Abstract
HIV assembly and replication proceed through formation of morphologically distinct immature and mature viral capsids that are organized by the Gag polyprotein (immature) and by the fully processed CA protein (mature). The Gag polyprotein is composed of three folded polypeptides (MA, CA, and NC) and three smaller peptides (SP1, SP2, and p6) that function together to coordinate membrane binding and Gag-Gag lattice interactions in immature virions. Following budding, HIV maturation is initiated by proteolytic processing of Gag, which induces conformational changes in the CA domain and results in assembly of the distinctive conical capsid. Retroviral capsids are organized following the principles of fullerene cones, and the hexagonal CA lattice is stabilized by three distinct interfaces. Recently identified inhibitors of viral maturation act by disrupting the final stage of Gag processing, or by inhibiting formation of a critical intermolecular CA-CA interface in the mature capsid. Following release into a new host cell, the capsid disassembles and host cell factors can potently restrict this stage of retroviral replication. Here, we review the structures of immature and mature HIV virions, focusing on recent studies that have defined the global organization of the immature Gag lattice, identified sites likely to undergo conformational changes during maturation, revealed the molecular structure of the mature capsid lattice, demonstrated that capsid architectures are conserved, identified the first capsid assembly inhibitors, and begun to uncover the remarkable biology of the mature capsid.
Keywords: HIV, Gag, CA
Introduction
HIV-1 assembles into morphologically distinct immature and mature virions. The virus initially assembles and buds from cells as a noninfectious, immature spherical particle that is organized by a layer of Gag proteins that are associated with the inner viral membrane (Fig. 1). Upon budding, immature particles dramatically rearrange to form mature, infectious virions. The mature capsids are mostly conical, though a small percentage of cylindrical capsids are also observed. The remarkably distinct morphologies of the immature Gag lattice and the capsid lattice presumably reflect specific requirements for organizing viral assembly in the producer cell and coordinating viral replication in the new host cell (Fig. 1a) (reviewed in [1-4]). The capsid and its contents are called the “core,” and this is the object that is released into the cytoplasm of a newly infected cell to initiate a new cycle of viral replication [3-8].
Figure 1.

(a) Summary of the HIV-1 replication cycle. (b) HIV-1 Gag polyprotein domain structure, showing the locations of MA, CANTD, CACTD, SP1, NC, SP2, and p6. (c) Structural model of the extended Gag polypeptide, derived from high-resolution structures and models of isolated domains. Unstructured and linker regions are represented by dashed lines. PR cleavage sites are indicated by the arrowheads in (b) and (c). (d, e) Schematic models of the immature (d) and mature (e) HIV-1 virions. (f, g) Central slices through cryo-EM tomograms of immature (f) and mature (g) HIV-1 particles. The spherical virions are approximately 130 nm in diameter.
The structural proteins of HIV-1 are all derived from the Gag polyprotein (Fig. 1b), which is myristoylated co-translationally. Gag molecules assemble at the plasma membrane, and immature virions acquire a lipid envelope as they bud (Fig. 1a, d, and f). The viral protease is activated during assembly, and it cleaves Gag to generate a set of new proteins and spacer peptides (SP), termed MA, CA, SP1, NC, SP2, and p6. These newly processed proteins then reassemble to form the distinct layers of the mature virion: MA remains associated with the inner viral membrane (the “matrix” layer), NC coats the viral RNA genome (the “nucleocapsid” layer), and CA assembles into the conical capsid that surrounds the nucleocapsid and its associated enzymes, reverse transcriptase (RT) and integrase (IN) (Fig. 1e and g). High-resolution tertiary structures of the MA, CA, and NC proteins from a number of different orthoretroviruses show that the folds of the individual Gag domains are highly conserved, despite limited primary sequence conservation (Fig. 1c and reviewed in [9,10]). It can therefore be inferred that they share the same basic architecture, despite significant variations in capsid shape that were historically used to define different retroviral genera.
Gag as an assembly machine
The immature virion is a roughly spherical shell of radially extended Gag molecules. The N-terminal Gag MA domains are bound to the inner viral membrane, and the C-termini of the Gag molecules project into the center of the virus (Fig. 1 and 2). To a first approximation, all of the information necessary for retroviral particle assembly resides in the Gag polypeptide. For example, Gag alone can form extracellular virus-like particles in the absence of other viral proteins [11] and Gag molecules can spontaneously assemble into spherical, immature virus-like particles in vitro [12-14]. Nevertheless, although Gag itself encodes the necessary tertiary and quaternary interactions, it must be emphasized that assembly requires nonspecific RNA interactions both in vivo and in vitro, and is assisted by host factors in vivo, including trafficking factors, assembly chaperones, and the ESCRT budding pathway, as reviewed elsewhere [15-18].
Figure 2. Structural features of the immature Gag lattice.
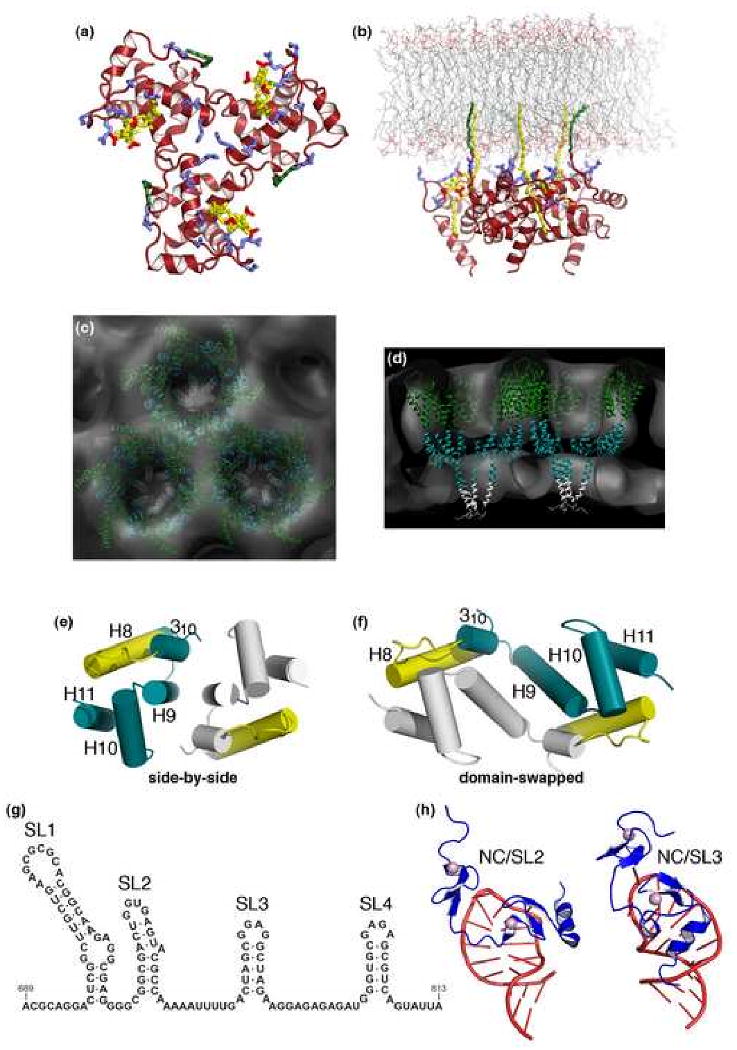
(a, b) Models for membrane binding by the N-terminal MA domain showing a top view (a) and a side view of the MA trimer bound to a lipid bilayer (b), with the myristoyl chain (colored in green) inserted into the inner leaflet, and basic residues (blue) interacting with acidic phospholipid headgroups, including PI(4,5)P (yellow) [25,40]. (c, d) Two views of a model based on electron cryotomography of the immature Gag lattice (gray) showing that Gag-Gag lattice interactions are mediated primarily by the CA and SP1 domains. Plausible positions of the CANTD (dark green), CACTD (blue green), and SP1 (gray) layers are shown for reference [24]. (e, f) Two crystallographically characterized HIV-1 CACTD dimers showing side-by-side (e) [68,69] and domain-swapped conformations (f) [72]. The monomers are shown in blue green and gray, and the MHR element in yellow. (g, h) Secondary structure of the HIV-1 Ψ packaging element (g), and high-resolution structures (h) of the isolated NC domain bound to the second (left) and third (right) Ψ stem loops, with NC in blue and RNA in red [85,86].
