Abstract
Forkhead box O (FOXO) transcription factors are involved in multiple signaling pathways and play critical roles in a number of physiological and pathological processes including cancer. The importance of FOXO factors ascribes them under multiple levels of regulation including phosphorylation, acetylation/deacetylation, ubiquitination and protein–protein interactions. As FOXO factors play a pivotal role in cell fate decision, mounting evidence suggests that FOXO factors function as tumor suppressors in a variety of cancers. FOXOs are actively involved in promoting apoptosis in a mitochondria-independent and -dependent manner by inducing the expression of death receptor ligands, including Fas ligand and tumor necrosis factor-related apoptosis-inducing ligand, and Bcl-2 family members, such as Bim, bNIP3 and Bcl-XL, respectively. An understanding of FOXO proteins and their biology will provide new opportunities for developing more effective therapeutic approaches to treat cancer.
Keywords: FOXO, cancer, apoptosis, Forkhead, death receptor ligand, Bcl-2 family members
Introduction
Forkhead box O (FOXO) genes are involved in multiple signaling pathways and play critical roles in a number of physiological and pathological processes. This review will focus on the role of these transcription factors in cancer as well as their regulation of programmed cell death.
The forkhead, or winged-helix family of transcription factors were named after the founding gene member in Drosophila (forkhead gene; for review, see Kaufmann and Knochel, 1996). The family members share a highly conserved 100 amino-acid DNA binding, or FOX domain encoding a winged-helix DNA-binding motif (Clark et al., 1993). Based on the homology within this region, the forkhead genes are grouped into 19 subclasses of FOX genes, FOXA–FOXS (Kaestner et al., 2000). While some are ubiquitously expressed in variety of tissue types, others are expressed in a restricted spatial-temporal manner (Clark et al., 1993).
FOXO factors belong to the ‘O’ (‘other’) class of the FOX superfamily (Kaestner et al., 2000). Although their FOXO forkhead box is clearly related to those found in other forkhead genes, they form the most divergent subfamily of Fox family due to a unique five amino-acid (GDSNS) insertion immediately prior to helix H3 within the forkhead domain that is directly involved in sequence-specific interaction with DNA-binding sites. FOXO proteins are conserved from worm to human. To date, only one FOXO species has been identified in invertebrates (called dauer formation-16 in Caenorhabditis elegans (Lin et al., 1997) and dFOXO in Drosophila). In mammals, four subfamily members have been identified: FOXO1 (previously known as FKHR; Galili et al., 1993), FOXO3 (previously known as FKHRL1; Hillion et al., 1997; Anderson et al., 1998), FOXO4 (previously known as AFX; Corral et al., 1993; Parry et al., 1994; Borkhardt et al., 1997) and FOXO6 (Jacobs et al., 2003). FOXO1, FOXO3 and FOXO4 mRNAs are expressed ubiquitously in varying levels in mammals (Anderson et al., 1998; Furuyama et al., 2000; Biggs et al., 2001). The FOXO1 transcript is highly expressed in adipose tissues, FOXO3 mRNA is abundant in the brain, heart, kidney and spleen, and the FOXO4 transcript is expressed at higher levels in skeletal muscle. In contrast, FOXO6 mRNA is predominantly present in the developing and adult brain, suggesting that FOXO6 may play an important role in the nervous system (Jacobs et al., 2003).
While the DNA-binding domain (forkhead box) of FOXO factors is located in the N-terminal portion of these proteins, the transactivation domain is located in the C-terminal region of the molecule. FOXO proteins can act not only as transcriptional activators but also as transcriptional repressors (Ramaswamy et al., 2002). Therefore, this family of transcription factors may exert positive and negative effects on gene expression depending on the promoter context and extracellular conditions. In addition, FOXO factors also contain a ‘nuclear export sequence’ and a ‘nuclear localization signal’ (Biggs et al., 1999; Brownawell et al., 2001), which allow nucleocytoplasmic shuttling.
FOXO proteins function predominantly as transcription factors in the nucleus and bind as monomers to their cognate DNA-targeting sequences. Selection of high-affinity DNA-binding sites from a pool of degenerate oligonucleotides has identified a consensus FOXO recognition element (FRE) as (G/C)(T/A)AA(C/T)AA (Biggs et al., 1999; Furuyama et al., 2000; Gilley et al., 2003), which diverges from that of other forkhead proteins. Functional FRE sites that match this consensus sequence have been identified in the promoters of FOXO target genes encoding Fas ligand (FasL), insulin-like growth factor-binding protein 1, the apoptotic regulator Bcl-2 interacting mediator of cell death (Bim) and others (for reviews, see Accili and Arden, 2004; Greer and Brunet, 2005). By comparative genomic approaches, many additional putative FOXO-target genes and their potential cis-regulatory sites have been identified (Xuan et al., 2005). Thus, FOXO transcription factors may preferentially interact with a distinct set of target sites in the genome and, thus, are involved in a number of signaling pathways and control diverse biochemical processes.
