Abstract
Many critical biologic processes involve dynamic interactions between proteins and nucleic acids. Such dynamic processes are often difficult to delineate by conventional static methods. For example, while a variety of nucleic acid polymerase structures have been determined at atomic resolution, the details of how some multi-protein transcriptase complexes actively produce mRNA, as well as conformational changes associated with activation of such complexes, remain poorly understood. The mammalian reovirus innermost capsid (core) manifests all enzymatic activities necessary to produce mRNA from each of the 10 encased double-stranded RNA genes. We used rapid freezing and electron cryo-microscopy to trap and visualize transcriptionally active reovirus core particles and compared them to inactive core images. Rod-like density centered within actively transcribing core spike channels was attributed to exiting nascent mRNA. Comparative radial density plots of active and inactive core particles identified several structural changes in both internal and external regions of the icosahedral core capsid. Inactive and transcriptionally active cores were partially digested with trypsin and identities of initial tryptic peptides determined by mass spectrometry. Differentially-digested peptides, which also suggest transcription-associated conformational changes, were placed within the known 3-dimensional structures of major core proteins.
Keywords: transcription, RNA, virus structure, electron microscopy, cryo-microscopy, image processing, mass spectrometry
1. Introduction
The capacity to replicate their nucleic acids, a process mediated by polymerases, is central to the long-term genetic survival of all biological organisms on Earth. Although the primary amino acid sequences are quite dissimilar, high-resolution structures of several polymerases show remarkable conservation in the “right hand” supramolecular design (for recent reviews see: (O'Reilly and Kao, 1998; Steitz, 2002; Tao et al., 2002; Love et al., 2004; Pan et al., 2007)). All known eukaryotic and prokaryotic organisms, as well as virtually all DNA viruses, contain DNA-dependent DNA polymerases and these are among the best studied.
RNA viruses constitute the largest and most diverse group of pathogenic organisms; at least 2/3 of the estimated 17 million yearly deaths attributed to infectious pathogens are caused by these viruses (Murray and Lopez, 1996; The World Health Report, 1996). An essential component shared by virtually all RNA viruses (except retroviruses) is an RNA-dependent RNA polymerase (RdRp; transcriptase), which is required for successful completion of the viral replication cycle. Since the RdRp is not found in host cells parasitized by the virus, it is a logical target for development of chemotherapeutic and antiviral agents (Love et al., 2004). Like DNA-dependent DNA polymerases, RdRp also have right-handed supramolecular organization (O'Reilly and Kao, 1998; Tao et al., 2002; Love et al., 2004; Pan et al., 2007). Many RdRp require co-factor proteins for full and specific activity. For example, the poliovirus 3Dpol protein also requires viral proteins 2B, 2C, and 3AB, as well as certain cellular proteins (such as Sam68; McBride et al., 1996) for full activity (reviewed in: Racaniello, 2007). Similarly, while the purified reovirus RdRp protein λ3 manifests limited polymerase activity (Starnes and Joklik, 1993), it is unable, by itself, to faithfully transcribe the reovirus genome (reviewed in Schiff et al., 2007). Thus, some details about how RNA-dependent RNA polymerase supramolecular multi-protein polymerase complexes carry out their functions, particularly those related to how mRNA transcription is carried out, as well as possible protein/nucleic acid conformational alterations that may accompany such functions, have not yet been fully elucidated.
Double-stranded (ds) RNA was discovered in reovirus (Gomatos and Tamm, 1963), which has served as a model for understanding RNA processing and viral pathogenesis. The mammalian orthoreoviruses (reovirus) are the prototype members of the virus family Reoviridae, genus Orthoreovirus. These viruses have a genome composed of 10 segments of dsRNA encased in multiple concentric icosahedral capsids composed of multiple, but non-equivalent copies, of 8 different structural proteins (for review, see: (Schiff et al., 2007)). During early stages of the viral replication process, the intact virus (virion) is converted by sequential proteolysis first into the Intermediate (or Infectious) Subviral Particle (ISVP), then into the core particle. The outer capsid contains 600 copies each of proteins μ1 (and/or its major cleavage product μ1C) and σ3 (Coombs, 1998; Dryden et al., 1993), and up to 36 copies of cell attachment protein σ1 (Larson et al., 1994; Lee et al., 1981). Conversion of virion to ISVP is accompanied by loss of σ3 and cleavage of μ1/μ1C into smaller peptides called δ and φ (reviewed in (Chandran and Nibert, 2003)). Both virion and ISVP are infectious. Conversion of ISVP to core involves loss of all remaining outer capsid proteins and peptides (Chandran et al., 2003; Nibert et al., 2001).
The mammalian reovirus core is composed of five proteins. The three major proteins are λ1 (120 copies) organized as 60 dimers that form a thin T = 1 icosahedral shell (Reinisch et al., 2000), 60 copies of core spike protein λ2, that forms prominent “turrets” on the core capsid (Luftig et al., 1972), and 150 copies of core nodule protein σ2 (Dryden et al., 1993; Reinisch et al., 2000)). In addition, there are two minor proteins (approximately 12 copies of λ3 and about 20 copies of μ2 (Coombs, 1998)). Purified cores manifest all enzymatic activities required for in vitro transcription of viral RNA (Chang and Zweerink, 1971; Drayna and Fields, 1982; Furuichi et al., 1975; Yin et al., 1996). The (-) strand of the parental dsRNA serves as template for synthesis of the capped (+) strand. Genetic and biochemical analyses have identified core protein λ3 as the RdRp catalytic subunit (Bruenn, 2003; Drayna et al., 1982; Morozov, 1989; Starnes and Joklik, 1993), protein μ2 as a polymerase cofactor (Kim et al., 2004; Yin et al., 1996), and protein λ2 as a guanylyltransferase and methyltransferase (Cleveland et al., 1986; Luongo et al., 2000; Mao and Joklik, 1991), which projects as pentameric turrets centered at each of the 12 five-fold vertex positions (Dryden et al., 1993; Luftig et al., 1972; Reinisch et al., 2000). In addition, photochemical cross-linking studies suggested that core shell protein λ1 (which possesses a zinc finger motif (Bartlett and Joklik, 1988), and nucleoside triphosphate phosphohydrolase, helicase, and RNA 5′-triphosphatase activities (Borsa, 1970; Kapuler, 1970; Bisaillon and Lemay, 1997; Noble and Nibert, 1997) may play functions in transcription (Powell et al., 1984).
Previous studies that employed photochemical cross-linking suggested mammalian reovirus core particles undergo conformational alterations during transcription (Powell et al., 1984). In addition, preliminary SDS-PAGE-based experiments performed by us that examined peptide patterns produced from 35S-Met/Cys-labeled T3D cores digested with various concentrations of various proteases (trypsin, α-chymotrypsin, pepsin, proteinase K, or staphylococcus V8 protease) revealed digestion differences between inactive and transcriptionally active particles (data not shown), implying also that there are conformational differences associated with reovirus transcription. However, cryoelectron microscopic studies with related rotaviruses suggested that rotavirus cores do not undergo significant structural reorganization upon transcriptase activation (Lawton et al., 1997). The mammalian reoviruses and rotaviruses represent two distinct genera within the Reoviridae family and a number of differences exist between them. Structurally, reoviruses possess striking turrets, built from core spike protein λ2, a feature absent from rotaviruses. Presence or absence of such turrets has significant ramifications with respect to outer capsid protein organization. The mammalian reoviruses construct their T=13licosahedral outer capsid from only 600 copies of σ3 (Dryden et al., 1993) whereas rotaviruses require 780 copies of outer capsid protein VP7 to accomplish the same task. Holes that would be present in the reovirus capsid that the “missing” 180 σ3 proteins would occupy are “plugged” by the λ2 turrets. Rotavirus mRNA is believed to be extruded from peripentonal channels rather than the channels located at the 5-fold vertices (Lawton et al., 1997) whereas reovirus is believed to use the channels located at the 5-fold vertices (reviewed in Zhang et al., 2003; Schiff et al., 2007). Additional enzymatic structure/function considerations that potentially arise from differences in supramolecular structure between these two virus genera include locations of various enzymes comprising the transcriptase and capping complexes. Thus, these significant structural differences may account for different conclusions reached about possible transcriptase-associated conformational changes for these two viruses. Alternatively, the different conclusions may be caused by different techniques used. To better address whether mammalian reovirus cores undergo structural reorganization during transcriptase activation, we examined differences in cryoEM image reconstructions of both inactive and transcriptionally active type 1 Lang (T1L) core particles. In addition, we used high-resolution mass spectrometry to identify tryptic peptides released from partially digested inactive and transcriptionally active T1L and type 3 Dearing (T3D) core particles. T1L and T3D peptides were localized within the known 3-dimensional core atomic structure.