Although it was appealing to imagine that the Gag molecules were organized with icosahedral symmetry, analyses of immature virions of HIV-1 [19-21], Rous sarcoma virus (RSV) [22] and murine leukemia virus (MLV) [23] revealed non-icosahedral paracrystalline lattices composed of hexameric Gag rings. The rings are close-packed, with characteristic inter-ring spacings of ∼80 Å. The immature virions of HIV-1 particles are often discontinuous, implying that the Gag lattice need not close fully during virus budding and that the membrane also plays an important role in stabilizing immature particles ([24], H.-G. Kräusslich, pers. comm.). The different regions of Gag must, of course, work together in concert to drive particle assembly, and the emerging picture is that many different interactions collaborate to create viral particles. Indeed, the requirement for multiple weak, but highly cooperative interactions probably provides a mechanism for ensuring that particle formation only occurs when all essential aspects of assembly such as membrane binding and RNA packaging have been met. Thus, while different regions of Gag contribute to membrane binding, Gag-Gag interactions and cofactor recruitment, we wish to emphasize that the Gag polypeptide is an assembly machine that simultaneously performs and integrates all of these different activities.
Membrane binding is mediated primarily by the MA domain
MA binds directly to the inner leaflet of the plasma membrane, and recent studies have helped elucidate how this domain targets Gag to the plasma membrane and triggers particle assembly at the correct time and place (Fig. 2a and b) [16]. HIV-1 MA (colored red in Fig. 1 and 2) is a helical domain that displays a conserved patch of basic residues on the same face as the N-terminal myristoyl modification. C-terminal MA residues form an extended α-helix that projects away from the globular domain, on the side opposite the membrane binding face [25,26]. Membrane binding is mediated by insertion of the myristoyl group into the lipid bilayer and by the basic patch, which binds acidic phospholipids, particularly PI(4,5)P2, a phosphoinosotide that is concentrated in the plasma membrane (Fig. 2a and b) [27-31]. Importantly, HIV-1 Gag is incorrectly directed to internal membranes upon depletion of cellular PI(4,5)P2, indicating that this phospholipid is required for proper targeting in vivo [28]. Binding studies indicate that ionic interactions dominate the energetics of HIV-1 MA membrane binding in vitro and that the myristoyl group makes only modest contributions to the binding interaction [32]. Indeed, some retroviral MA proteins lack myristoyl (or similar) modifications altogether, and in those cases, membrane binding is mediated exclusively by ionic interactions [33,34]. Cholesterol also plays an important role in Gag targeting and particle assembly [35], but the molecular mechanism by which Gag senses cholesterol is not yet known.
MA membrane binding appears to be positively coupled to Gag oligomerization and assembly in several different ways. Locally, the MA domain exhibits a two-state, “myristoyl switching” mechanism in which the myristoyl group is sequestered in a groove along the body of the globular domain in the soluble form of the protein and is then exposed for insertion into the bilayer [36-39]. Myristoyl exposure is favored by PI(4,5)P2 binding, providing a mechanism for coupling plasma membrane recognition and membrane binding [40]. Stable membrane binding and myristoyl exposure are also favored by Gag oligomerization, [32,37,41,42], and inositol phosphate binding promotes trimerization of Gag fragments even in the absence of the myristoyl moiety [43]. Thus, downstream Gag-Gag interactions contribute to the affinity of MA-membrane interactions (and vice versa). The MA domain may also negatively regulate Gag assembly, probably by “folding back” onto the NC region, providing yet another potential mechanism for coupling membrane binding with the formation of the lateral Gag-Gag interactions essential for immature virion assembly [43,44].
Gag-Gag lattice interactions in the immature virion are mediated primarily by CA and SP1
The immature virion is stabilized by lateral interactions distributed throughout the Gag polypeptide, and the historical view of a discrete interaction site (or “I” domain) is therefore an oversimplification. Nevertheless, specific Gag regions are particularly important for immature particle formation, whereas other regions, such as MA and p6, are largely dispensable. Although MA couples membrane binding and assembly, it does not form a continuous regular lattice in the immature virion [22] and is dispensable for particle formation [45]. Similarly, p6 recruits the cellular machinery required for virus budding [46], but does not make important Gag-Gag contacts. In contrast, critical contacts are made by the C-terminal domain of CA (CACTD), the adjacent SP1 spacer, the NC region, and, to a lesser extent, the N-terminal domain of CA (CANTD) [42,47-52].
A recent electron cryotomographic analysis of immature HIV-1 virions confirmed the radial “beads on a string” arrangement of the different Gag domains and showed that the CA and SP1 regions formed a close-packed lattice of cup-shaped hexamers, in which the walls and bottom of the cups appeared to correspond to the CA and SP1 layers, respectively (Fig. 2c and d) [24]. In contrast, the MA and NC layers lacked hexagonal order. Although the resolution was insufficient to position domains unambiguously, this reconstruction can now be used to guide possible models for Gag-Gag interactions, as constrained by relevant biochemical and high resolution structural data.
High resolution structures show that the free MA and CANTD domains are connected by a flexible linker that could easily span the distance between the MA and CANTD layers of the immature virion [53,54]. The MA domain does not change structure when tethered to CANTD, consistent with the idea that the membrane-binding “heads” of MA are connected via flexible linkers to the CANTD hexamers below. The oligomeric state of these membrane-bound MA proteins has not yet been established, however, since the matrix layer does not form a continuous lattice [22], and MA proteins can form both trimers [25,40,43,55] and hexamers [56] in vitro. Interestingly, the MA and CA domains of RSV Gag are separated by an extended linker, called p10, which contributes to Gag assembly by forming an α-helical bundle with the neighboring subunit in the Gag hexamer ([57], V. Vogt, pers. comm.).
The CANTD domain (colored dark green in Figs. 1-3) is composed of seven α-helices packed in the shape of an arrowhead (CA helices 1-7), with an extended loop connecting helices 4 and 5 that binds the prolyl isomerase, cyclophilin A. CANTD interfaces within the immature lattice are still a matter of speculation, but biochemical studies have identified two regions that are important for immature virion formation: one encompassing helices 1 and 2, and a second surrounding helices 4 and 7 [42,47,58,59]. These observations are consistent with models in which the hexameric rings of CANTD in the immature lattice are stabilized by six-fold symmetric interactions involving helices 1 and 2, and contact(s) across the local two- or three-fold axes between neighboring hexamers involving helices 4 and 7 [24]. Indeed, it was recently proposed that similar interhexamer contacts seen in two different MLV CANTD crystal forms might mimic the three-fold symmetric contacts between Gag hexamers in the immature lattice, and this proposal was supported by mutational analyses [60,61]. Nevertheless, further studies will be required to confirm these models and provide molecular details of the intra- and interhexamer contacts in the immature HIV-1 Gag lattice. In this regard, considerable insight will likely be gleaned from higher resolution structural studies of immature HIV-1, MLV, and MPMV (Mazon-Pfizer monkey virus) Gag assemblies formed in vitro [62-65], and from deuterium-exchange protection experiments that map protein-protein interfaces in the immature lattice (P. Prevelige, pers. comm.).
