Abstract
Self tolerance loss is fundamental to autoimmunity. While understanding of immune regulation is expanding rapidly, the mechanisms causing loss of tolerance in most autoimmune diseases remain elusive. Autoimmunity is believed to develop when genetically predisposed individuals encounter environmental agents that trigger the disease. Recent advances in the genetic and environmental contributions to autoimmunity suggest that interactions between genetic elements and epigenetic changes caused by environmental agents may be responsible for inducing autoimmune disease. Genetic loci predisposing to autoimmunity are being identified through multi-center consortiums, and the number of validated genes is growing rapidly. Recent reports also indicate that the environment can contribute to autoimmunity by modifying gene expression through epigenetic mechanisms. This article will review current understanding of the genetics and epigenetics of lupus, rheumatoid arthritis, multiple sclerosis and type 1 diabetes, using systemic lupus erythematosus as the primary example. Other autoimmune diseases may have a similar foundation.
Keywords: Epigenetics, Genetics, Lupus, Multiple Sclerosis, Rheumatoid Arthritis
1. Introduction
Self tolerance is essential for normal immune function, and loss of self tolerance can result in autoimmunity. Why self tolerance breaks down is incompletely understood. Susceptibility to auto-immunity is associated with multiple risk factors. A generally higher disease concordance rate in monozygotic relative to dizygotic twins or other family members indicates a genetic contribution for some autoimmune diseases [1]. However, autoimmune disease concordance in identical twins is often incomplete, indicating a requirement for additional factors, presumably from the environment [2]. The nature of the environmental component is poorly understood.
Recent evidence indicates that environmentally-induced epigenetic changes, and in particular altered patterns of DNA methylation, contribute to the environment–host interaction in some forms of autoimmunity. A failure to maintain epigenetic homeostasis, due to environmental influences, can lead to aberrant gene expression in specific cells causing loss of tolerance, and the modified cells then contribute to the development of autoimmunity in genetically predisposed individuals [3]. Thus a combination of genetic and epigenetic mechanisms appears to be important in understanding causation of some autoimmune diseases.
Many autoimmune diseases occur more frequently in women, and the reason for this female preponderance is also incompletely understood. Functional studies suggest a role for female sex hormones [4,5], with higher Th1-mediated immune responses [6] and altered T cell homing [7] as possible contributors to the female predominance in autoimmune diseases. Others have suggested that women have a genetic predisposition to autoimmunity due to their second X chromosome [8,9]. Since one X chromosome is silenced by epigenetic mechanisms in women, it is possible that epigenetic mechanisms also contribute to the female predisposition to autoimmunity through effects on the inactive X.
This article reviews the current status of genetic contributions to human lupus, rheumatoid arthritis, multiple sclerosis and type 1 diabetes, and how epigenetic mechanisms may contribute to the development of autoimmunity in genetically predisposed hosts. Systemic lupus erythematosus (SLE) is used as an example where some predisposing genes are known, cells affected by epigenetic alterations have been identified, and genetic/epigenetic interactions are reported. Evidence supporting a role for epigenetic mechanisms in the female predisposition to lupus is also discussed. The status of epigenetic research in rheumatoid arthritis (RA), multiple sclerosis (MS) and type 1 diabetes (T1DM) is also summarized, with the goal of stimulating further study in these areas.
2. Genetics and epigenetics
2.1. Genetics
Genetic polymorphisms are heritable alterations in the DNA sequence. Genetic polymorphisms contribute to phenotypic variation, and sometimes to disease susceptibility, through effects on gene expression and function. Identifying predisposing genetic polymorphisms in rheumatic and other autoimmune diseases provides clues to understand the pathogenic mechanisms involved. Recent advances in gene expression analyses, high-throughput single nucleotide polymorphism (SNP) genotyping, and association studies have identified genetic loci or genes that dictate immune abnormalities in autoimmune diseases. Genome wide studies have already identified genes predisposing to human SLE, RA, MS and T1DM (vide infra). Further, spontaneous and genetically engineered animal models have been used to characterize the function of some susceptibility genes in autoimmune models. However, as noted above, twin studies demonstrate that genes alone are often insufficient to cause autoimmunity, and other factors are required. Some of these are epigenetic.
2.2. Epigenetics
In contrast to genetic alterations, an epigenetic change is defined as a heritable change in gene expression that does not involve a change in the DNA sequence. Epigenetic mechanisms play an essential role in eukaryotic gene regulation by modifying chromatin structure, which in turn modulates gene expression. How epigenetic mechanisms regulate gene expression is perhaps best explained from an evolutionary standpoint.
Prokaryotic organisms have a limited number of genes, comparatively small amounts of DNA, and no nucleus. Prokaryotic gene transcription is primarily regulated by transcription factors binding to promoter elements in the DNA. In contrast, eukaryotes have multiple cell types, more genes, and correspondingly more DNA. Eukaryotes also have nuclei, containing the DNA packaged as chromatin. Chromatin serves in part to package the DNA into the nucleus, and in part to regulate gene expression [10].