2. Materials and Methods
2.1 Reovirus virion and core purification
Reovirus serotype 1 Lang (T1L) and serotype 3 Dearing (T3D) are laboratory stocks that were plaque purified and grown in mouse L929 cell monolayers in Joklik modified minimal essential medium (GIBCO, Grand Island, NY) supplemented to contain 2.5% fetal calf serum (GibcoBRL Life Technologies, Burlington, Ontario), 2.5% VSP neonate bovine serum (Biocell, Carson, CA), 2mM glutamine, 100U penicillin per ml, 100μg streptomycin sulfate per ml, and 1μg amphotericin-B per ml, as described (Mendez et al., 2000). Large amounts of virus were grown in spinner culture, extracted with Vertrel-XF®, and purified in cesium chloride gradients as described (Mendez et al., 2000). Virus bands were harvested, dialyzed extensively against Dialysis Buffer (D buffer: 150mM NaCl, 15mM MgCl2, 10mM Tris, pH 7.4) and stored at 4°C until used. Aliquots of purified virions were adjusted to appropriate concentrations (Nibert, 1998) and digested with 200μg/ml α-chymotrypsin to generate Infectious Subviral Particles (ISVPs) and cores, which were separately re-purified in cesium chloride gradients (Coombs, 1998). Purified cores were dialyzed against Core Buffer (1M NaCl, 100mM MgCl2, 25mM Hepes, pH 8.0), glycerol was added to a final concentration of 25%, and aliquots frozen at -80°C. Freezing had no effect on core structure, as determined by comparing sedimentation rates and transcriptase characteristics.
2.2 Transcriptase activation
Aliquots of cores were thawed and diluted into additional Core Buffer, diethylpyrocarbonate-treated distilled H2O, and reaction buffer such that final reaction components were: 2mM each of ATP, CTP, GTP, and UTP; 3.3mM phosphoenol pyuvate; 100ng/μl pyruvate kinase; 0.8 units/μl RNAsin (Boehringer Mannheim, Laval, Québec); 9.5mM MgCl2; and 100mM Hepes, pH 8.0 (Yin et al., 1996). Activated cores were then incubated at 50°C for 5 minutes (for subsequent mass spectrometry), or incubated at 37°C for 12 minutes (for cryoEM), then chilled on ice prior to subsequent analyses. Non-activated cores were diluted into additional Core Buffer and diethylpyrocarbonate-treated distilled H2O such that final concentrations were also 9.5mM MgCl2 and 100mM Hepes, pH 8.0, then heated at 37°C for 12 minutes (for cryoEM) or 50°C for 5 minutes (for mass spectrometry), then chilled on ice prior to subsequent analyses.
2.3 Transcriptase activation for cryoEM
The reovirus transcriptase exhibits a striking temperature dependence, with maximal activity at ∼50°C and a Q10 of ∼14 between 30-40°C (Gillies et al., 1971; Kapuler, 1970; Yin et al., 1996). Preliminary experiments demonstrated that in vitro transcription at 50°C generated so much RNA that the core particle boundaries were obscured. Thus, as indicated above, reovirus serotype 1 strain Lang core particles were incubated for 12 min at 37°C with all necessary transcription components. Aliquots were applied to holey carbon films, plunged into a liquid ethane slush and examined by low dose transmission electron cryo-microscopy as previously described (Dryden et al., 1993). Micrographs were digitized at a 25μm sampling interval corresponding to 7.14Å resolution at the specimen. A linear background gradient was subtracted from masked particle images, and the contrast was normalized. Within the instrumental error of the microscope, the inactive and active core particles had the same shell diameter. Hence, back projections of a three-dimensional reconstruction of the inactive reovirus core particle (Dryden et al., 1993) were scaled to have the same diameter as the corrected active core particles. Correlation methods that employed the inactive core particle as a starting model were then used to derive particle orientations (Baker and Cheng, 1996), and the three-dimensional reconstruction was derived by Fourier-Bessel inversion using the best 45 images from a single micrograph (Crowther, 1971). The data set was randomly divided in half, and two maps were compared by calculating correlation coefficients in Fourier-space shells. The resolution was estimated to be 25Å, at which point the correlation coefficient fell to 0.5. Corrections were not made for effects of the contrast transfer function because the first node was beyond 25Å resolution. Projection density maps of icosahedral shells were computed as described (Bottcher and Crowther, 1996). Since the inactive and active core particles were scaled to have the same shell diameter, we assume that the size of the core shell does not change with transcription. For calculation of difference density maps, the two maps were scaled to have the same maximum and minimum density values, which respectively correspond to the solid protein shell and solvent regions. By this scaling we assume that the mass density of these regions does not change with transcription. The radial and density scaling procedures had the effect of minimizing differences between the two maps. AVS software was used for rendering the maps (Sheehan et al., 1996). For each map in Fig. 3, the same isosurface values were used for rendering the protein shell and the interior RNA.
Figure 3.
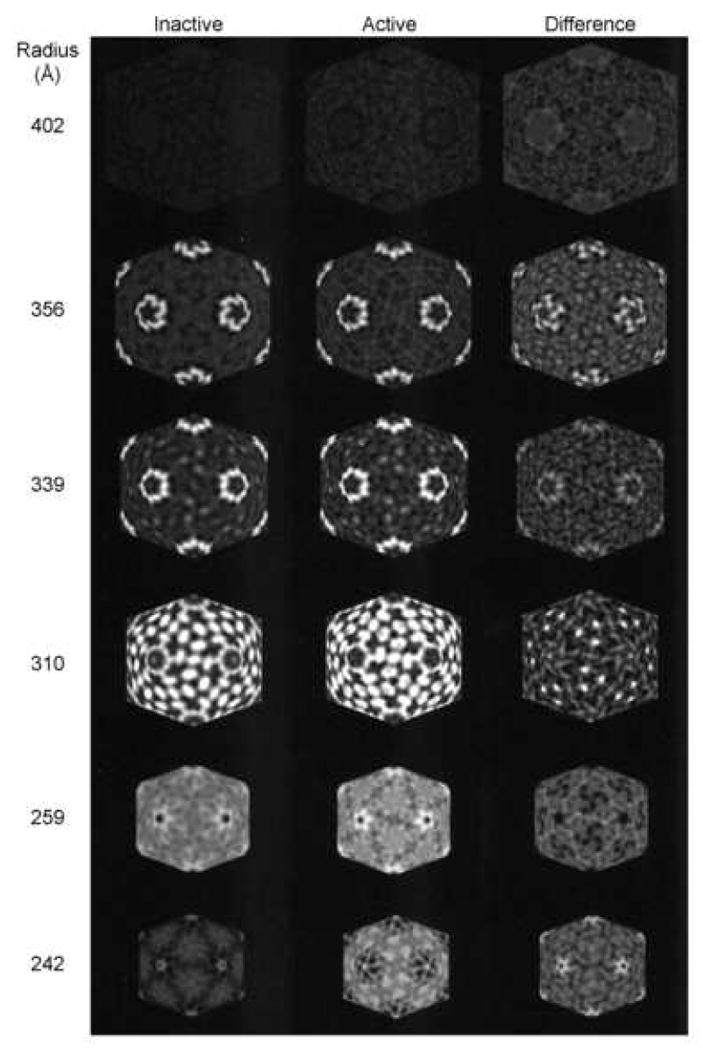
Projection density maps, at indicated particle radii, of icosahedral shells for the inactive (left) and active (middle) reovirus core particle; three-dimensional reconstructions viewed down a two-fold symmetry axis. Icosahedral sections from the difference map are displayed in the right column. Radius 402Å is beyond the perimeter of the particle and is included to show the background level of density. The difference maps at 356 and 339Å show that RNA transcription is associated with conformational changes in the λ2 pentamer. Radius 310Å shows density on the 5-fold symmetry axis of the active particle map that is ascribed to exiting (+) strand RNA. In addition, a shift in density is seen in the surface nodules clustered around the three-fold symmetry axes. Radius 259Å, which corresponds to a radius within the core capsid shell, is included to show a region with no significant shifts in density. Radius 242Å corresponds to a radius at the RNA-protein interface (see Fig. 5a in Dryden et al., 1993). RNA transcription is associated with a concentration of density around the five-fold symmetry axis where the transcriptase is located, and is presumably due to (-) strand template RNA, nascent (+) strand RNA, and/or conformational changes in the transcriptase.