Figure 3. Structure of the mature HIV-1 capsid.
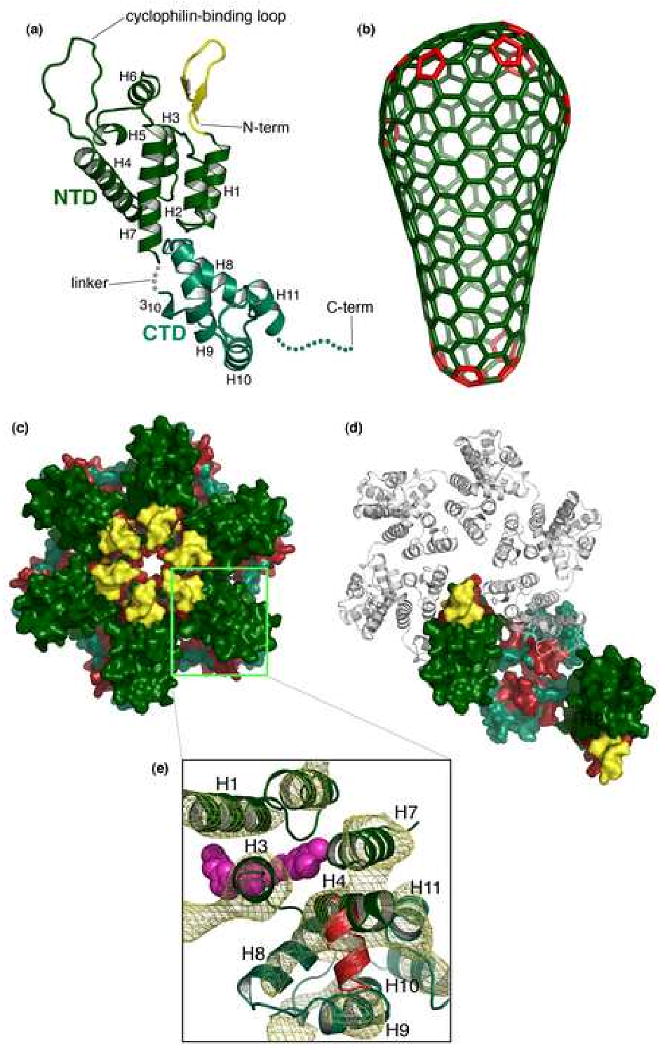
(a) Tertiary structure of the mature, processed CA protein, with the N-terminal domain colored in dark green and the C-terminal domain in blue-green. The β-hairpin is colored yellow. The final 11 residues of CA, indicated by the dashed line are typically disordered in crystal structures. (b) Fullerene model for the conical capsid, with CA hexamers (green) and pentameric declinations (red) [89]. (c, d) Surface representation of the pseudoatomic model of the mature HIV-1 CA hexamer [102], emphasizing the CANTD hexamer (c) and CACTD dimer (d), viewed from the outer surface of the capsid. Color coding is the same as in (a), except with CA residues colored red to denote sites of significant protection from deuterium exchange in the hexagonal CA lattice [103]. Note that the exposed red patch in (d) corresponds to the CANTD-binding site on the CACTD, which is exposed when the neighboring CACTD domain is removed. (e) Top view of the CACTD-CANTD intermolecular interface. The pseudoatomic model is shown in ribbon representation, fitted to the experimental density map derived by electron cryocrystallography (gold mesh) contoured at 1.6σ [102]. Binding sites of the CA-I (red) [123] and CAP-1 (magenta) [121] maturation inhibitors are indicated.
The CACTD is also critical for immature particle assembly, and it appears to make both inter- and intrahexamer contacts in the Gag lattice [24,50-52,66,67]. The globular CACTD domain (colored blue-green in Fig. 1-3) is composed of a short 310 helix followed by an extended strand and four α-helices (CA helices 8-11) [68,69]. The strand/turn/helix 8 element is termed the major homology region (MHR) because its sequence is highly conserved across most retroviruses and retrotransposons [2,70]. Mutations in conserved MHR residues inhibit the assembly of immature particles, indicating that the MHR plays an important role in Gag lattice formation [47,50,70,71].
HIV-1 CA dimerizes in solution, and CACTD constructs have been crystallized as two distinct, but related dimers. One dimer is a side-by-side complex that is stabilized primarily by parallel hydrophobic packing of helix 9 against a symmetry mate (Fig. 2e) [68,69]. The RSV CACTD forms a similar dimer at low pH, albeit with a more hydrophilic interface and a somewhat different interdomain angle (R.L. Kingston, pers. comm.). The second HIV-1 CACTD dimer is a “domain-swapped” analog in which the MHR segments from each monomer associate to create a more extended dimer interface (Fig. 2f) [72]. In this case, the structure was determined for a CACTD mutant that lacked a single amino acid residue in the helix 8/9 linker, which favored the domain-swapped conformation. Mutations expected to disrupt intermolecular helix 9 packing interfaces seen in both types of dimer reduced Gag dimerization (and trimerization) in vitro, and inhibited the production of immature particles, supporting the biological relevance of the interface(s) [42,43,47]. Possible roles for the two different CACTD dimers are discussed in greater detail below.
Residues located at the C-terminal end of CA and at the N-terminal end of the SP1 spacer are also essential for immature particle formation [48,73]. In the tomographic reconstruction of frozen-hydrated immature particles, density ascribed to the CACTD-SP1 region was located immediately beneath the central pore of each hexamer (i.e., the bottom of the cup), suggesting that this region stabilizes the hexamer. NMR studies of isolated Gag fragments indicate that the helical propensity for the CA-SP1 junction is weak [74], but sequence analyses and mutational studies support the idea that this region may adopt an ordered helical structure in assembled virions [73,75,76]. The CA-SP1 junction was therefore modelled as a six-helix bundle in the tomographic study (Fig. 2c and d). MLV Gag, which lacks a spacer peptide between its CA and NC domains, may contain an equivalent region at the C-terminus of the CA domain, termed the “charged assembly helix”, which is also required for immature particle assembly [77].
Finally, the NC region of Gag (colored blue in Fig. 1b) forms the innermost layer of the immature virion and also plays essential roles in particle formation [21,23,51,78-81]. NC/RNA complex(es) do not follow the hexagonal symmetry of the CA and SP1 regions, and most experiments indicate that NC primarily “tethers” Gag molecules together, probably via RNA bridges, although additional NC-NC interactions may also occur [79,80,82,83]. NC/RNA tethers presumably increase the effective concentration of assembling Gag molecules, which could also play a more active role in orienting or otherwise facilitating CA-CA interactions.