Several modes of action have been proposed by which nuclear FOXO proteins interact with DNA and partner proteins to regulate the transcription of specific target genes (for reviews, see Barthel et al., 2005). (1) FOXO can bind directly to specific promoters and recruit transcriptional co-activators or co-DNA-binding transcription factors to activate transcription. (2) FOXO factors may repress transcription by competing with other transcription factors for a common binding site in a gene promoter. (3) FOXO factors may also cooperate with or titrate away specific transcription factors or cofactors, thereby regulating (activating or repressing) transcription through promoters that lack FOXO-binding sites.
As a convergence point of various signaling pathways, FOXO factors are subject to multiple levels of regulation. While regulation of subcellular FOXO localization is a major event, modulation of FOXO transactivation properties also provides additional control. Furthermore, FOXO factor abundance is regulated by degradation and/or through the control of FOXO gene expression.
Regulation of the subcellular localization and transcriptional activity of FOXO protein is achieved primarily by posttranslational modifications, such as phosphorylation and acetylation. Two main classes of stimuli trigger FOXO phosphorylation with opposing effects on FOXO localization. In response to growth and survival factors such as insulin and insulin-like growth factor 1, FOXO factors become phosphorylated and localized to the cytoplasm. By contrast, phosphorylation of FOXO proteins in response to oxidative stress, for example, by Jun-N-terminal kinase or mammalian Ste 20-like kinase, results in retaining FOXO proteins in the nucleus even in the presence of growth factors.
Recent studies indicate that FOXO transcription factors are acetylated by p300 and CREB-binding protein, co-activators possessing acetyltransferase activity, at several conserved lysine residues, many of which are located in the DNA-binding domain (Fukuoka et al., 2003; van der Horst et al., 2004). Acetylation of FOXO factors appears to exert inhibitory effects on their transactivation activity, possibly by reducing their DNA-binding activity. Deacetylation of FOXO proteins has been shown to result from the activity of SIRT1, a nicotinamide adenine dinucleotide-dependent deacetylase (Brunet et al., 2004; Motta et al., 2004; van der Horst et al., 2004). The effects of SIRT1 on FOXO function are complex and vary depending upon the FOXO target genes. It has been shown that SIRT1 promotes transcription of FOXO target genes involved in stress resistance, while decreasing transcription of genes involved in apoptosis (Greer and Brunet, 2005). Thus, SIRT1 appears to shift the FOXO-dependent response away from cell death toward stress resistance. This is in agreement with the concept that acetylation/deacetylation of FOXO protein may switch target specificity.
In addition, modulation of FOXO protein–protein interaction with accessory proteins or cofactors through covalent modifications (phosphorylation and acetylation) might alter their inherent transactivation potential as well as determine which target genes are regulated by specific FOXOs (Perrot and Rechler, 2003; Puigserver et al., 2003). FOXO-binding partners can modulate FOXO transcription activity either positively, for example, β-catenin (Essers et al., 2005) or negatively, for example, PPARγ or the androgen receptor (Dowell et al., 2003; Li et al., 2003).
FOXO proteins are also regulated by the ubiquitin-proteasome system. Upon polyubiquitination, FOXO proteins are degraded by proteolysis through the proteasome pathway (Vogt et al., 2005). Phosphorylation by Akt at serine 256 in FOXO1 creates a binding site forSkp2, the substrate-binding component of the Skp1/culin 1/F-box protein (SCFSkp2) E3 ligase complex (Matsuzaki et al., 2003; Huang et al., 2005). For FOXO3, the critical kinase is IκB kinase-β (IKKβ) rather than Akt. It targets the residue serine 644 and interacts with an as yet unidentified ubiquitin ligase for ubiquitination-dependent proteasomal degradation (Hu et al., 2004).
Furthermore, mRNA levels of the different FOXO gene subtypes vary from tissue to tissue in mammals (Furuyama et al., 2000) and can be modulated under some circumstances (Furuyama et al., 2000; Richards et al., 2002). These findings suggest that the transcription of FOXO genes is subject to regulation. Altogether, multiple layers of regulation offer a large spectrum of options for the fine-tuning of FOXO function.
