007)]; however, a study design utilizing phenotypically screened controls would generally be desirable in future studies.
The association analyses between human MPDZ variation and alcoholism investigated here were an exploratory component of this study. A replication in an independent sample is necessary to confirm the obtained positive results. In addition, the possibility and nature of potential associations between the MPDZ gene variations, alcohol withdrawal, and/or other alcoholism-related phenotypes need to be further explored. If any of such associations were to be confirmed, it could be a useful genetic marker for increased risk of the development of alcoholism or related phenotypes.
In conclusion, our sequencing of the MPDZ gene in samples obtained from alcohol-dependent subjects and nonalcoholic control samples from Coriell Institute (all with EA ancestry) revealed 67 new, mostly rare variations. None of these variations was located in the sites identical to those associated with AWS sensitivity in mice. Exploratory haplotype and single SNP association analyses suggest possible association between the MPDZ gene and alcohol dependence and/or related phenotypes. Further investigation of MPDZ variations, including those that could alter MPDZ mRNA stability, translational efficiency, and/or alternative splicing (e.g., rs13297480 SNP in exon 7, rs41265290 in intron 15, and 7303G/A in intron 23) could provide important insights into the mechanism of this association and reveal valuable genetic markers of alcoholism and related phenotypes.
Supplementary Material
Table S1. List of Primers for Genomic DNA Fragment Amplication and Resequencing
Acknowledgments
This study was supported by grants from the Samuel C. Johnson Genomics of Addiction Program (VMK, DSC, JMB, DAM, and EDW), the National Institutes of Health (AA015164, DSC), Mayo-Thompson Fellowship on Basic Research in Alcoholism (VMK), Decker-Denko Foundation (VMK), and Educational Foundations of America (VMK). We thank Dina Drubach, Maureen Drews, Vickie Courson, and Tracy Pietrzak for expert study coordination and data management.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology. 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Bettler B, Kaupmann K, Mosbacher J, Gassmann M. Molecular structure and physiological functions of GABA(B) receptors. Physiol Rev. 2004;84:835–867. doi: 10.1152/physrev.00036.2003. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano R, Clark CL, Greenfield TK. Prevalence, trends, and incidence of alcohol withdrawal symptoms: analysis of general population and clinical samples. Alcohol Health ResWorld. 1998;22:73–79. [PMC free article] [PubMed] [Google Scholar]
- Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahchour A, De Witte P. Excitatory and inhibitory amino acid changes during repeated episodes of ethanol withdrawal: an in vivo microdialysis study. Eur J Pharmacol. 2003;459:171–178. doi: 10.1016/s0014-2999(02)02851-0. [DOI] [PubMed] [Google Scholar]
- De Witte P, Pinto E, Ansseau M, Verbanck P. Alcohol and withdrawal: from animal research to clinical issues. Neurosci Biobehav Rev. 2003;27:189–197. doi: 10.1016/s0149-7634(03)00030-7. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centi-morgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991–1992 and 2001–2002. Drug Alcohol Depend. 2004;74:223–234. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Griffon N, Jeanneteau F, Prieur F, Diaz J, Sokoloff P. CLIC6, a member of the intracellular chloride channel family, interacts with dopamine D(2)-like receptors. Brain ResMol Brain Res. 2003;117:47–57. doi: 10.1016/s0169-328x(03)00283-3. [DOI] [PubMed] [Google Scholar]
- Heinz A, Siessmeier T, Wrase J, Hermann D, Klein S, Grusser SM, Flor H, Braus DF, Buchholz HG, Grunder G, Schreckenberger M, Smolka MN, Rosch F, Mann K, Bartenstein P. Correlation between dopamine D(2) receptors in the ventral striatum and central processing of alcohol cues and craving. Am J Psychiatry. 2004;161:1783–1789. doi: 10.1176/appi.ajp.161.10.1783. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Kashima H. New findings on the genetic influences on alcohol use and dependence. Curr Opin Psychiatry. 2006;19:253–265. doi: 10.1097/01.yco.0000218595.54054.7a. [DOI] [PubMed] [Google Scholar]
- Kalivas PW, Volkow ND. The neural basis of addiction: A pathology of motivation and choice. Am J Psychiatry. 2005;162:1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Greenspan RJ. The nature of genetic influences on behavior: lessons from simpler organisms. Am J Psychiatry. 2006;163:1683–1694. doi: 10.1176/ajp.2006.163.10.1683. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A silent polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43:563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Petrakis IL, Mason G, Trevisan L, D’Souza DC. N-methyl-D-aspartate glutamate receptors and alcoholism: reward, dependence, treatment, and vulnerability. Pharmacol Ther. 2003;99:79–94. doi: 10.1016/s0163-7258(03)00054-8. [DOI] [PubMed] [Google Scholar]
- Liu IC, Blacker DL, Xu R, Fitzmaurice G, Lyons MJ, Tsuang MT. Genetic and environmental contributions to the development of alcohol dependence in male twins. Arch Gen Psychiatry. 2004;61:897–903. doi: 10.1001/archpsyc.61.9.897. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Alcohol withdrawal severity in inbred mouse (Mus musculus) strains. Behav Neurosci. 2005;119:911–25. doi: 10.1037/0735-7044.119.4.911. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Matsushita S, Nishiguchi N, Kimura M, Yoshino A, Higuchi S. Association of a polymorphism of the 5HT2A receptor gene promoter region with alcohol dependence. Mol Psychiatry. 1999;4:85–88. doi: 10.1038/sj.mp.4000474. [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- Parker LL, Backstrom JR, Sanders-Bush E, Shieh BH. Agonist-induced phosphorylation of the serotonin 5-HT2C receptor regulates its interaction with multiple PDZ protein 1. J Biol Chem. 2003;278:21576–21583. doi: 10.1074/jbc.M210973200. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Goodwin DA, Winokur G. A study of alcoholism in half siblings. Am J Psychiatry. 1972;128:1132–1136. doi: 10.1176/ajp.128.9.1132. [DOI] [PubMed] [Google Scholar]
- Shirley RL, Walter NA, Reilly MT, Fehr C, Buck KJ. Mpdz is a quantitative trait gene for drug withdrawal seizures. Neurosci Biobehav Rev. 2004;7:699–700. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- Tyndale RF. Genetics of alcohol and tobacco use in humans. Ann Med. 2003;35:94–121. doi: 10.1080/07853890310010014. [DOI] [PubMed] [Google Scholar]
- Ullmer C, Schmuck K, Figge A, Lubbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of Primers for Genomic DNA Fragment Amplication and Resequencing