While NC can bind RNA nonspecifically, packaging of the genome requires specific recognition of the dimeric, full-length viral RNA transcript. HIV-1 genome packaging requires an RNA element located near the 5′ end of the gag gene (termed “Ψ”), which is composed of four stem-loops that can dimerize through self-complementary base pairing interactions of loop 1 residues (Fig. 2g) (reviewed in [84]). Structures of NC in complex with two of the isolated stem loops from HIV-1 Ψ show that the CCHC zinc fingers of NC make sequence-specific, but remarkably different, contacts with exposed loop residues (Fig. 2h) [85,86]. The NC/RNA interactions visualized in these structures undoubtedly contribute to the specificity of genomic RNA packaging, although the full mechanism by which HIV-1 Gag packages dimeric RNA genomes has yet to be delineated. Even greater progress on the mechanism of genomic RNA packaging has been made for RSV and MLV, where complexes of NC with minimal Ψ RNA elements are now available [87,88]. In the MLV case, the high-affinity NC binding site is initially sequestered by base pairing in the monomeric form of the viral RNA, and then becomes exposed in the dimeric RNA, providing an elegant mechanism for coupling genome dimerization and encapsidation [87].
As is clear from the above discussion, structural studies of the immature virion are still at an early stage, and important future goals include: (1) determining high resolution structures of Gag in its soluble dimeric and trimeric conformations [43], (2) extending the resolution of models for the assembled hexagonal Gag lattice, and (3) understanding the mechanism by which the Gag lattice curves into a spherical assembly (e.g., through the inclusion of pentameric declinations and/or through irregular juxtapositions of hexagonal sheets). These are challenging problems, as retroviruses are irregular, yet highly organized assemblies, and progress will therefore continue to require the coordinated application of multiple different structural methods.
Mature retroviral capsids are fullerene structures
Following Gag processing, ∼1,500 copies of the mature CA protein reassemble to form the mature capsid (Fig. 3) [89-91]. Retroviral capsids appear to adopt preferred shapes: cones (e.g., HIV), cylinders (e.g., MPMV), or “spheres” (e.g., MLV, RSV). However, recent tomographic analyses of mature HIV-1 and RSV particles have revealed significant capsid polymorphism, including amorphous shapes, multiple and nested capsids, and incompletely closed shells [92-94]. These observations suggest that mature retroviral capsids assemble through favored, but not strictly defined pathways.
The conical HIV-1 capsid is a curved hexagonal assembly called a fullerene cone [89-91,95] (Fig. 3b). Fullerene cones require twelve pentameric declinations (“pentons”) to close and can adopt one of five allowed cone angles. The narrowest allowed cone angle (19.2°) predominates in both authentic HIV-1 capsids and in “synthetic” capsids assembled from pure recombinant proteins [89,93,96]. These narrow cones are closed by inclusion of five and seven pentons at their narrow and wide ends, respectively (Fig. 3b). The tubular and spherical shapes of other retroviral capsids can also be explained as related fullerene assemblies [89]. Specifically, spherical shapes are formed when the pentons are dispersed more evenly throughout a curved hexagonal lattice, and tubular capsids are formed by a cylindrical hexagonal lattice that is closed by inclusion of six pentons at either end of the tube [89,90,97,98].
Tomographic studies of frozen-hydrated mature HIV-1 particles support the fullerene organization of the viral capsid and reveal regions of high density immediately inside the broad end of the cone, which presumably represent sites of close contact with the genomic ribonucleoprotein complex [92,93]. The broad end of the capsid appears to reside at a constant (11-12 nm) distance from the matrix, indicating that the two layers may interact. The narrow end of the cone has been reported to approach the membrane even more closely than the broad end, and in some cases, appeared connected to the membrane/matrix, which may represent nucleation sites where capsid assembly is initiated [92]. Direct capsid/matrix linkages were also observed in a recent tomographic study of mature RSV particles [94], and the linkage sites frequently sat beneath Env spikes and linked to capsid vertices (i.e., pentons). This may imply that Env assemblies help initiate capsid assembly and/or promote formation of pentameric declinations, although it should be noted that specific Env linkages cannot be absolutely required for functional capsid formation, since retroviruses remain infectious when pseudotyped with heterologous envelope proteins.
Structure of the hexameric CA lattice
Pure recombinant HIV-1 CA proteins and CA-SP1-NC/RNA complexes can form cylindrical and conical shells in vitro, implying that all of the information necessary to form a conical capsid resides within the CA polypeptide [89,99-101]. Mature HIV-1 and RSV CA proteins form hexagonal arrays of hexameric CANTD rings, with each ring connected to its six nearest neighbors via CACTD dimer interactions [90,91,97]. A crystal structure of the N-MLV CANTD hexamer revealed that the subunits associate via an 18-helix bundle formed by helices 1-3 [61]. Recently, electron cryocrystallography of well-ordered two-dimensional crystals of full-length HIV-1 CA yielded a structure of the complete hexagonal lattice at 9 Å resolution [102]. At this resolution, it was possible to resolve rods of density that corresponded to all of the CA secondary structural elements, allowing unequivocal positioning of the CANTD and CACTD crystal structures. The resulting pseudoatomic model showed that the mature HIV-1 capsid lattice is stabilized by three different intermolecular CA-CA interfaces (Fig. 3c-e): (1) intrahexamer CANTD-CANTD interactions are mediated by CA helices 1-3, which associate as an 18-helix bundle in the center of the hexamer (as seen for the N-MLV CANTD hexamer), (2) intrahexamer CANTD-CACTD interactions are formed by asymmetric insertion of the CANTD helix 4 into a groove of CACTD from an adjacent molecule within the hexamer, allowing the six CACTD domains to form a “belt” surrounding the CANTD core, and (3) interhexamer CACTD-CACTD interactions are mediated by symmetric, parallel dimerization of helix 9 from CACTD subunits of adjacent hexamers (as seen in the side-by-side CACTD dimer) (Fig. 2c) [68,69]. Gratifyingly, this pseudoatomic model of the hexagonal capsid lattice is consistent with a large body of biochemical and genetic data, including deuterium exchange, crosslinking, mutagenesis, and second-site suppressor studies (e.g., see Fig. 3c and d) [47,58,98,103,104].
Although a molecular model for the hexagonal CA lattice is now in hand, we still lack a comprehensive, high-resolution structural model of the mature HIV-1 capsid. Obtaining such a model will require: (1) extending studies of the hexameric CA lattice to atomic resolution, (2) determining how this lattice accommodates conical curvature, (3) obtaining a structural description of the pentameric declinations, (4) determining precisely how the ends of the capsid interact with the matrix shell, and (5) defining how the core components, particularly the replication-initiation complex and associated enzymes, are organized within the capsid.
Retroviral maturation
Gag processing by the viral protease (PR) initiates the essential process of virion maturation, in which the liberated NC/RNA complex condenses at the center of the core, the genomic RNA dimer becomes more stable [105], and the processed CA protein forms the conical capsid. HIV-1 PR and other viral enzymes encoded within Gag-Pol fusion proteins are generated when ∼5% of the ribosomes shift to the -1 frame while translating the C-terminal end of gag, and the resulting Gag-Pol proteins are apparently packaged into virions via Gag-Gag lattice interactions. PR is liberated by autoproteolysis at a late stage of viral assembly, which ensures that Gag proteins are not processed before they assemble. All five different HIV-1 Gag processing sites are essential for infectivity, and the sites are processed at different rates, in the order: SP1/NC > MA/CA and SP2-p6 > NC-SP2 > CA-SP1 [106].