2.4 Proteolysis and mass spectrometry
Gradient-purified core particles that remained inactive, or had been activated by incubating in transcription reaction buffer at 50°C for 5 minutes, as described earlier, were chilled on ice, mixed with a 1:2000 concentration of trypsin, and incubated at 37°C for various periods of time. Digestions were stopped by the addition of specific inhibitors, pH adjustment, and/or chilling. Digestion products were diluted with five volumes of distilled H2O to reduce the salt concentrations to levels suitable for MALDI-Qq-TOFMS analysis. Five-μl aliquots of diluted digestions were mixed with an equal volume of 160 mg/ml 2,5-dihydroxybenzoic acid (dissolved in a 1:1 mixture of acetonitrile and water) and 0.5 μl of the mixture placed onto a gold target probe and allowed to dry. Single MS spectra were acquired on the University of Manitoba prototype MALDI-Qq-TOF (Chernushevich et al., 1999; Shevchenko et al., 1997) by irradiating the samples with a nitrogen laser (Laser Science Inc. model 337 ND) at a repetition rate of 7 Hz for uninterrupted time intervals of approximately one minute. Selected ions were fragmented using argon as the collision gas and tandem mass spectra (MS/MS) were acquired at a laser repetition rate of 9 Hz. The collision energy for MS/MS was set by applying the accelerating voltage by the rule 0.5 V/Da and then adjusted manually to obtain desirable fragmentation of the parent ion. Spectra data acquisition and analysis was performed using software developed in-house (Tofma). To identify the peptide fragments, the m/z of monoisotopic ions were used to search databases using the computational program ProMac (Sciex, Concord, ON) or to search databases on the Internet by the MS-Tag and MS-Fit program (http://prospector.ucsf.edu/).
2.5 Manipulation of molecular structures
Molecular graphics images of the MRV λ1/λ2/σ2 complex (PDB # 1EJ6; Reinisch et al., 2000) were manipulated with PyMOL (DeLano, 2004; http://www.pymol.org) and the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (Pettersen et al., 2004; supported by NIH P41 RR-01081).
3. Results
3.1 Genomic RNA exhibits differential packing in inactive and transcriptionally active cores and nascent mRNA is extruded through the λ2 spike turrets
A growing number of atomic structures have been determined for biomolecules by X-ray crystallography. This method provides a wealth of detailed structural information but is often limited to stable and relatively static structures; thus it is not generally amenable to elucidation of structural alterations that may take place while a macromolecular complex is engaged in dynamic processes. To begin to better delineate structure/function alterations associated with the dynamic process of transcription, we used rapid freezing and cryoEM to trap and visualize inactive and transcriptionally active reovirus core particles. We analyzed cores derived from reovirus serotype 1 Lang (T1L), the same serotype used in previous cryo-EM studies (Dryden et al., 1993). The reovirus transcriptase exhibits a striking temperature dependence, with maximal activity at ∼50°C and a Q10 of ∼14 between 30-40°C (Gillies et al., 1971; Kapuler, 1970; Yin et al., 1996). Thus, we examined cores that had been treated a variety of ways; cores that were maintained at <10°C after purification but prior to flash-freezing for cryo-EM served as standard (Dryden et al., 1993); cores that were heated to either 50°C for 5 minutes, or 37°C for 12 minutes, in the absence of activation components served as controls for effects of heating; and cores were activated by either heating to 50°C for 5 minutes, or heating to 37°C for 12 minutes, in the presence of activation components. As described earlier in Materials and Methods, preliminary reconstruction experiments demonstrated that in vitro transcription at 50°C generated so much RNA that the core particle boundaries were obscured. Thus, for capsid reconstructions, T1L core particles were incubated for 12 min at 37°C with all necessary transcription components.
Transcriptionally active T1L cores display numerous fibrillar strands on their surface (Fig. 1A), which were not observed in inactive core particles (Dryden et al., 1993; and data not shown). Examination of cores that had been heated to either 37 or 50°C without activation components also did not display these fibrillar strands (data not shown). We interpret these fibrillar strands as newly synthesized mRNA since they can be removed by RNAse treatment (data not shown). Transcriptionally active particles prepared by the Kleinschmidt technique (Gillies et al., 1971) or by negative-staining (data not shown) also demonstrated that multiple RNA strands are transcribed simultaneously. Reconstructions of activated core particles were generated from images such as that depicted in Fig.1A. Two-dimensional central sections from reconstructions of the inactive and active core particles show many similarities, but there also appear to be significant differences. First, there is density difference within the λ2 spike turrets (Fig. 1B and 1C, arrows). As discussed below, this may correspond to mRNA being extruded through the spike channels. There also are apparent differences in the particle interiors (compare Fig 1B to 1C). We interpret these differences to mean reorganization of material inside the respective capsids. Since the genomic dsRNA is located inside the core capsid (Joklik, 1983; Schiff et al., 2007) it seems likely that this internal reorganization represents changes in the genomic RNA associated with activation of transcription.
Figure 1.
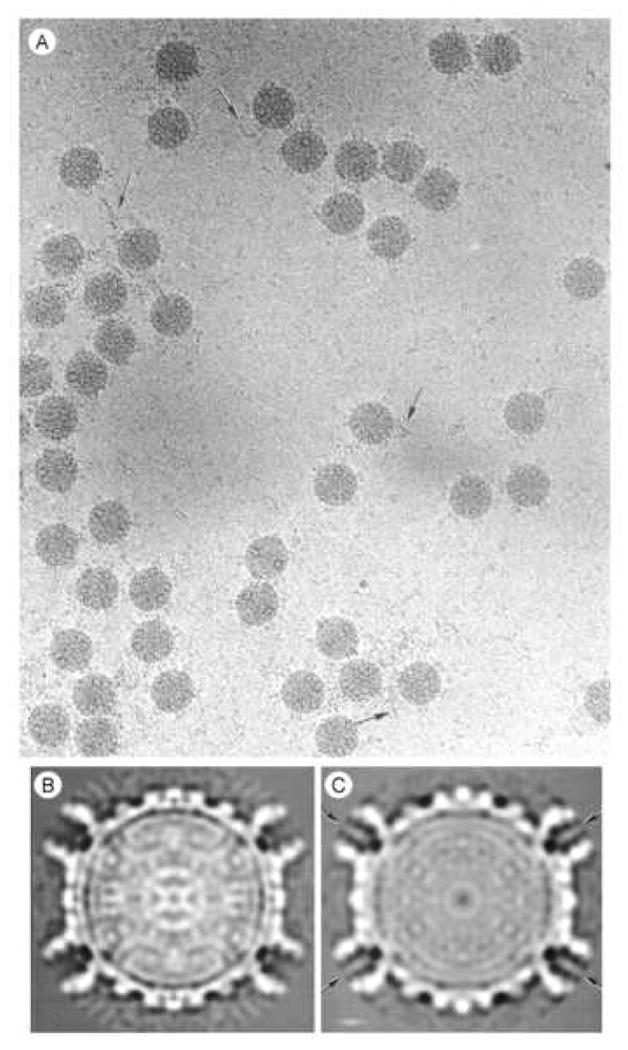
A. Images of frozen-hydrated actively transcribing reovirus core particles recorded by minimal-dose electron cryo-microscopy. Arrows identify fiber-like densities on the perimeter of the particles and in the background that we attribute to newly synthesized (+) strand RNA. B and C. Two-dimensional central sections from the 3-dimensional maps of B, the inactive; and C, the transcriptionally active reovirus core particle. Density visualized in the center of the λ2 turrets (arrows) is attributed to exiting (+) strand RNA.