FOXO in cancer
FOXO factors play pivotal role in cell fate decisions. Their functions are regulated by multiple signaling pathways including Akt and SGK. Many of these pathways are known to be dysregulated in cancer, suggesting that FOXO factors may play a tumor suppressor role in a variety of cancer. A role of FOXO factors in tumorigenesis was initially suggested by the observation that three of the four known FOXO genes were found at chromosomal breakpoints in certain types of tumor (rhabdomyosarcomas for FOXO1, and acute myeloid leukemias for FOXO3 and FOXO4; Barr, 2001). These chromosomal translocations all result in a chimeric protein in which the DNA-binding domain of other transcriptional regulators (Pax3 or Pax7 for FOXO1 and mixed-lineage leukemia gene for FOXO 3 and FOXO4) are fused to the transactivation domain of FOXOs (Galili et al., 1993; Parry et al., 1994; Borkhardt et al., 1997; Hillion et al., 1997; Anderson et al., 1998). Interestingly, these translocations occur at the identical position, immediately N terminus to the recognition helix (H3) of the FOXO family members and these fusions are no longer controlled by Akt and are constitutively retained in the nucleus (del Peso et al., 1999).
Although the FOXO proteins are physically affected by chromosomal translocations, the mechanism by which these genetic alterations are related to their functions in tumorigenesis is unclear. Two models have been proposed to explain the role of FOXO1 in tumor formation of alveolar rhabdomyosarcoma (ARMS). One model is based upon the assumption that tumors arise from a gain of function of FOXO1. These fusions alter the transactivation properties of the respective proteins and are thereby thought to contribute to carcinogenesis. The main evidence supporting this notion is that the Pax3–FOXO1 fusion protein is a more potent transcription activator than normal Pax3 protein in increasing the transcription of Pax3 target gene (Xia et al., 2002). The fusion protein is able to induce oncogenic transformation in cell culture (Scheidler et al., 1996; Kempf and Vogt, 1999; Xia and Barr, 2004), suggesting that the fusion protein may function as an oncogenic transcription factor by enhancing activation of normal PAX3 target genes. However, in transgenic or knock-in murine models, expression of PAX3–FOXO1 fusion proteins fails to induce tumors (Anderson et al., 2001; Lagutina et al., 2002; Relaix et al., 2003; Keller et al., 2004b).
Therefore, a second model has been proposed in which, instead of an altered transcription of fusion gene, the disruption of one FOXO allele in ARMS results in a depletion of FOXO protein and subsequent loss of function is a critical event in tumorigenesis (Accili and Arden, 2004). As FOXO factors play a pivotal role in cell fate decisions by keeping cells in check by inhibiting cell proliferation and promoting cell death, disruption of their function leads to evasion of normal limitation on cell proliferation and transformation (Burgering and Kops, 2002). Indeed, immunoblots indicate that ARMS tumor and tumor cell lines harboring the PAX3– FOXO1 fusion gene do not express FOXO1 (Bois and Grosveld, 2003). Recent findings suggest that the two models are not mutually exclusive and both may contribute to tumorigenesis. Indeed, ectopic expression of the Pax3–FOXO1 fusion protein results in an elevated level of Skp2 (Zhang and Wang, 2003). Moreover, ARMS cell lines that bear the t(2;13) chromosomal translocation exhibit higher levels of Skp2 when compared to normal skeletal muscle cells (Zhang and Wang, 2003). Based on the observation that FOXO1 is degraded via ubiquitin-dependent proteasome pathway through interacting with Skp2 (Huang et al., 2005) and that FOXO protein degradation often accompanies cell transformation (Hu et al., 2004; Huang et al., 2005), it is tempting to speculate that oncogenic Pax3–FOXO1 fusion protein not only transactivates expression of tumor-promoter genes, but also leads to a complete loss of the function of FOXOs via ubiquitin-mediated proteasomal degradation. Furthermore, mice with homozygous knock in of Pax3–FOXO1 develop ARMS in an Ink4a- orp53 -deficient background (Keller et al., 2004a), further supporting this concept.