Retroviral capsids do not assemble via a direct, concerted condensation of the immature lattice, but rather are likely reassembled from a subset of free CA subunits, following specific nucleation events that help restrict the overall capsid assembly pathway [20,58,92-94]. CA processing must therefore destabilize the immature Gag lattice while promoting formation of the mature capsid lattice. Proteolytic processing appears to trigger conformational changes at both ends of the CA protein that favor capsid assembly. Processing at the N-terminal end of CA (the MA-CA junction in HIV-1 Gag) converts the first 13 CA residues from an extended conformation into a folded β-hairpin structure (colored yellow in Fig. 3) [53,66,107]. This structure is stabilized by formation of a partially buried salt bridge between a highly conserved aspartate residue within CA helix 3 and the newly processed N-terminus [66,108]. β-hairpin formation functions as a switch that favors mature CA hexamer formation, though its effects appear subtle. In the mature CA hexamer, the β-hairpin sits immediately above the interface created by CA helices 1-3 but does not make extensive intermolecular interactions [61,102]. Rather, β-hairpin formation appears to help position CANTD helices 1-3 to form the mature capsid hexamer (and may also help disrupt immature lattice interactions).
Proteolysis at the C-terminal end of CA (the CA-SP1 junction of HIV-1 Gag) also appears to trigger a second molecular switch that favors mature CA lattice formation. In support of this idea, removal of SP1 can cause recombinant HIV-1 Gag proteins to switch from immature-like spherical assemblies to mature-like cylinders in vitro [13]. As discussed above, the CA-SP1 region appears to stabilize the immature Gag hexamer, and proteolysis at the CA-SP1 junction therefore presumably destabilizes the Gag lattice. This switch may also involve large rearrangements in the orientation of the CACTD dimer. As noted above, CACTD has been crystallized as both side-by-side and domain-swapped, dimers (Fig. 2e and f). One attractive model is that the domain-swapped dimer exists within the immature virion and then converts into a side-by-side dimer in the mature capsid [72,109]. This would allow the more extended domain-swapped dimer to stabilize the assembly-competent immature virion, whereas the smaller side-by-side dimer interface could contribute to the lability of the disassembly-competent mature capsid. In principle, interactions within the SP1-NC region could also contribute to formation of domain-swapped dimers, rationalizing how proteolytic processing at the CA-SP1-NC junctions could promote the conformational switch. Moreover, the constraints of having to switch between monomers (when Gag is first made), to domain-swapped dimers (in the immature virion), to side-by-side dimers (in the mature capsid), offer an appealing explanation for the extreme conservation of the MHR. Consistent with this idea, conserved MHR residues form extensive hydrophobic interactions and hydrogen bonding networks in all three structures [68,69,72,110]. While this speculative model requires rigorous testing, it is not unreasonable to expect that CA may undergo major conformational changes during maturation, given the dramatic conformational rearrangements seen in other well characterized viral maturation processes [111,112].
Novel maturation inhibitors block Gag processing and CA assembly
The clinical success of HIV-1 PR inhibitors validates maturation as an attractive therapeutic target, and non-protease maturation inhibitors have recently been identified. One class of maturation inhibitors is exemplified by 3-O-(3′,3′-dimethylsuccinyl)betulinic acid (also called PA-457) [113,114], which inhibits HIV-1 replication in tissue culture and animal model systems, and is currently in Phase 2 clinical trials [115]. PA-457 inhibits Gag processing at the CA-SP1 junction and appears to bind directly to this site because the inhibitor is incorporated into assembling virions and because mutations at the CA-SP1 junction induce drug resistance [115-118]. PA-457 does not bind free Gag, however, and therefore presumably recognizes SP1 in its assembled conformation. Cells treated with PA-457 release noninfectious virions with aberrant capsids that resemble those seen for viruses with mutations that inhibit CA-SP1 processing [115]. However, it appears that PA-457 does not completely block, but simply delays, proteolysis of the CA-SP1 junction [119], suggesting that Gag proteolysis and maturation must occur within a defined temporal window.
A second class of inhibitors binds the processed CA protein and inhibits capsid assembly. The founding members of this class are methylphenylurea compounds, such as CAP-1, which bind in a deep pocket in CANTD, formed at the junction of helices 1, 2, 4, and 7 [120,121]. Interestingly, this pocket is not present in the static structure of free CANTD, but rather is created when the conserved CA Phe32 residue swings out into solution. Another inhibitor of this type is CA-I, a 12-residue peptide identified by phage display, which binds between CACTD helices 8 and 11 [122,123]. Both CA-I and CAP-1 bind within (or adjacent to) the mature CANTD-CACTD interface (Fig. 3e), suggesting that they inhibit capsid assembly by disrupting this interaction [102,121,123]. CA-I and CAP-1 bind too weakly to be clinically useful, but the independent discovery of two different inhibitors that bind at the CANTD-CACTD interface makes this site an attractive target for further inhibitor development.
Capsid disassembly
The fate of the core particle in the cytoplasm of newly infected cells represents perhaps the most interesting, yet poorly understood stage of the retroviral life cycle. In essence, we still need to learn why retroviruses go to the trouble of building the mature core particle, and how host proteins can positively and negatively regulate its different functions. Following fusion of the viral and cell membranes, the HIV-1 core is released into the cytoplasm, the capsid uncoats, the single-stranded RNA genome is reverse-transcribed into double-stranded DNA, and the proviral DNA genome is trafficked into the nucleus and integrated into host chromosomes (Fig. 1a) (reviewed in refs [124-126]). The HIV-1 preintegration complex travels to the nuclear envelope via the microtubule network [127], and is subsequently actively transported through the nuclear pore, although the cis- and trans-acting factors required for nuclear uptake are still debated [128]. Genetic analyses indicate that properly assembled capsids are required for the successful completion of reverse transcription (and other early events) [47,49,50,67,71,129,130] and also for later events that accompany nuclear localization of the preintegration complex [130,131]. Moreover, mutations that either increase or decrease capsid stability can reduce viral infectivity, suggesting that the timing (or extent) of capsid disassembly is probably important for successful completion of the first half of the viral life cycle [129].
Capsid restriction
One of the most exciting recent developments in retrovirology is the realization that cells possess a number of intrinsic antiviral responses that can potently inhibit, or restrict, viral replication (reviewed in [132]). One very important family of cellular restriction factors is the tripartite motif 5 (TRIM5) proteins, which mediate species-specific early blocks to retrovirus infection. TRIM5 proteins come in two distinct forms (Fig. 4a): TRIM5α (expressed by most primates) [133] and TRIM-Cyp (expressed by owl monkeys) [134] (see [135-137] for recent reviews). Both TRIM alleles recognize the viral capsid, but do so in different ways. TRIM5α restriction is species- and virus-specific, and the specificity of restriction is largely determined by interactions between the C-terminal SPRY/B30.2 domain and the intact retroviral capsid. Although structural details are still lacking, mutational studies and sequence analyses of different SPRY alleles have provided insights into the determinants of capsid recognition (e.g., see [138-141]). In TRIM-Cyp, the SPRY domain of TRIM5α is replaced by cyclophilin A (CypA), a well-known peptidyl prolyl isomerase. In this case, the mechanism of capsid recognition is better understood because CA/CypA interactions have been characterized extensively. CypA makes sequence-specific contacts with the extended loop that connects CA helices 4 and 5, burying the CA Pro90 sidechain in the enzyme's active site [141,142]. Pro90 is efficiently isomerized by CypA [143], although the biological relevance of this activity remains to be determined.
Figure 4. TRIM restriction factors.