Three-dimensional reconstructions of the inactive and active cores at 25Å resolution demonstrate that they display very similar structural organization, with 150 ellipsoidal σ2 surface nodules and pentameric λ2 turrets at the icosahedral vertices (Fig. 2). Closer inspection of active particles shows a rod-like density centered within the cavity of each λ2 pentamer (compare Fig. 2E with 2F). Even though the genome is comprised of 10 dsRNA strands, icosahedral image processing averages the density so that it appears within the λ2 spike channels at all 12 icosahedral vertices. Previous Kleinschmidt analyses of transcriptionally active cores demonstrated that RNA was extruded from cores near or at the icosahedral five-fold vertices (Gillies et al., 1971). Similarly, cryoEM reconstructions of transcriptionally active rotavirus cores also demonstrated that RNA was extruded from cores near, but not at, the icosahedral five-fold vertices (Lawton et al., 1997). Our results unequivocally localize the site of reovirus RNA extrusion to the λ2 spike channels and provide further evidence that reovirus mRNA transcription takes place at the icosahedral vertices of the core particle.
Figure 2.
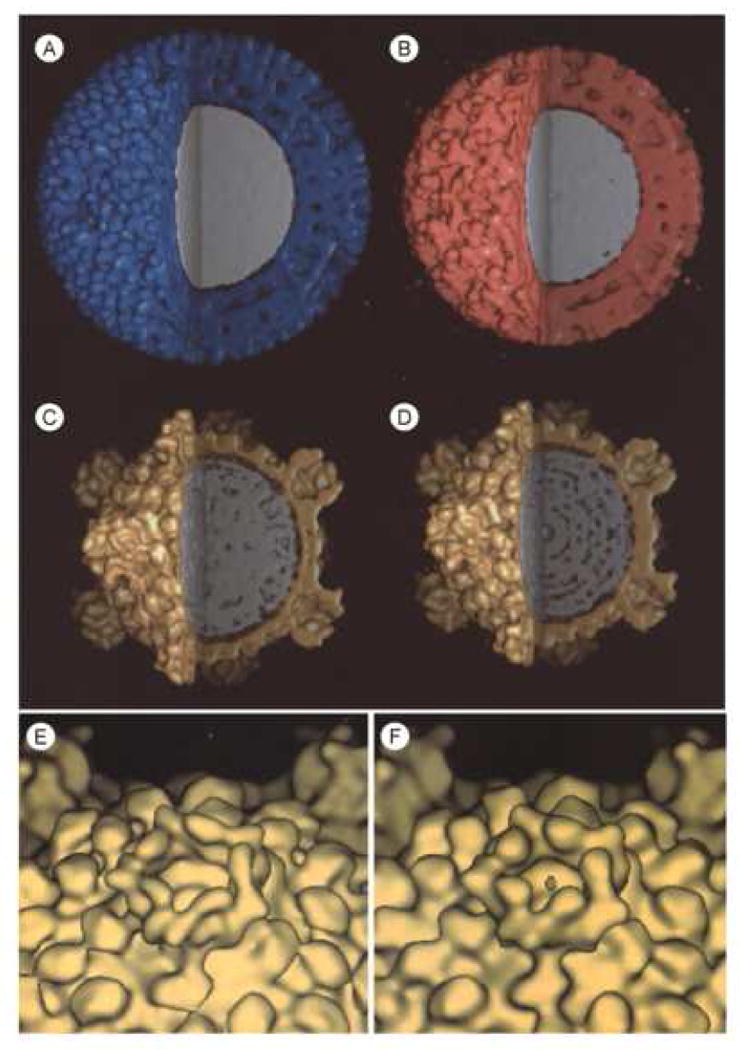
Surface-shaded, three-dimensional reconstructions of A, the native reovirus virion (blue), B, the infectious subvirion particle (red), C, the inactive (gold), and D, the transcriptionally active core particle (gold). The maps have been cropped to reveal the interior density (gray) corresponding to the segmented dsRNA genome. Note the fenestrated interior density in the transcriptionally active core particle. E and F are close-up, oblique views of λ2 spike turrets located on each of the particles' icosahedral vertices of the inactive and active cores, respectively.
The fingerprint-like appearance of close-to-focus images (Dryden et al., 1993) and a 27Å reflection in low angle x-ray scattering profiles (Harvey et al., 1981) suggest tight, liquid-crystalline packing of dsRNA segments within reovirus, similar to what has been seen in bluetongue virus (Gouet et al., 1999). One might expect the dsRNA to have a similar organization in the native virion, infectious subvirion particle (ISVP), and the inactive core particle. The interior density within native virion and ISVP is quite uniform (Fig. 2A and 2B), consistent with close-packing of dsRNA strands. In contrast, the core particle interiors (Fig. 2C and 2D) show less uniform density. Previous cryoEM reconstructions demonstrated reorganization of the λ2 spike turrets in cores compared to virions and ISVPs (Dryden et al., 1993) and it is possible that some genomic reorganization also takes place during the ISVP – core transition, which could lead to apparent differences in interior density in these two inactive particles (Fig. 2B and 2C). Additionally, the active core particle interior (Fig. 2D) shows a more fenestrated appearance than any of the other particles (Fig. 2A – C), which is consistent with a more dramatic reorganization of the genome during events associated with transcription.
3.2 Comparative cryo-EM of inactive and active cores demonstrates conformational changes associated with activation of transcription
Comparative radial density plots of the inactive and transcriptionally active core particles show numerous differences, which suggest conformational changes on the T1L core capsid surface and in the interior of the particle (Fig. 3). Differences at a radius of 356Å, centered on the icosahedral vertices, correspond to the λ2 spike tips, and suggest shifts in the conformation of these regions of the λ2 pentamers. These regions of λ2 correspond to the “flap” (or “lid”) motifs (Breun et al., 2001), so these density shifts might be associated with exit of nascent (+) mRNA and/or capping. There also are density shifts detected at radius 310Å, at sites corresponding to the σ2 surface nodules that surround the icosahedral three-fold symmetry axes. The most prominent density shifts at this radius correspond to the σ2(ii) (also known as σ2.3) molecules centered on the 3-fold axes. There are no detectable changes in density between radii ∼255 to ∼290Å, which corresponds to the λ1 core capsid shell. The comparative radial density plots also show density differences centered at the five-fold vertices at radius 242Å, which, based on radial density plots of the inactive core particle (Dryden et al., 1993) and of empty genome-deficient particles (Dryden et al., 1998), as well as the atomic structure of the core (Reinisch et al., 2000), corresponds to the boundary between the RNA and the core capsid. Since the transcriptase complexes, composed of λ3, and also possibly μ2, reside at these sites (Dryden et al., 1998; Zhang et al., 2003), it is quite likely these density differences may be caused by template (-) strand RNA that is moving through the catalytic site, nascent (+) strand RNA, and/or protein conformational changes associated with transcription.
3.3 Comparative partial proteolysis identifies differences in exposed regions of inactive and transcriptionally active cores
To more precisely map regions of various core proteins that might be undergoing conformational changes, we then digested a variety of inactive and transcriptionally active cores with trypsin for brief periods of time (1 – 10 minutes) and the released peptides were identified by matrix assisted laser desorption ionization quadrupole time of flight mass spectrometry (MALDI-Qq-TOFMS) (Mendez et al., 2003). We compared the peptides released from non-heated inactive T1L and serotype 3 Dearing (T3D) cores to heated (50°C) inactive cores and to heated activated cores. Comparisons of the times at which particular peptides were initially released from digested T1L and T3D particles are indicated in Table 1. All identified peptides were also detected at subsequent times of digestion (data not shown). In addition, much larger numbers of peptides were detected after 7½ minutes of digestions of each particle type, and there were no substantive differences between inactive and active peptides identified at later time points (data not shown). There were no significant differences in the peptides released from inactive cores, whether those cores had been heated or not (data not shown).
Table 1.