A role for FOXO inactivation in cellular transformation can also be inferred from the fact that FOXO negatively regulates cell survival and cell cycle progression in mammalian cells and that FOXO members are regulated by the PTEN tumor suppressor. Indeed, FOXO1 is cytosolic in PTEN-negative renal and prostate carcinoma cells (Nakamura et al., 2000). Overproduction of FOXO proteins in PTEN-negative tumor-cells has the same effect on cell survival and cellular proliferation as the overexpression of PTEN in these cells. In addition, PTEN-null cells induce tumorigenesis in nude mice, which can be overridden by the expression of a constitutively active form of FOXO1 (Ramaswamy et al., 2002). Furthermore, FOXO factors are dysregulated in several tumor types including breast cancer(Hu et al., 2004), prostate cancer (Modur et al., 2002), glioblastoma (Seoane et al., 2004), rhabdomyosarcoma (Galili et al., 1993) and leukemia (Parry et al., 1994). Nuclear exclusion of FOXO3 correlates with expression of IKKβ or phosphorylated Akt in many primary tumors and links with poor survival of the patients with breast tumors (Hu et al., 2004). Cell proliferation and tumorigenicity in nude mice induced by IKKβ expression can be overridden by FOXO3 (Hu et al., 2004). Similarly, the expression of a constitutively active form of Foxo4 suppresses oncogene HER2-mediated tumorigenesis in nude mice (Yang et al., 2005). Taken together, these observations suggest that FOXOs are mediators of tumor suppression.
FOXOs can associate with tumor suppressors or oncogenes, which suggests additional mechanisms by which FOXOs play a role in tumorigenesis. In response to stress stimuli or to nutrient deprivation, FOXO3 has been found to interact with the tumor suppressor p53 in vitro (Brunet et al., 2004). Given that FOXOs share similar target genes including p21, GADD45, WIP1 and PA26 with p53, it suggests that these two proteins may coordinate tumor suppression. In addition, FOXO factors form a complex with SMAD transcription factors (which also act as tumor suppressors), resulting in transforming growth factor-β-dependent activation of p21Cip1 and subsequent G1 arrest (Seoane et al., 2004). However, in glioblastomas FOXO factors are repressed by an overactive phosphoinositide 3-kinase (PI3K)–Akt pathway as well as by association with the oncogene FOXG, another FOX family member, to prevent FOXO from binding to the p21Cip1 promoter, thereby promoting cancer cell proliferation (Seoane et al., 2004). Furthermore, the oncogene β-catenin binds to FOXO factors (Essers et al., 2005), thereby enhancing the ability of FOXO proteins to inhibit cell cycle progression (Essers et al., 2005). Given that β-catenin associates with T-cell factor (TCF) and converts it from transcription repressor to activator, which has been implicated in cancer progression, in particular in colon cancer, it is possible that FOXO factors could counteract tumor progression by sequestering β-catenin away from TCF, thereby inhibiting cell cycle progression. The transcription factor and candidate tumor suppressor gene, RUNX3, which mediates apoptosis and cell growth inhibition in gastric epithelial cells, is frequently lost in gastric cancer cells. The physical interaction of RUNX3 and FOXO3 on the promoter of the proapoptotic factor Bim activates transcription of Bim, thereby inducing apoptosis in gastric cancer, suggesting that it may play an important role in tumor suppression in gastric cancer (Yamamura et al., 2006).
FOXO in the regulation of apoptosis
Apoptosis plays a critical role in tumorigenesis. Factors regulating the expression levels of survival proteins are likely to play a crucial role in tumor formation. Overactivation of PI3K–AKT signaling is a hallmark of many human cancers. FOXOs have emerged as important effector arms of PI3K–AKT signaling.
In mammals, the PI3K–Akt signaling pathway is activated by insulin and growth factors. This regulates diverse cellular processes, such as cell proliferation and survival (Datta et al., 1999; Cantley, 2002; Vivanco and Sawyers, 2002). Binding of insulin or growth factors to their receptors activates PI3K, resulting in the production of phosphatidylinositol 3,4,5-triphosphate, which creates a membrane-binding site for the serine-threonine kinase Akt, an important regulator of cell survival. The translocation of Akt to the plasma membrane leads to its activation via phosphorylation by 3′-phosphoinositide- dependent kinase 1 (Datta et al., 1999; Cantley, 2002; Vivanco and Sawyers, 2002). Activated Akt then phosphorylates key regulatory proteins at serine and threonine residues that lie in (RXRXX(S/T)) motif (Alessi et al., 1996). FOXOs are phosphorylated by Akt at three consensus Akt sites, corresponding to Thr24, Ser256 and Ser319 of FOXO1 (Kops et al., 1999; Nakae et al., 1999; Rena et al., 1999; Takaishi et al., 1999; Tang et al., 1999; Brunet et al., 2004). This leads to the interaction of FOXOs with 14-3-3 proteins and the nuclear export of the FOXO-14-3-3 complex mediated by chromosomal region maintenance 1 (CRM1) and Ran GTPase. Translocation of FOXOs to the cytoplasm results in inhibition of target gene transcription (Burgering and Kops, 2002). Growth factor withdrawal leads to the PI3K–Akt pathway inactivation, FOXO dephosphorylation at its Akt sites, nuclear translocation and target gene activation. Within the nucleus, FOXO triggers apoptosis by inducing the expression of death genes such as the FasL gene, and thereby participates actively in the process of apoptosis (Brunet et al., 2004).