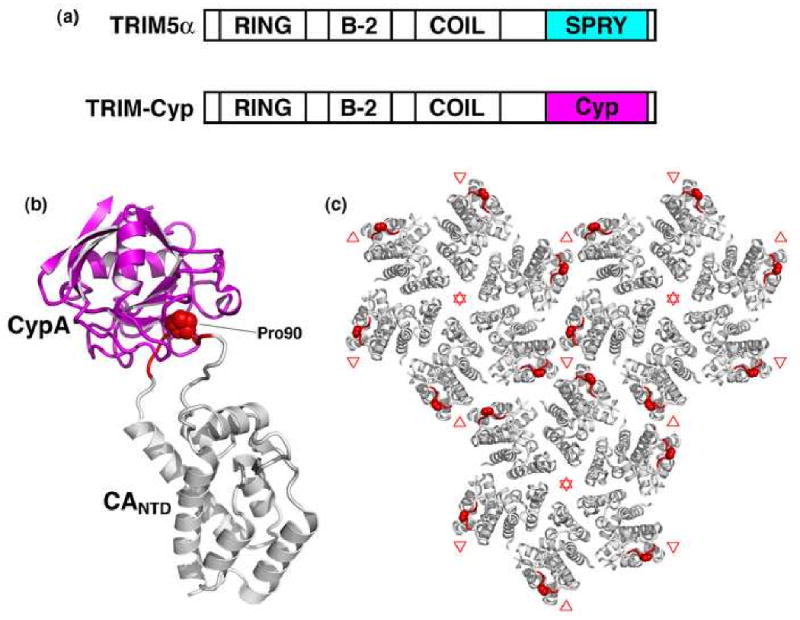
(a) Domain structures of TRIM5α and TRIM-Cyp highlighting the differences in their C-terminal capsid binding domains. (b) Structure of cyclophilin A (magenta) bound to CANTD (gray) [142]. Cyclophilin-binding residues are colored red, with CA Pro90 shown explicitly. (c) Top view of the mature lattice, with three full hexamers [102]. Note that the cyclophilin-binding loops (red) surround local three-fold symmetry axes (denoted by red triangles).
As illustrated in Figure 4c, the cyclophilin-binding loop sits on the surface of the capsid, on the outer edge of the CA hexamer. TRIM5 proteins are apparently trimeric, and it is therefore likely that they bind over one of the two sites of local three-fold symmetry on the capsid surface, most probably on the site between three adjacent hexamers (red in Fig. 4c) [145]. Although the subsequent events that lead to restriction are not yet well understood, TRIM5α and TRIM-Cyp typically prevent accumulation of reverse transcripts (although later-stage blocks are observed under some restricting conditions [146,147]). TRIM5α restriction correlates with accelerated rates of retroviral capsid dissociation, leading to the proposal that TRIM5α (and its cofactors) destabilize the capsid before it can perform essential functions [138,148]. Alternatively, TRIM5α may inactivate cores by diverting them to cytoplasmic “TRIM bodies” and/or to the proteasome (e.g., see ref. [147]). This is a very active area of research, and ongoing functional studies will likely soon set the stage for structural studies on the mechanism of TRIM5 restriction.
Virus assembly in vitro and in vivo
This review has summarized our growing understanding of the structures of immature and mature retroviral capsids. It is quite remarkable that accurate mimics of both structures can be assembled in vitro using pure recombinant proteins and non-specific nucleic acids. Indeed, these (and other viral systems) provide dramatic extensions of Anfinsen's original hypothesis by showing that primary polypeptide sequences can encode all of the information necessary to dictate the assembly of megadalton virus-like particles, even when those particles lack any formal symmetry. Of course, eukaryotic cells contain elaborate pathways that are used by viruses to control the localization, timing, and efficiency of particle assembly, and the next frontier is to understand how retroviruses assemble in the complex environment of the cell.
Acknowledgments
Work in the authors' laboratories are supported by NIH grants RO1 GM066087 (M.Y.), R37 AI45405 (W.I.S.), and P50 GM082545 (W.I.S. & M.Y.). B.G.-P. is a postdoctoral fellow of the George E. Hewitt Foundation for Medical Research. We thank Kelly Dryden, Owen Pornillos, Michael Summers and Elizabeth Wright for help with figure preparation, Volker Vogt, Peter Prevelige, Hans-Georg Kräusslich, and Richard Kingston for helpful discussions and communicating unpublished data; and Owen Pornillos, Volker Vogt, Hans-Georg Kräusslich and Alan Rein for critical reading of the manuscript.
Abbreviations
- Env
envelope protein
- CACTD
C-terminal domain of the CA protein
- CANTD
N-terminal domain of the CA protein
- CA-I
12-residue peptide that inhibits CA assembly
- CAP-1
N-(3-chloro-4-methylphenyl)-N′-{2-[({5-[(dimethylamino)-methyl]-2-furyl}-methyl)-sulfanyl]ethyl}-urea), an inhibitor of CA assembly
- CypA
cyclophilin A
- Env
membrane envelope glycoprotein
- Gag
the Gag polyprotein is the major structural protein of retroviruses
- HIV
human immunodeficiency virus
- MHR
major homology region
- MLV
murine leukemia virus
- M-PMV
Mason-Pfizer monkey virus
- N-MLV
N-tropic murine leukemia virus
- PA-457
3-O-(3′,3′-dimethylsuccinyl)betulinic acid, an inhibitor of capsid assembly
- PR
retroviral protease
- RING
really interesting new gene, a ubiquitin E3 ligase domain
- RSV
Rous sarcoma virus
- SP1
spacer peptide 1
- SP2
spacer peptide 2
- SPRY
conserved domain from the SPL A kinase and ryanodine receptor
- TRIM5α
tripartite motif protein 5α
- TRIM-Cyp
tripartite motif protein-cyclophilin A fusion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of particular interest
- 1.Kräusslich HG. Morphogenesis and Maturation of Retroviruses. Springer; 1996. [Google Scholar]
- 2.Wills JW, Craven RC. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Freed EO. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 4.Göttlinger HG. The HIV-1 assembly machine. AIDS. 2001;15 5:S13–20. doi: 10.1097/00002030-200100005-00003. [DOI] [PubMed] [Google Scholar]
- 5.Adamson CS, Jones IM. The molecular basis of HIV capsid assembly – five years of progress. Rev Med Virol. 2004;14:107–121. doi: 10.1002/rmv.418. [DOI] [PubMed] [Google Scholar]
- 6.Morikawa Y. HIV capsid assembly. Curr HIV Res. 2003;1:1–14. doi: 10.2174/1570162033352084. [DOI] [PubMed] [Google Scholar]
- 7.Vogt V. Retroviral Virions and Genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 8.Frankel AD, Young JA. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 10.Turner BG, Summers MF. Structural biology of HIV. J Mol Biol. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 11.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 12.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Kräusslich HG. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci USA. 2001;98:10875–10879. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein KC, Reed JC, Lingappa JR. Intracellular destinies: degradation, targeting, assembly, and endocytosis of HIV Gag. AIDS Rev. 2007;9:150–161. [PubMed] [Google Scholar]
- 16.Resh MD. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 2005;7:84–91. [PubMed] [Google Scholar]
- 17.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 18.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 19.Wilk T, Gross I, Gowen BE, Rutten T, de Haas F, Welker R, Kräusslich HG, Boulanger P, Fuller SD. Organization of immature human immunodeficiency virus type 1. J Virol. 2001;75:759–771. doi: 10.1128/JVI.75.2.759-771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *20.Briggs JA, Simon MN, Gross I, Kräusslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]; A cryoEM and scanning transmission EM study of immature HIV-1 virions showing that the hexagonal Gag lattice exhibits an inter-ring spacing of ∼80 Å, estimating the subunit stoichiometry in immature and mature capsids, and showing that not all available CA subunits are used to assemble the capsid.