Identification of tryptic peptides released from inactive- and transcriptionally-active core particlesa
| Type 1 Lang | Type 3 Dearing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | Inactive Cores | Active Cores | Inactive Cores | Active Cores | ||||||||
| (min) | λ1 | λ2 | σ2 | λ1 | λ2 | σ2 | λ1 | λ2 | σ2 | λ1 | λ2 | σ2 |
| 1.0 | 55-64 | 9-21 | 136-168 | 38-54 | 9-21 | 378-393 | ||||||
| 65-84 | 55-64 | 394-418 | ||||||||||
| 136-168 | 136-168 | |||||||||||
| 1.5 | 32-54 | 549-556 | 315-322 | 38-54 | ||||||||
| 112-135 | 769-778 | 394-418 | 55-64 | |||||||||
| 2.0 | 32-54 | 9-21 | 315-322 | 136-168 | 769-778 | 394-418 | ||||||
| 65-84 | 549-556 | 394-418 | ||||||||||
| 112-135 | 769-778 | |||||||||||
| 3.0 | 938-947 | 769-778 | ||||||||||
| 5.0 | 12-37 | 326-349 | 370-377 | 23-54 | 350-371 | 315-322 | 38-54 | 315-322 | ||||
| 23-54 | 350-371 | 109-135 | 417-443 | 55-64 | 378-393 | |||||||
| 109-135 | 508-548 | 508-548 | ||||||||||
| 1237-1241 | 938-947 | |||||||||||
| 1205-1236 | ||||||||||||
| 1237-1241 | ||||||||||||
Values listed in column 1 correspond to initial times at which specific peptides listed in columns 2 – 13 were identified by MALDI-Qq-TOFMS, and confirmed by tandem MS/MS, as described (Mendez et al., 2003). Indicated peptides were also identified in each subsequent analysis time.
Inspection of the identities of peptides released from inactive and active cores of both serotypes after brief proteolysis revealed many similarities. For example, the λ1 peptide Ala55 – Arg64, the λ2 peptide Leu769 – Arg778, and the σ2 peptide Thr394 – Ile418 were all detected within 5min of digestion, irrespective of whether the core was inactive or active, or derived from T1L or T3D (Table 1). However, more T1L peptides from each of the two major lambda class core proteins (λ1 and λ2) were observed during the first 5min of digestion than from these same proteins in T3D cores, despite the fact that λ1 and λ2 share >99 and ∼93% sequence identities between the T1L and T3D serotypes (Harrison et al., 1999; Breun et al., 2001), and despite the fact that in silico digestions predict all of these observed peptides should be produced by both T1L and T3D (data not shown). The corresponding T3D peptides were detected after 5min in T3D core digestion reactions. These serotype-dependent kinetically different results imply that, despite the high degree of sequence identity between these T1L and T3D proteins, they are folded slightly differently, as also implied by observable differences in spike morphology (Metcalf et al., 1991; Dryden et al., 1993) and differences in capacities of these cores to be crystallized (Coombs et al., 1990). In addition, the order in which specific peptides were initially detected depended upon the strain of virus as well as the activation state. For example, the T1L and T3D λ1 Ala55 – Arg64 peptides were released from inactive cores after 1min of trypsin digestion, but the corresponding peptide was not detected until 30 seconds later in digestions of transcriptionally active T1L cores, or until 4min later from active T3D cores. Similarly, the majority of peptides that were detected during the first 5min of digestions were observed either at the same time, or earlier, in the inactive core digests compared to the transcriptionally active particles. This difference in initial detection of specific peptides between inactive and active cores may result from differences in protein conformation between the different core states. Alternatively, since most identified peptides were initially released later from active cores than from inactive cores, it is possible that the nascent RNA interfered with protease digestion. If the later, it is striking that some peptides (the T1L λ2 Leu417 – Arg443, the T1L λ2 Asp1205 – Arg1236, and the T3D λ2 Leu769 – Arg778 peptides) were consistently released earlier from active cores than from the corresponding inactive cores, which suggests activation-dependent conformational changes in these λ2 spike proteins. Conformational changes may also have taken place in other core proteins as a result of transcription activation, but, because of the possibility that nascent RNA interfered with digestion kinetics, such other possible changes could not be unequivocally determined from differences in apparent protease sensitivities.
4. Discussion
The dsRNA viruses are unique in that transcription of their genomes takes place within an icosahedral capsid that remains intact throughout the viral replication cycle (Schiff et al., 2007). The high-resolution structure of the mammalian reovirus RdRp, the λ3 protein, has been reported (Tao et al., 2002) and fitting of this protein into both a high-resolution atomic structure (Reinisch et al., 2000) and slightly lower-resolution cryoEM structures (Dryden et al., 1998; Zhang et al., 2003) has allowed generation of a model that suggests that the RdRp is fixed to the inner surface of the core shell, offset slightly from the 5-fold axis to allow nascent RNA to exit the RdRp and wind through holes in the λ1 shell before the RNA molecule is directed through the λ2 spike for mRNA capping (see Fig. 6 in Zhang et al., 2003). A fixed polymerase implies that the RNA template must be capable of movement, both within the RdRp itself (and appropriately-sized channels have been identified in the high-resolution structure of λ3 (Tao et al., 2002)), and within the icosahedral capsid. However, direct evidence of RNA movement within transcriptionally active particles has been sparse. The 27Å reflection in low angle x-ray scattering profiles (Harvey et al., 1981) and the fingerprint-like appearance of close-to-focus images (Dryden et al., 1993) suggest tight, liquid-crystalline packing of dsRNA segments within reovirus. The interior density within native virion and ISVP is quite uniform (Fig. 2A and 2B), consistent with close-packing of dsRNA strands. Inactive core particle interiors (Fig. 2C) shows less uniform density, which may correspond to some genomic reorganization during the ISVP – core transition. The λ2 spike turret tips also undergo structural reorganization during the ISVP – core transition (Dryden et al., 1993). The active core particle interior (Fig. 2D) shows a more fenestrated appearance compared to the other particles (Fig. 2A – C), implying a more dramatic reorganization of the genome during events associated with transcription. The interior volume available for packaging the ∼23.5 kilobase pairs of reovirus genome is ∼5.6 × 107Å3 (Nibert, 1998). Thus, using a genome molecular weight of 1.51 × 107 Daltons, the packaging density of the reovirus genomic material, expressed either as Vm (Matthews, 1968), or ρpack (Purohit et al., 2005), can be calculated as Vm ≈3.7 Å3/Da or ρpack ≈0.419. The Vm for anhydrous RNA is 0.91 Å3/Da and the Vm of the genomes of other virus particles, such as human rhinovirus 14 and RNA2 of cowpea mosaic virus, are 1.69 and 3.5 Å3/Da, respectively (Johnson and Rueckert, 1997). The Vm for ssDNA viruses, such as minute virus of mouse, has been determined as 1.7 Å3/Da (Carrasco et al., 2006) and the ρpack for several dsDNA viruses, such as bacteriophages T4, T7, and Φ29, are 0.442, 0.541, and 0.461, respectively (Purohit et al., 2005). Thus, the genome in reovirus cores is packed less densely than any of the above examples, which indicates the solvent volume within reovirus particles would be sufficient to allow substantial conformational rearrangements. This also implies increased freedom of movement, as would be required for simultaneous transcription of all ten genomic dsRNA segments by the λ3 RdRp complexes that are fixed at the icosahedral vertices (Dryden et al., 1998; Zhang et al., 2003).