The tumor suppressor PTEN, a lipid 3′-phosphatase, functions as the key PI3K–Akt pathway antagonist through a negative effect on Akt. In PTEN-deficient tumor cell lines, FOXO1 proteins are constitutively phosphorylated and hence constitutively cytoplasmic (Nakamura et al., 2000). In contrast, when PTEN is exogenously expressed, FOXO1 relocates to the nucleus and restores transcriptional activation (Nakamura et al., 2000). In addition, a constitutively active form of FOXO1 that cannot be phosphorylated by Akt induces apoptosis in PTEN-null cells, which has the same effect upon PTEN reconstitution (Nakamura et al., 2000). These data suggest that FOXOs are key mediators of tumor suppression downstream of PTEN and play a critical role in regulating apoptosis.
Besides Akt, CDK2 interacts with and phosphorylates FOXO1 at serine 249, leading to nuclear export and thereby inhibiting FOXO1 activity (Huang et al., 2006). In response to DNA damage, the phosphorylation event was abolished through the CHK2-dependent cell cycle checkpoint pathway (Huang et al., 2006). Thus, functional interaction between CDK2 and FOXO1 provides a mechanism that regulates programmed cell death after DNA strand breakage.
In addition, androgens provide an Akt-independent cell survival signal in prostate cancer through interaction with FOXO1. In prostate cancer, the androgen receptor and FOXO1 form a complex, which blocks the binding of FOXO1 to its DNA response element, thereby interfering with its ability to induce apoptosis and cell cycle arrest of prostate cancer cells (Li et al., 2003).
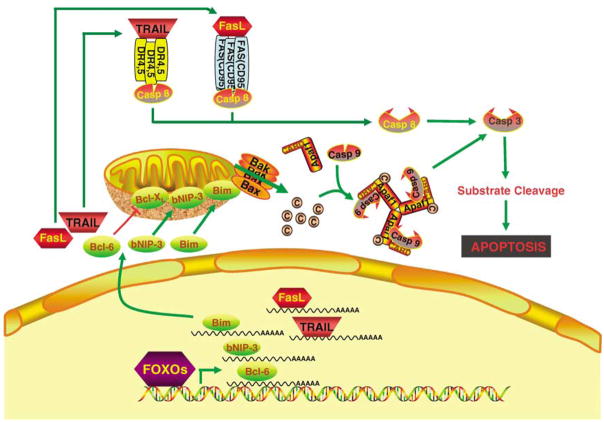
Three consensus FRE sequences are found in the promoter of the proapoptotic gene FasL, which encodes a protein that activates the death receptor Fas/CD95/APO-1 and thereby promotes mitochondria-independent apoptosis (Brunet et al., 1999). Binding of FOXO3 to those FREs leads to FOXO3-dependent transcription (Figure 1). In addition, a constitutively active form of FOXO3 induces the activity of the FasL native promoter (Brunet et al., 1999). Furthermore, expression of a constitutively active form of FOXO3 triggers apoptosis in cerebellar granule neurons through the Fas signaling cascade (Brunet et al., 1999).
Figure 1.
Forkhead box O (FOXO) can induce apoptosis through mitochondria-dependent and -independent pathways. Upregulation of Fas ligand (FasL) or tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) results in cross-linking and activation of death receptors, which thereby activates caspase-8 and subsequent apoptosis. Direct transcriptional activation of the proapoptotic Bcl-2 family members, such as Bim and bNIP3, or indirectly suppressing the expression of the prosurvival Bcl-2 family member Bcl-XL by regulating expression of the transcriptional repressor Bcl-6, leads to increase mitochondrial permeability and promote apoptosis. These pathways triggered by FOXOs may operate independently or cooperatively a cell- and/or tissue type-specific manner.