- 21.Fuller SD, Wilk T, Gowen BE, Kräusslich HG, Vogt VM. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 22.Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM. Cryo-electron microscopy reveals conserved and divergent features of Gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J Mol Biol. 2006;355:157–168. doi: 10.1016/j.jmb.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Yeager M, Wilson-Kubalek EM, Weiner SG, Brown PO, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the three-dimensional structure of individual immature virions revealing patches of hexagonal Gag arrays composed of ordered CA and SP1 domains. Real space averaging of well-defined unit cells yielded a model for the immature lattice.
- 25.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 27.Göttlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that PI(4,5)P2 plays a key role in targeting Gag to the plasma membrane in vivo.
- 29.Spearman P, Wang JJ, Vander Heyden N, Ratner L. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 1994;68:3232–3242. doi: 10.1128/jvi.68.5.3232-3242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between HIV-1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag-membrane binding. J Virol. 2007 doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt VM. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J Virol. 2007;81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erdie CR, Wills JW. Myristylation of Rous sarcoma virus Gag protein does not prevent replication in avian cells. J Virol. 1990;64:5204–5208. doi: 10.1128/jvi.64.10.5204-5208.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalton AK, Murray PS, Murray D, Vogt VM. Biochemical characterization of Rous sarcoma virus MA protein interaction with membranes. J Virol. 2005;79:6227–6238. doi: 10.1128/JVI.79.10.6227-6238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 36.Paillart JC, Göttlinger HG. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W, Resh MD. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the NMR structure of myristoylated MA with PI(4,5)P2 and proposes an appealing mechanism for Gag targeting to enriched membranes. The study reveals that PI(4,5)P2 not only anchors Gag to enriched membranes, through MA binding, but that it also can trigger myristate exposure.
- 41.Ono A, Demirov D, Freed EO. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J Virol. 2000;74:5142–5150. doi: 10.1128/jvi.74.11.5142-5150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Dou J, Ding L, Spearman P. Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. J Virol. 2007;81:12899–12910. doi: 10.1128/JVI.01280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Datta SA, Zhao Z, Clark PK, Tarasov S, Alexandratos JN, Campbell SJ, Kvaratskhelia M, Lebowitz J, Rein A. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J Mol Biol. 2007;365:799–811. doi: 10.1016/j.jmb.2006.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that HIV-1 Gag exhibits a monomer-dimer equilibrium in solution and that addition of IP6 converts this to a monomer-trimer equilibrium.
- *44.Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. Conformation of the HIV-1 Gag protein in solution. J Mol Biol. 2007;365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]; Experiments in this paper support a model in which monomeric Gag subunits fold back into an “autoinhibited” conformation in which the MA and NC domains are in close contact (companion to reference 43).
- 45.Reil H, Bukovsky AA, Gelderblom HR, Göttlinger HG. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kräusslich HG, Facke M, Heuser AM, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Göttlinger HG. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mammano F, Ohagen A, Hoglund S, Göttlinger HG. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Accola MA, Strack B, Göttlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borsetti A, Ohagen A, Göttlinger HG. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 54.Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. Implications for viral capsid assembly from crystal structures of HIV-1 Gag1-278 and CAN133-278. Biochemistry. 2006;45:11257–11266. doi: 10.1021/bi060927x. [DOI] [PubMed] [Google Scholar]
- 55.Rao Z, Belyaev AS, Fry E, Roy P, Jones IM, Stuart DI. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 56.Alfadhli A, Huseby D, Kapit E, Colman D, Barklis E. Human immunodeficiency virus type 1 matrix protein assembles on membranes as a hexamer. J Virol. 2007;81:1472–1478. doi: 10.1128/JVI.02122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nandhagopal N, Simpson AA, Johnson MC, Francisco AB, Schatz GW, Rossmann MG, Vogt VM. Dimeric Rous sarcoma virus capsid protein structure relevant to immature Gag assembly. J Mol Biol. 2004;335:275–282. doi: 10.1016/j.jmb.2003.10.034. [DOI] [PubMed] [Google Scholar]
- **58.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE., Jr Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2004;11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]; Using hydrogen/deuterium exchange, the intersubunit interactions involved in assembly and maturation were characterized.
- 59.Auerbach MR, Brown KR, Singh IR. Mutational analysis of the N-terminal domain of Moloney murine leukemia virus capsid protein. J Virol. 2007;81:12337–12347. doi: 10.1128/JVI.01286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortuza GB, Dodding MP, Goldstone DC, Haire LF, Stoye JP, Taylor IA. Structure of B-MLV capsid amino terminal domain reveals key features of viral tropism, Gag assembly and core formation. J Mol Biol. 2008;376:1493–1508. doi: 10.1016/j.jmb.2007.12.043. [DOI] [PubMed] [Google Scholar]
- **61.Mortuza GB, Haire LF, Stevens A, Smerdon SJ, Stoye JP, Taylor IA. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]; This study reports the first high-resolution structure of a hexameric retroviral CANTD domain.
- 62.Huseby D, Barklis RL, Alfadhli A, Barklis E. Assembly of human immunodeficiency virus precursor Gag proteins. J Biol Chem. 2005;280:17664–17670. doi: 10.1074/jbc.M412325200. [DOI] [PubMed] [Google Scholar]
- 63.Mayo K, Huseby D, McDermott J, Arvidson B, Finlay L, Barklis E. Retrovirus capsid protein assembly arrangements. J Mol Biol. 2003;325:225–237. doi: 10.1016/s0022-2836(02)01176-2. [DOI] [PubMed] [Google Scholar]
- 64.Mayo K, McDermott J, Barklis E. Hexagonal organization of Moloney murine leukemia virus capsid proteins. Virology. 2002;298:30–38. doi: 10.1006/viro.2002.1452. [DOI] [PubMed] [Google Scholar]
- 65.Nermut MV, Bron P, Thomas D, Rumlova M, Ruml T, Hunter E. Molecular organization of Mason-Pfizer monkey virus capsids assembled from Gag polyprotein in Escherichia coli. J Virol. 2002;76:4321–4330. doi: 10.1128/JVI.76.9.4321-4330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reicin AS, Ohagen A, Yin L, Hoglund S, Goff SP. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 69.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 70.Strambio-de-Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Craven RC, Leure-duPree AE, Weldon RA, Jr, Wills JW. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **72.Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci USA. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the structure of a “domain swapped” HIV-1 CACTD dimer and proposes that the dimer helps stabilize the immature Gag lattice.
- 73.Accola MA, Hoglund S, Göttlinger HG. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Newman JL, Butcher EW, Patel DT, Mikhaylenko Y, Summers MF. Flexibility in the p2 domain of the HIV-1 Gag polyprotein. Protein Sci. 2004;13:2101–2107. doi: 10.1110/ps.04614804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang C, Hu J, Russell RS, Roldan A, Kleiman L, Wainberg MA. Characterization of a putative α-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J Virol. 2002;76:11729–11737. doi: 10.1128/JVI.76.22.11729-11737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morellet N, Druillennec S, Lenoir C, Bouaziz S, Roques BP. Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci. 2005;14:375–386. doi: 10.1110/ps.041087605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheslock SR, Poon DT, Fu W, Rhodes TD, Henderson LE, Nagashima K, McGrath CF, Hu WS. Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination. J Virol. 2003;77:7058–7066. doi: 10.1128/JVI.77.12.7058-7066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bowzard JB, Bennett RP, Krishna NK, Ernst SM, Rein A, Wills JW. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sandefur S, Smith RM, Varthakavi V, Spearman P. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J Virol. 2000;74:7238–7249. doi: 10.1128/jvi.74.16.7238-7249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma YM, Vogt VM. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J Virol. 2003;76:52–60. doi: 10.1128/JVI.78.1.52-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D'Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 85.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Ψ-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 86.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- **87.D'Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]; The NMR structure of NC in complex with a 101-nucleotide segment of the MLV Ψ site elucidates an elegant mechanism for genome encapsidation in MLV.