We attempted to better understand activation-dependent structure/function alterations in the reovirus core by placing the peptides released from briefly-digested particles into the 3-dimensional reovirus core crystal structure. Currently, the atomic structure is known for the F18 core (PDB #1EJ6; Reinisch et al., 2000). F18 is a reassortant hybrid virus that contains the λ2 protein derived from T3D but all other proteins derived from T1L (Nibert et al., 1996). It is likely that F18 accurately represents the atomic structures of both T1L and T3D because amino acid sequence identity is >92% between all the major T1L and T3D core proteins. Thus, despite the caveat that there may be subtle differences in protein folding, as implied by differences in peptide release from T1L and T3D cores (Table 1), we attempted to use the F18 structure as a framework to localize T1L and T3D peptides (Figure 4). All of the λ1 peptides identified within the first 5min of digestion are located within the amino-terminal 168 amino acids. This region of the λ1 protein has different structure, and also possibly different location, within the core, depending upon the particular λ1 molecule. Half of the 120 λ1 molecules that constitute the core shell are designated λ1-A (or λ1.5 because they approach the 5-fold icosahedral axes), whereas the other 60 λ1 molecules are designated λ1-B (or λ1.3 because they approach the 3-fold symmetry axes). Five λ1-A monomers are associated in a pinwheel conformation at each vertex position and the λ1-B molecules are organized as trimers on the icosahedral faces interdigitating between three adjacent λ1-A pinwheels (Reinisch et al., 2000). The amino-terminal 240 amino acid residues in λ1-A are disordered and not visible in the crystallographic structure; hence peptides originating from this region would not be visible (Fig. 4). Most of these same residues in the λ1-B molecules form long thread-like arms that are presumed to interact with other λ1-B molecules, although mutational analyses show the amino-terminal 230 amino acids are not required for formation of a stable core shell (Kim et al., 2002). The λ1 protein has a number of functions presumed associated with transcription, including nucleoside triphosphate phosphohydrolase, helicase, and RNA 5′-triphosphatase activities (Borsa, 1970; Kapuler, 1970; Bisaillon & Lemay, 1997; Noble & Nibert, 1997). Three of the six putative RNA helicase motifs also are present in the amino-terminal 168 amino acids of λ1, but the disordered nature of these residues in λ1-A, and extended conformation of this region in λ1-B, argues that these do not have enzymatic function. All of the first λ1 peptides detected after brief digestion of inactive cores are thought, from the crystal structure, to lie either inside the core shell (Ala38 – Lys54 in T3D, Ala55 – Arg64 in both T1L and T3D, and Ala65 – Lys84 in T1L) or relatively buried between adjacent λ1 monomers (Ser136 – Arg168 in both T1L and T3D; with the possible exception of Arg168 which is not visible in the structure but may lie underneath a σ2 nodule; Fig. 4) and would therefore not be expected to be accessible to proteases until after the core was partially digested. The ability to detect these regions first suggests that they are not buried within the structure, but are accessible to the surface of the core. Accessibility of crystal-determined “buried” residues in viral capsid proteins has also been demonstrated by brief proteolysis of Flock house virus (Bothner et al., 1998) and of hepatitis B virus (Hilmer et al., 2007) and has been interpreted to mean dynamic “breathing” of such capsids. Thus, similar to what has been observed for these other viruses, the reovirus core appears to be a dynamic macromolecular structure, even before its transcriptase is activated.
Figure 4.
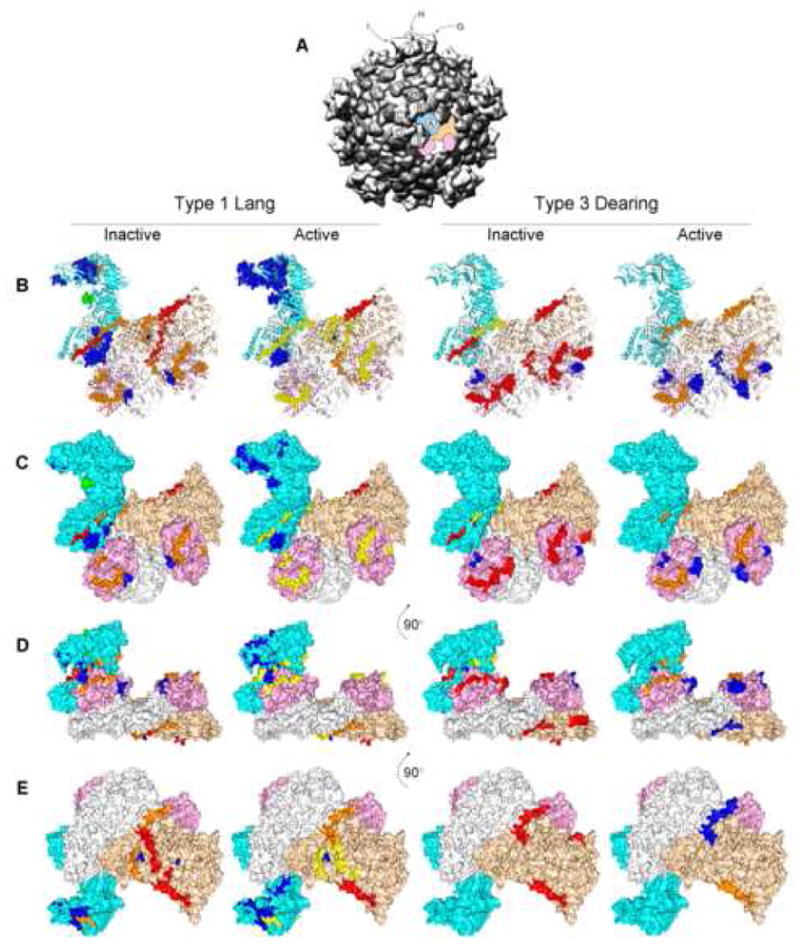
Tryptic peptides released after limited digestion from inactive and actively transcribing T1L and T3D core particles identified by mass spectrometry. The T3D core crystal structure (PDB # 1EJ6, (Reinisch et al., 2000)) was manipulated with PyMOL (DeLano, 2004) and with Chimera (Pettersen et al., 2004). A, Low-resolution overview of the reovirus core structure, viewed down a 5-fold axis (prepared with Chimera from the VIPER database; Pettersen et al., 2004). One asymmetric unit is color-highlighted, consisting of 1 core spike λ2 protein (cyan), 2 core shell λ1 proteins (white, wheat), and 2 core clamp σ2 proteins (pink), to correspond to the molecular models in (B – E). λ2 molecules labeled with small letters (A – J) correspond to similarly-labeled enlarged views in Fig. 5. B, “Open” ribbon diagram of asymmetric unit in approximately same orientation as in (A) (= top view); C, Surface representation of asymmetric unit in same orientation as in (B); D, Surface representation of asymmetric unit rotated 90° with respect to orientation in (C) (= side view); E, Surface representation of asymmetric unit rotated 90° with respect to orientation in (D) (= bottom view, looking from inside particle outwards). Tryptic peptides detected after 1 minute of digestion (as well as in subsequent digestion times; see Table) are shown in red spheres; peptides identified after 1½ minutes of digestion (as well as in subsequent digestion times) are depicted as orange spheres; peptides identified after 2 minutes of digestion are shown in orange spheres; peptides identified after 3 minutes of digestion are depicted as green spheres; and peptides identified after 5 minutes of digestion are shown as blue spheres. Some of the identified peptides (for example, all the λ1 peptides in the white λ1 molecule) are not seen in this figure because those particular regions of this molecule (designated λ1-A; these λ1 molecules approach the 5-fold axes in the core structure, as opposed to the wheat-colored λ1 molecule, designated λ1-B that approache the 3-fold axes in the core structure) are not resolved (Reinisch et al., 2000).
The λ2 protein has been identified as a guanylyltransferase, methyltransferase, and SAM-binding protein (Cleveland et al., 1986; Luongo et al., 2000; Mao and Joklik, 1991), all functions of which are presumed to be intimately involved in transcription. The various λ2 peptides identified within the first 5min of digestion are located throughout the molecule and represent regions of each of the 6 identified motifs (amino acids 2-380 [a cup-shaped GTase], 381-443 [extended region], 434-691 [MTase I], 692-803 [spacer region of unknown function], 804-1022 [MTase II], and 1023-1289 [flaps]; Breun et al., 2001). This protein also is the only one of the major core proteins in which several peptides are released from the active core particles before the same peptides are released from inactive cores (Figure 5). These preferentially-released peptides are found in the “extended region” (Leu417 – Arg443), the “spacer region” (Leu769 – Arg778), and the “flaps” (Asp1205 – Arg1236). The “flaps” region also appears to undergo conformational rearrangement, as determined above by comparative radial density plots of inactive and active core particles (Fig. 3, radius ∼ 356Å). The various σ2 peptides identified within the first 5min of digestion are located within the carboxyl-terminal ∼100 amino acids (residues 315-322, 370-377, and 394-418). These regions are located on the outer distal surfaces of this globular “nodule” protein, which is presumed to function primarily as a “clamp” to help hold the core shell together (Xu et al., 1993; Coombs et al., 1994; Kim et al., 2002). This is the only reovirus core protein that has not yet been implicated in any enzymatic activities associated with reovirus transcription.