In PTEN-deficient prostate carcinoma cell lines, FOXO1 and FOXO3 are cytoplasmically sequestered and inactive (Modur et al., 2002). In addition, the expression level of tumor necrosis factor family member TRAIL (tumor necrosis factor-related apoptosis-inducing ligand; Modur et al., 2002), a proapoptotic effector, is decreased (Modur et al., 2002). Overexpression of FOXO1 and FOXO3 in the prostate cancer cell line results in apoptosis and increased expression of TRAIL. FREs of FOXO3 exist in the TRAIL promoter, indicating that TRAIL is a direct target of FOXO3 (Modur et al., 2002). It is conceivable that the loss of PTEN contributes to tumor cell survival through decreased transcriptional activity of FOXOs followed by decreased TRAIL expression and apoptosis. Thus, FOXO proteins regulate cell survival by modulating the expression of death receptor ligands (for example, FasL and TRAIL) that function in autocrine and paracrine pathways (Figure 1).
In addition to the death receptor ligands, FOXO proteins have been shown to be involved in the transactivation of the Bcl-2 family, which has both pro- and antiapoptotic members and plays a critical role in regulating cell survival (Figure 1). One family member, the Bim, contains only a protein interaction motif known as the BH3 domain, which functions in the intrinsic, mitochondrial apoptotic pathway. FOXO proteins induce Bim expression in hematopoietic cells deprived of growth factors (Dijkers et al., 2000; Stahl et al., 2002). In paclitaxel-sensitive breast cancer, upregulation of FoxO3a by paclitaxel results in increased levels of Bim mRNA and protein, leading to apoptosis in breast cancer cells and contributing to the tumor response to paclitaxel (Sunters et al., 2003). Two functional FRE sites have been located in the Bim promoter (Gilley et al., 2003). In addition, another proapoptotic member of the Bcl-2 family, bNIP3, a BH3-only protein, is a FOXO target gene (Tran et al., 2002). Furthermore, FOXO4 indirectly suppresses the expression of the prosurvival Bcl-2 family member Bcl-XL by regulating expression of the transcriptional repressor Bcl-6 (Tang et al., 2002). Thus, FOXO factors can also trigger apoptosis by modulating the ratio of proapoptotic and prosurvival members of the Bcl-2 family. Taken together, FOXO transcription factors can induce cell death through mitochondria-dependent (Bcl-2 family members) and -independent (death cytokine) mechanisms.
Perspective
One distinguishing feature of FOXO family members is their overlapping but different patterns of expression, indicating that they may have redundant as well as distinct functions. In cell culture-based systems, FOXO1, FOXO3 and FOXO4 behave similarly. However, knockout murine models reveal unique roles for different FOXO proteins. Foxo1-null mice die on embryonic day 10.5 with defects in vascular development (Furuyama et al., 2004; Hosaka et al., 2004). Foxo3-null mice are viable, showing age-dependent infertility and have abnormal ovarian follicular development (Castrillon et al., 2003). No histological abnormalities have been identified in Foxo4-null mice (Hosaka et al., 2004). Interestingly, while individual disruption of each of the three Foxo genes has not revealed a direct role of FOXO family members in cancer, disruption of all three Foxos (Foxo1, Foxo3 and Foxo4) leads to tumorigenesis in mice in a context-dependent manner(Paik et al., 2007; Tothova et al., 2007), further supporting the notion that Foxos function as tumor suppressors and their isoforms display functional redundancy and diversification.
In addition, signaling downstream of FOXOs is both cell type specific and tissue-type specific within the same cell type. While activation of FOXOs suppresses growth in a variety of cell types, they induce apoptosis in cells of the immune system. For example, whereas FOXO3 upregulation of p27kip1 in A14 cells, mouse embryo fibroblasts, DLD-1 and 786-0 cells (Medema et al., 2000; Nakamura et al., 2000) results in a potent cell cycle arrest, it causes apoptosis in Ba/F3 cells (Dijkers et al., 2000). However, the mechanism that confers cell-type specificity and determines why some cells become cell cycle arrested and others become apoptotic is still undetermined. One possible explanation is that activation of FOXOs results in cell type-specific gene regulation, which is regulated by interaction of FOXOs with other transcription factors or accessory proteins. Furthermore, cell types might respond differently to FOXO-mediated expression of the same gene product. Indeed, FOXOs control unique biological consequences in different cells or tissue contexts via modulation of distinct downstream targets.