- 88.Zhou J, Bean RL, Vogt VM, Summers M. Solution structure of the Rous sarcoma virus nucleocapsid protein:μΨ RNA packaging signal complex. J Mol Biol. 2007;365:453–467. doi: 10.1016/j.jmb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 90.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 91.Briggs JA, Wilk T, Welker R, Kräusslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *92.Briggs JA, Grunewald K, Glass B, Forster F, Kräusslich HG, Fuller SD. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- *93.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *94.Butan C, Winkler DC, Heymann JB, Craven RC, Steven AC. RSV capsid polymorphism correlates with polymerization efficiency and envelope glycoprotein content: implications that nucleation controls morphogenesis. J Mol Biol. 2008;376:1168–1181. doi: 10.1016/j.jmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 92-94 report independent electron cryotomographic studies of mature HIV-1 [92-93] and RSV [94] particles. These studies demonstrate the large structural variation in mature capsid morphology and propose mechanistic models for capsid formation.
- 95.Jin Z, Jin L, Peterson DL, Lawson CL. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 96.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Kräusslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mayo K, Vana ML, McDermott J, Huseby D, Leis J, Barklis E. Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J Mol Biol. 2002;316:667–678. doi: 10.1006/jmbi.2001.5354. [DOI] [PubMed] [Google Scholar]
- 98.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ehrlich LS, Agresta BE, Carter CA. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gross I, Hohenberg H, Kräusslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 101.Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **102.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]; Using electron crystallography, this study describes the structure of hexameric assemblies of full-length HIV-1 CA and characterizes three important protein-protein assembly interfaces required for capsid formation.
- 103.Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE., Jr Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J Mol Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 104.Bowzard JB, Wills JW, Craven RC. Second-site suppressors of Rous sarcoma virus CA mutations: evidence for interdomain interactions. J Virol. 2001;75:6850–6856. doi: 10.1128/JVI.75.15.6850-6856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 107.Gross I, Hohenberg H, Huckhagel C, Kräusslich HG. N-Terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 109.Kingston RL, Vogt VM. Domain swapping and retroviral assembly. Mol Cell. 2005;17:166–167. doi: 10.1016/j.molcel.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 110.Alcaraz LA, del Alamo M, Barrera FN, Mateu MG, Neira JL. Flexibility in HIV-1 assembly subunits: solution structure of the monomeric C-terminal domain of the capsid protein. Biophys J. 2007;93:1264–1276. doi: 10.1529/biophysj.106.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, Johnson JE, Tsuruta H, Steven AC. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100:253–263. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- 112.Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- 113.Fujioka T, Kashiwada Y, Kilkuskie RE, Cosentino LM, Ballas LM, Jiang JB, Janzen WP, Chen IS, Lee KH. Anti-AIDS agents, 11. Betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. J Nat Prod. 1994;57:243–247. doi: 10.1021/np50104a008. [DOI] [PubMed] [Google Scholar]
- 114.Kashiwada Y, Hashimoto F, Cosentino LM, Chen CH, Garrett PE, Lee KH. Betulinic acid and dihydrobetulinic acid derivatives as potent anti-HIV agents. J Med Chem. 1996;39:1016–1017. doi: 10.1021/jm950922q. [DOI] [PubMed] [Google Scholar]
- 115.Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, et al. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci USA. 2003;100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou J, Huang L, Hachey DL, Chen CH, Aiken C. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J Biol Chem. 2005;280:42149–42155. doi: 10.1074/jbc.M508951200. [DOI] [PubMed] [Google Scholar]
- 117.Zhou J, Chen CH, Aiken C. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. J Virol. 2006;80:12095–12101. doi: 10.1128/JVI.01626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li F, Zoumplis D, Matallana C, Kilgore NR, Reddick M, Yunus AS, Adamson CS, Salzwedel K, Martin DE, Allaway GP, et al. Determinants of activity of the HIV-1 maturation inhibitor PA-457. Virology. 2006;356:217–224. doi: 10.1016/j.virol.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 119.Zhou J, Yuan X, Dismuke D, Forshey BM, Lundquist C, Lee KH, Aiken C, Chen CH. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol. 2004;78:922–929. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Sun Y, Huang M, Summers MF. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol. 2003;327:1013–1020. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- *121.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373:355–366. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]; This papers reports the structure of HIV-1 CANTD in complex with CAP-1 using a combination of NMR spectroscopy and X-ray crystallography. The structure reveals that the CANTD changes conformation to create a deep binding pocket for CAP-1.
- *122.Sticht J, Humbert M, Findlow S, Bodem J, Muller B, Dietrich U, Werner J, Kräusslich HG. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 2005;12:671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- *123.Ternois F, Sticht J, Duquerroy S, Kräusslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]; References 122 and 123 describe the identification, biochemical behavior, and structural characterization of CA-I, a peptide inhibitor of HIV-1 assembly. The works demonstrate that CA-I binds to the CACTD, inhibits mature and immature capsid assembly, but does not affect dimerization.
- 124.Dvorin JD, Malim MH. Intracellular trafficking of HIV-1 cores: journey to the center of the cell. Curr Top Microbiol Immunol. 2003;281:179–208. doi: 10.1007/978-3-642-19012-4_5. [DOI] [PubMed] [Google Scholar]
- 125.Lehmann-Che J, Saib A. Early stages of HIV replication: how to hijack cellular functions for a successful infection. AIDS Rev. 2004;6:199–207. [PubMed] [Google Scholar]
- 126.Gomez C, Hope TJ. The ins and outs of HIV replication. Cell Microbiol. 2005;7:621–626. doi: 10.1111/j.1462-5822.2005.00516.x. [DOI] [PubMed] [Google Scholar]
- 127.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yamashita M, Emerman M. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 2005;1:e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80:3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- **133.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]; In this study, TRIM5α was identified as the restriction factor that blocks HIV-1 infection in Old world monkeys.
- **134.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]; This paper identifies TRIM-Cyp as the restriction factor for HIV-1 infection in owl monkeys.
- 135.Sokolskaja E, Luban J. Cyclophilin, TRIM5, and innate immunity to HIV-1. Curr Opin Microbiol. 2006;9:404–408. doi: 10.1016/j.mib.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 136.Perez O, Hope TJ. Cellular restriction factors affecting the early stages of HIV replication. Curr HIV/AIDS Rep. 2006;3:20–25. doi: 10.1007/s11904-006-0004-3. [DOI] [PubMed] [Google Scholar]
- 137.Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80:6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 142.Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 143.Howard BR, Vajdos FF, Li S, Sundquist WI, Hill CP. Structural insights into the catalytic mechanism of cyclophilin A. Nat Struct Biol. 2003;10:475–481. doi: 10.1038/nsb927. [DOI] [PubMed] [Google Scholar]
- 144.Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc Natl Acad Sci USA. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 146.Yap MW, Dodding MP, Stoye JP. Trim-cyclophilin A fusion proteins can restrict human immunodeficiency virus type 1 infection at two distinct phases in the viral life cycle. J Virol. 2006;80:4061–4067. doi: 10.1128/JVI.80.8.4061-4067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol. 2007;81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