Figure 5.
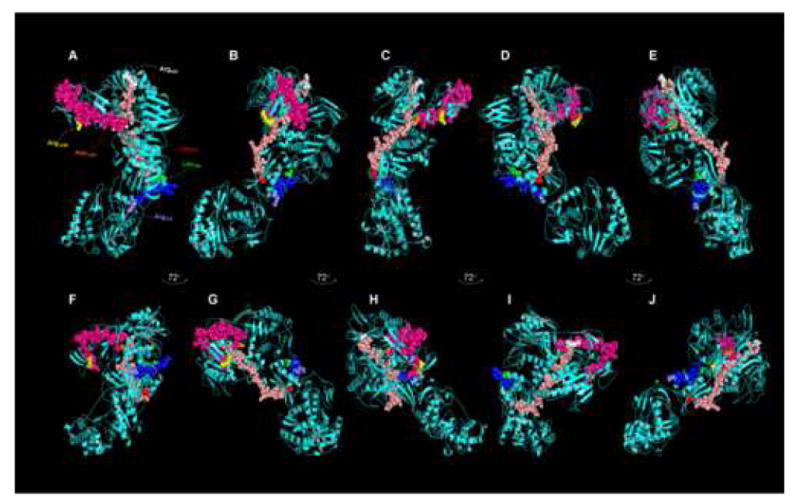
Composite representation of λ2 spike protein tryptic peptides released preferentially after limited digestion from actively transcribing T1L and T3D core particles identified by mass spectrometry. The T3D core crystal structure (PDB # 1EJ6, (Reinisch et al., 2000)) was manipulated with PyMOL (DeLano, 2004) and with Chimera (Pettersen et al., 2004). A, Top view of cyan-colored λ2 molecule in Fig. 4A (labeled with small letter ‘A’); B – E, Represent successive 72° rotations around vertical axis (to correspond approximately to similarly-labeled λ2 molecules around 5-fold axis in Fig. 4A; F, Side view of cyan-colored λ2 molecule in Fig. 4A (view corresponds approximately to λ2 molecule in Fig. 4A, labeled “F’; G – J, Represent successive 72° rotations around vertical axis (to correspond approximately to similarly-labeled λ2 molecules in Fig. 4A). The T1L Leu417 – Arg443 peptide is shown in salmon, with Leu417 in red and Arg443 in white, the T1L Asp1205 – Arg1236 peptide is shown in hot pink, with Asp1205 in white and Arg1236 in yellow, and the T3D Leu769 – Arg778 peptide is shown in blue, with Leu769 in green and Arg778 in slate. These residues are labeled only in ‘A’ for clarity.
In summary, structural reorganization of major core protein λ2 is inferred from both comparative radial density plots (Fig. 3) and from comparative mass spectrometric analyses (Fig. 4, 5). In addition, although we currently do not have unequivocal corroborating mass spectrometry data, structural reorganization in major core protein σ2 is also inferred from comparative radial density plots (Fig. 3). Finally, comparative radial density plots of the inner layers of the capsid shell around the five-fold axes at radius ∼240Å (Fig. 3; where the RdRp protein λ3 is believed to reside) suggest structural reorganizations also take place in this protein. The complementary assays employed in this study suggest structural differences between inactive and transcriptionally active reovirus cores. These data imply that the dynamic process of genomic transcription carried out by the mammalian reovirus core is associated with measurable conformational rearrangements in the core capsid proteins.
Acknowledgments
We thank Mariana Tihova for preparation of Figure 3. This work has been supported by grants from the National Institutes of Health (MY), a Grant-in-Aid from the American Heart Association, National Center (MY), the Gustavus and Louise Pfeiffer Research Foundation (MY), the Donald E. and Delia B. Baxter Foundation (MY), Canadian Institutes of Health Research (KC) and the Manitoba Health Research Council (KC). During some of this work KC was a Manitoba Health Research Council scholar, IM was supported by an MHRC studentship, and MY was an Established Investigator of the American Heart Association and Bristol-Myers Squibb. MY is now supported by a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund.
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. Journal of Structural Biology. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Bartlett JA, Joklik WK. The sequence of the reovirus serotype 3 L3 genome segment which encodes the major core protein lambda 1. Virology. 1988;167:31–37. doi: 10.1016/0042-6822(88)90051-7. [DOI] [PubMed] [Google Scholar]
- Bisaillon M, Bergeron J, Lemay G. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus lambda1 protein. J Biol Chem. 1997;272:18298–18303. doi: 10.1074/jbc.272.29.18298. [DOI] [PubMed] [Google Scholar]
- Bothner B, Dong XF, Bibbs L, Johnson JE, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- Bottcher B, Crowther RA. Difference imaging reveals ordered regions of RNA in turnip yellow mosaic virus. Structure. 1996;4:387–394. doi: 10.1016/s0969-2126(96)00044-5. [DOI] [PubMed] [Google Scholar]
- Borsa J, Grover J, Chapman JD. Presence of nucleoside triphosphate phosphohydrolase activity in purified virions of reovirus. J Virol. 1970;6:295–302. doi: 10.1128/jvi.6.3.295-302.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breun LA, Broering TJ, McCutcheon AM, Harrison SJ, Luongo CL, Nibert ML. Mammalian reovirus L2 gene and lambda2 core spike protein sequences and whole-genome comparisons of reoviruses type 1 Lang, type 2 Jones, and type 3 Dearing. Virology. 2001;287:333–348. doi: 10.1006/viro.2001.1052. [DOI] [PubMed] [Google Scholar]
- Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucl Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco C, Cariera A, Schaap IAT, Serena PA, Gómez-Herrero J, Mateu MG, dePablo PJ. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc Natl Acad Sci (USA) 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Nibert ML. Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol. 2003;11:374–382. doi: 10.1016/s0966-842x(03)00178-1. [DOI] [PubMed] [Google Scholar]
- Chang CT, Zweerink HJ. Fate of parental reovirus in infected cell. Virology. 1971;46:544–555. doi: 10.1016/0042-6822(71)90058-4. [DOI] [PubMed] [Google Scholar]
- Chernushevich IV, Ens W, Standing KG. Orthogonal-injection TOFMS for analyzing biomolecules. Analytical Chemistry. 1999;71:452A–461A. doi: 10.1021/ac990524l. [DOI] [PubMed] [Google Scholar]
- Cleveland DR, Zarbl H, Millward S. Reovirus guanylyltransferase is L2 gene product lambda 2. J Virol. 1986;60:307–311. doi: 10.1128/jvi.60.1.307-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs KM. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology. 1998;243:218–228. doi: 10.1006/viro.1998.9061. [DOI] [PubMed] [Google Scholar]
- Coombs KM, Fields BN, Harrison SC. Crystallization of the reovirus type 3 Dearing core. Crystal packing is determined by the lambda 2 protein. J Mol Biol. 1990;215:1–5. doi: 10.1016/s0022-2836(05)80089-0. [DOI] [PubMed] [Google Scholar]
- Coombs KM, Mak SC, Petrycky-Cox LD. Studies of the major reovirus core protein sigma 2: reversion of the assembly-defective mutant tsC447 is an intragenic process and involves back mutation of Asp-383 to Asn. J Virol. 1994;68:177–186. doi: 10.1128/jvi.68.1.177-186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos Trans R Soc Lond B Biol Sci. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. World Wide Web 2004 [Google Scholar]
- Drayna D, Fields BN. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden KA, Farsetta DL, Wang G, Keegan JM, Fields BN, Baker TS, Nibert ML. Internal structures containing transcriptase-related proteins in Top Component particles of mammalian orthoreovirus 1. Virology. 1998;245:33–46. doi: 10.1006/viro.1998.9146. [DOI] [PubMed] [Google Scholar]
- Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Muthukrishnan S, Shatkin AJ. Reovirus messenger RNA contains a methylated, blocked 5′-terminal structure: m-7G(5′)ppp(5′)G-MpCp- Proc Natl Acad Sci U S A. 1975;72:362–366. doi: 10.1073/pnas.72.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S, Bullivant S, Bellamy AR. Viral RNA polymerases: electron microscopy of reovirus reaction cores. Science. 1971;174:694–696. doi: 10.1126/science.174.4010.694. [DOI] [PubMed] [Google Scholar]
- Gomatos PJ, Tamm I. Animal and plant viruses with double-helical RNA. Proc Natl Acad Sci U S A. 1963;50:878–885. doi: 10.1073/pnas.50.5.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Diprose JM, Grimes JM, Malby R, Burroughs JN, Zientara S, Stuart DI, Mertens PP. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- Harrison SJ, Farsetta DL, Kim J, Noble S, Broering TJ, Nibert ML. Mammalian reovirs L3 gene sequences and evidence for a distinct amino-terminal region of the λ1 protein. Virology. 1999;258:54–64. doi: 10.1006/viro.1999.9707. [DOI] [PubMed] [Google Scholar]