FOXO transcription factors are emerging as master signaling regulators, which control a plethora of physiological and pathological processes, including cancer protection. Moreover, their functions are tightly regulated at multiple levels, which include phosphorylation, ubiquitylation, acetylation and protein–protein interactions. Therefore, FOXOs represent an interesting potential target to develop novel therapeutic approaches for cancer.
For instance, as inactivation of FOXOs appears to be a crucial step in tumorigenesis, restoring activity of these factors represents a potential effective therapeutic strategy. Mounting evidence indicates that constitutively active FOXO mutants, which restrain FOXOs in the nucleus, restore FOXO functions. Accordingly, developing chemical molecules that mimic FOXO mutants that act to restore the function of defective FOXO genes could be a potential choice for cancer-related drug design. In addition, modulation of subcellular translocation could be another possibility. As FOXO is actively transported from the nucleus by a CRM1-dependent manner, nuclear export inhibitors including CRM1 inhibitors (for example, leptomycin B) could be considered. A high throughput, chemical genetic screen for inhibitors of FOXO1 nuclear export has been reported and other candidate molecules are under further investigation (Kau et al., 2003). Recently, a bromotyrosine derivative, psammaplysene A, has shown to cause relocalization of FOXO1 to the nucleus in PTEN-deficient cells (Schroeder et al., 2005), and could be another candidate. Furthermore, as Akt-dependent phosphorylation of FOXOs results in nuclear export and interaction with Skp2 and thereby consequent degradation, both PI3K–Akt pathway and Skp2 could be potential targets for cancer therapy.
Although substantial progress in understanding the function and regulation of FOXO proteins has been made, much remains to be discovered. Whether FOXO1, FOXO3, FOXO4 and FOXO6 have different subsets of target genes or share similar target genes is still not determined. In light of the observation that FOXOs rely heavily on cell type and tissue context to trigger different, even opposite, functions, it will be important to determine the mechanisms by which FOXO factors specify precise programs of gene expression and execute appropriate cellular functions accordingly. Thus, a detailed understanding of FOXO proteins and their biology will provide new opportunities for developing more effective therapeutic approaches to treat cancer.
Acknowledgments
This work was supported in part by grants from NIH (CA121277 and CA125747) and the TJ Martell Foundation (to DT). ZF is a recipient of a Ruth L Kirschstein NRSA individual Fellowship from NIH.
References
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- Anderson MJ, Shelton GD, Cavenee WK, Arden KC. Embryonic expression of the tumor-associated PAX3–FKHR fusion protein interferes with the developmental functions of Pax3. Proc Natl Acad Sci USA. 2001;98:1589–1594. doi: 10.1073/pnas.98.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20:5736–5746. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Biggs WH, III, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome. 2001;12:416–425. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- Biggs WH, III, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois PR, Grosveld GC. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J. 2003;22:1147–1157. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkhardt A, Repp R, Haas OA, Leis T, Harbott J, Kreuder J, et al. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23) Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- Brownawell AM, Kops GJ, Macara IG, Burgering BM. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol Cell Biol. 2001;21:3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Kops GJ. Cell cycle and death control: long live Forkheads. Trends Biochem Sci. 2002;27:352–360. doi: 10.1016/s0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Corral J, Forster A, Thompson S, Lampert F, Kaneko Y, Slater R, et al. Acute leukemias of different lineages have similar MLL gene fusions encoding related chimeric proteins resulting from chromosomal translocation. Proc Natl Acad Sci USA. 1993;90:8538–8542. doi: 10.1073/pnas.90.18.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- del Peso L, Gonzalez VM, Hernandez R, Barr FG, Nunez G. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene. 1999;18:7328–7333. doi: 10.1038/sj.onc.1203159. [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem. 2003;278:45485–45491. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Daitoku H, Hatta M, Matsuzaki H, Umemura S, Fukamizu A. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int J Mol Med. 2003;12:503–508. [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, III, Emanuel BS, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Hillion J, Le Coniat M, Jonveaux P, Berger R, Bernard OA. AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood. 1997;90:3714–3719. [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, III, Tieu D, Boyer AD, Varki NM, Cavenee WK, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci USA. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278:35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kau TR, Schroeder F, Ramaswamy S, Wojciechowski CL, Zhao JJ, Roberts TM, et al. A chemical genetic screen identifies inhibitors of regulated nuclear export of a Forkhead transcription factor in PTEN-deficient tumor-cells. Cancer Cell. 2003;4:463–476. doi: 10.1016/s1535-6108(03)00303-9. [DOI] [PubMed] [Google Scholar]
- Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004a;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 2004b;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf BE, Vogt PK. A genetic analysis of PAX3–FKHR, the oncogene of alveolar rhabdomyosarcoma. Cell Growth Differ. 1999;10:813–818. [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- Lagutina I, Conway SJ, Sublett J, Grosveld GC. Pax3-FKHR knock-in mice show developmental aberrations but do not develop tumors. Mol Cell Biol. 2002;22:7204–7216. doi: 10.1128/MCB.22.20.7204-7216.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Lee H, Guo S, Unterman TG, Jenster G, Bai W. AKT-independent protection of prostate cancer cells from apoptosis mediated through complex formation between the androgen receptor and FKHR. Mol Cell Biol. 2003;23:104–118. doi: 10.1128/MCB.23.1.104-118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. Daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci USA. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–47937. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nakae J, Park BC, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Ramaswamy S, Vazquez F, Signoretti S, Loda M, Sellers WR. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol Cell Biol. 2000;20:8969–8982. doi: 10.1128/mcb.20.23.8969-8982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry P, Wei Y, Evans G. Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer. 1994;11:79–84. doi: 10.1002/gcc.2870110203. [DOI] [PubMed] [Google Scholar]
- Perrot V, Rechler MM. Characterization of insulin inhibition of transactivation by a C-terminal fragment of the forkhead transcription factor Foxo1 in rat hepatoma cells. J Biol Chem. 2003;278:26111–26119. doi: 10.1074/jbc.M212750200. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- Relaix F, Polimeni M, Rocancourt D, Ponzetto C, Schafer BW, Buckingham M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev. 2003;17:2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G, Guo S, Cichy SC, Unterman TG, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- Richards JS, Sharma SC, Falender AE, Lo YH. Expression of FKHR, FKHRL1, and AFX genes in the rodent ovary: evidence for regulation by IGF-I, estrogen, and the gonadotropins. Mol Endocrinol. 2002;16:580–599. doi: 10.1210/mend.16.3.0806. [DOI] [PubMed] [Google Scholar]
- Scheidler S, Fredericks WJ, Rauscher FJ, III, Barr FG, Vogt PK. The hybrid PAX3–FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci USA. 1996;93:9805–9809. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder FC, Kau TR, Silver PA, Clardy J. The psammaplysenes, specific inhibitors of FOXO1a nuclear export. J Nat Prod. 2005;68:574–576. doi: 10.1021/np049624z. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, et al. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc Natl Acad Sci USA. 1999;96:11836–11841. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ED, Nunez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- Tang TT, Dowbenko D, Jackson A, Toney L, Lewin DA, Dent AL, et al. The forkhead transcription factor AFX activates apoptosis by induction of the BCL-6 transcriptional repressor. J Biol Chem. 2002;277:14255–14265. doi: 10.1074/jbc.M110901200. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2 (SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Vogt PK, Jiang H, Aoki M. Triple layer control: phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle. 2005;4:908–913. doi: 10.4161/cc.4.7.1796. [DOI] [PubMed] [Google Scholar]
- Xia SJ, Barr FG. Analysis of the transforming and growth suppressive activities of the PAX3-FKHR oncoprotein. Oncogene. 2004;23:6864–6871. doi: 10.1038/sj.onc.1207850. [DOI] [PubMed] [Google Scholar]
- Xia SJ, Pressey JG, Barr FG. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- Xuan Z, Zhao F, Wang J, Chen G, Zhang MQ. Genome-wide promoter extraction and analysis in human, mouse, and rat. Genome Biol. 2005;6:R72. doi: 10.1186/gb-2005-6-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura Y, Lee WL, Inoue K, Ida H, Ito Y. RUNX3 cooperates with FoxO3a to induce apoptosis in gastric cancer cells. J Biol Chem. 2006;281:5267–5276. doi: 10.1074/jbc.M512151200. [DOI] [PubMed] [Google Scholar]
- Yang H, Zhao R, Yang HY, Lee MH. Constitutively active FOXO4 inhibits Akt activity, regulates p27 Kip1 stability, and suppresses HER2-mediated tumorigenicity. Oncogene. 2005;24:1924–1935. doi: 10.1038/sj.onc.1208352. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang C. PAX3-FKHR transformation increases 26 S proteasome-dependent degradation of p27Kip1, a potential role for elevated Skp2 expression. J Biol Chem. 2003;278:27–36. doi: 10.1074/jbc.M205424200. [DOI] [PubMed] [Google Scholar]