- Harvey JD, Bellamy AR, Earnshaw WC, Schutt C. Biophysical studies of reovirus type 3. IV Low-angle x-ray diffraction studies. Virology. 1981;112:240–249. doi: 10.1016/0042-6822(81)90629-2. [DOI] [PubMed] [Google Scholar]
- Hilmer JK, Zlotnik A, Bothner B. Conformational equilibria and rates of localized motion within hepatitis B virus capsids. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.10.044. available online 22 Oct 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joklik WK. The reovirus particle. In: Joklik WK, editor. The Reoviridae. Plenum Press; New York: 1983. pp. 9–78. [Google Scholar]
- Johnson J, Rueckert R. Packaging and release of the viral genome. In: Chiu W, Burnett RM, Garcea RL, editors. Structural Biology of Viruses. Oxford University Press; New York: 1997. pp. 269–287. [Google Scholar]
- Kapuler AM. An extraordinary temperature dependence of the reovirus transcriptase. Biochemistry. 1970;9:4453–4457. doi: 10.1021/bi00824a029. [DOI] [PubMed] [Google Scholar]
- Kim J, Parker JSL, Murray KE, Nibert ML. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. Journal of Biological Chemistry. 2004;279:4394–4403. doi: 10.1074/jbc.M308637200. [DOI] [PubMed] [Google Scholar]
- Kim J, Zhang X, Centonze VE, Bowman VD, Noble S, Baker TS, Nibert ML. The hydrophobic amino-terminal arm of reovirus core shell protein λ1 is dispensable for particle assembly. J Virol. 2002;76:12211–12222. doi: 10.1128/JVI.76.23.12211-12222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson SM, Antczak JB, Joklik WK. Reovirus exists in the form of 13 particle species that differ in their content of protein sigma 1. Virology. 1994;201:303–311. doi: 10.1006/viro.1994.1295. [DOI] [PubMed] [Google Scholar]
- Lawton JA, Estes MK, Prasad BVV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nature Structural Biology. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- Lee PW, Hayes EC, Joklik WK. Protein sigma 1 is the reovirus cell attachment protein. Virology. 1981;108:156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- Love RA, Maegley KA, Yu X, Ferre RA, Lingardo LK, Diehl W, Parge HE, Dragovich PS, Fuhrman SA. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure. 2005;12:1533–1544. doi: 10.1016/j.str.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Luftig RB, Kilham SS, Hay AJ, Zweerink HJ, Joklik WK. An ultrastructural study of virions and cores of reovirus type 3. Virology. 1972;48:170–181. doi: 10.1016/0042-6822(72)90124-9. [DOI] [PubMed] [Google Scholar]
- Luongo CL, Reinisch KM, Harrison SC, Nibert ML. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein lambda2. J Biol Chem. 2000;275:2804–2810. doi: 10.1074/jbc.275.4.2804. [DOI] [PubMed] [Google Scholar]
- Mao ZX, Joklik WK. Isolation and enzymatic characterization of protein lambda 2, the reovirus guanylyltransferase. Virology. 1991;185:377–386. doi: 10.1016/0042-6822(91)90785-a. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McBride AE, Schlegel A, Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc Natl Acad Sci (USA) 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez II, Hermann LL, Hazelton PR, Coombs KM. A comparative analysis of freon substitutes in the purification of reovirus and calicivirus. J Virol Meth. 2000;90:59–67. doi: 10.1016/s0166-0934(00)00217-2. [DOI] [PubMed] [Google Scholar]
- Mendez II, She YM, Ens W, Coombs KM. Digestion pattern of reovirus outer capsid protein sigma3 determined by mass spectrometry. Virology. 2003;311:289–304. doi: 10.1016/S0042-6822(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf P, Cyrklaff M, Adrian M. The three-dimensional structure of reovirus obtained by cryo-electron microscopy. EMBO J. 1991;10:3129–3136. doi: 10.1002/j.1460-2075.1991.tb04874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov SY. A possible relationship of reovirus putative RNA polymerase to polymerases of positive-strand RNA viruses. Nucleic Acids Res. 1989;17:5394. doi: 10.1093/nar/17.13.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJL, Lopez AD. The Global Burden of Disease. A comprehensive assessment of mortality and disability from diseases, injuries, and risk factors in 1990 and projected to 2020. Harvard School of Public Health; Boston: 1996. p. 990. [Google Scholar]
- Nibert ML. Structure of mammalian orthoreovirus particles. Curr Topics Microbiol Immunol. 1998;233(I):1–30. doi: 10.1007/978-3-642-72092-5_1. [DOI] [PubMed] [Google Scholar]
- Nibert ML, Margraf RL, Coombs KM. Nonrandom segregation of parental alleles in reovirus reassortants. J Virol. 1996;70:7295–7300. doi: 10.1128/jvi.70.10.7295-7300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Nibert ML. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein lambda1. J Virol. 1997;71:2182–2191. doi: 10.1128/jvi.71.3.2182-2191.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly EK, Kao CC. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- Pan J, Vakharia VN, Tao YJ. The structure of birnavirus polymerase reveals a distinct active site topology. Proc Natl Acad Sci (USA) 2007;104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Powell KF, Harvey JD, Bellamy AR. Reovirus RNA transcriptase: evidence for a conformational change during activation of the core particle. Virology. 1984;137:1–8. doi: 10.1016/0042-6822(84)90002-3. [DOI] [PubMed] [Google Scholar]
- Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondeu J, Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello VR. Picornaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th. Lippencott Williams & Wilkins; Philadelphia: 2007. pp. 795–838. [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 Ǻ resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Schiff LA, Nibert ML, Tyler KL. Orthoreoviruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th. Lippencott Williams & Wilkins; Philadelphia: 2007. pp. 1853–1915. [Google Scholar]
- Sheehan B, Fuller SD, Pique ME, Yeager M. AVS software for visualization in molecular microscopy. Journal of Structural Biology. 1996;116:99–106. doi: 10.1006/jsbi.1996.0017. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Chernushevich I, Ens W, Standing KG, Thomson B, Wilm M, Mann M. Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1997;11:1015–1024. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Starnes MC, Joklik WK. Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology. 1993;193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- Steitz TA. Structures of the Central Dogma of molecular biology. Structures and Mechanisms: from Ashes to Enzymes. 2002;827:231–248. [Google Scholar]
- Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage – Structural studies of reovirus polymerase lambda3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- The World Health Report. Fighting Disease, Fostering Development. World Health Organization; Geneva: 1996. [PubMed] [Google Scholar]
- Xu P, Miller SE, Joklik WK. Generation of reovirus core-like particles in cells infected with hybrid vaccinia viruses that express genomic segments L1, L2, L3, and S2. Virology. 1993;197:726–731. doi: 10.1006/viro.1993.1648. [DOI] [PubMed] [Google Scholar]
- Yin P, Cheang M, Coombs KM. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J Virol. 1996;70:1223–1227. doi: 10.1128/jvi.70.2.1223-1227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 angstrom. Nature Structural Biology. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]