The basic chromatin subunit is the nucleosome, consisting of two turns of DNA wrapped around a histone octamer made from 2 molecules each of histones H2A, H2B, H3 and H4. The nucleosomes are then arranged into higher order structures to form chromatin fibers. By this mechanism the 2–3 m of human DNA is packaged into the nucleus of each cell. Chromatin in its native form is tightly compacted and inaccessible to transcription factors and the transcription initiation machinery. However, histone “tails” protrude from the nucleosome, and are covalently modified by acetylation, methylation, phosphorylation, ubiquitination, and sumoylation. These modifications serve as signals, referred to as the “histone code”, that initiate a number of processes including the localized remodeling of chromatin from a compact, transcriptionally silent configuration to a more open structure accessible to the transcription initiation machinery. One example of this is histone acetylation. N-acetylation of Lys in histone tails prevents the ε-amino group from binding DNA, promoting an open, transcriptionally active chromatin configuration, while deacetylation of Lys permits DNA binding and transcriptional silencing. Other histone modifications are involved in regulating chromatin structure and gene expression as well [11]. Eukaryotic cells use chromatin structure not only as a mechanism to package DNA into the nucleus, but also as a mechanism to prevent expression of genes not essential or detrimental to cellular function, but for which the cell expresses transcription factors which could bind to their recognition elements within the genes, referred to as “transcriptional noise” [10].
Histone acetylation reflects a balance of the histone acetyltransferases and the histone deacetylases, and is thus sensitive to environmental influences that modify these reactions, potentially altering gene expression. For example, acetyl-CoA is the acetyl donor for the histone acetylation process, and acetyl-CoA pools can be influenced by lipid and protein metabolism, and pyruvate dehydrogenase reactions. Similarly, the histone deacetylase inhibitor sulforaphane (SFN), found in cruciferous vegetables such as broccoli and Brussels sprouts, has been shown to promote histone H3 and H4 acetylation in peripheral blood mononuclear cells [12]. The other histone modifications are similarly sensitive to the environment, creating an inherent instability in this mechanism of gene regulation.
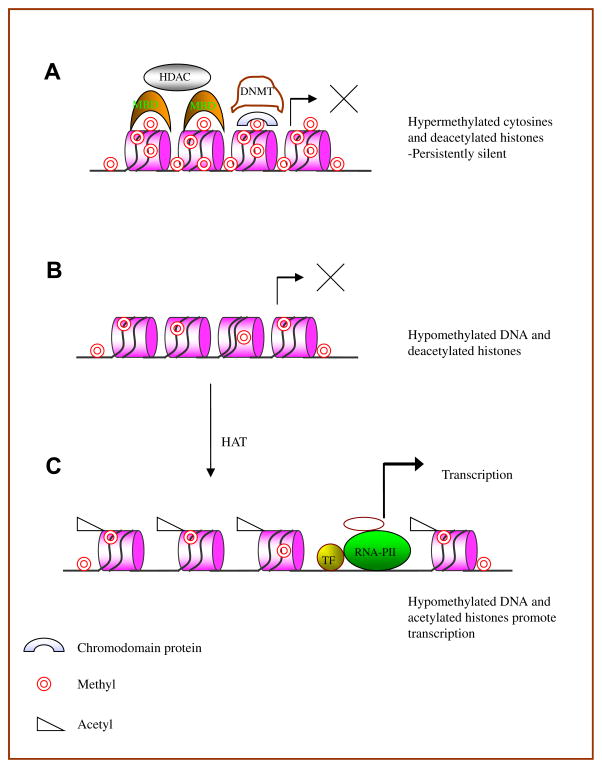
Vertebrates, with quantitatively more DNA than the lower eukaryotes, use DNA methylation to provide a more stable epigenetic mark for gene suppression [10]. DNA methylation in vertebrates refers to the methylation of deoxycytosine (dC) bases uniquely in CG pairs, forming deoxymethylcytosine (dmC). Methylcytosine binding proteins such as MBD1, MBD2 and MeCP2 bind dmC then attract and tether chromatin inactivation complexes including the histone deacetylases, promoting localized chromatin condensation into a transcriptionally inactive configuration [13]. The effects of DNA methylation and histone acetylation on chromatin structure are illustrated in Fig. 1.
Fig. 1.
Role of DNA methylation and histone modifications in gene regulation. A. Methylation of cytosines in CpG pairs recruits proteins containing a methyl-CpG-binding domain (MBD) such as MeCP2. Once bound, the MDB’s form a complex with histone deacetylases (HDAC) or directly block transcription factor binding. In addition, methylated histone tails may recruit DNMTs through chromodomain proteins, to methylate DNA for long term gene silencing. B. MBD cannot bind hypomethylated DNA. Thus the recruitment of HDAC is impaired. C. Acetylation of positively charged lysine amino groups in histones by histone acetyltransferases (HAT) neutralizes the charge and disrupts binding to the negatively charged phosphates in DNA. The “relaxed” DNA formed facilitates binding of transcription factors and RNA polymerase II promoting active transcription.
DNA methylation and histone methylation may be functionally linked. Methylated H3-K9 can bind chromodomain-containing proteins such as HP1 and recruit DNA methyltransferases to methylate adjacent CpG sequences and suppress gene expression [14]. However, methylation of H3K4 and arginine residues on H3 and H4 can result in transcriptional activation.
DNA methylation patterns for any given cell type are established during development by the de novo DNA methyltransferases Dnmt3a and Dnmt3b, then replicated during mitosis by the maintenance of methyltransferase Dnmt1. Dnmt1 binds proliferating cell nuclear antigen (PCNA) in the replication fork, and catalyzes the transfer of the methyl group from S-adenosylmethionine (SAM) to dC bases in the newly synthesized DNA only if the parent strand is methylated at that position, thereby replicating methylation patterns. Importantly, the replication of DNA methylation patterns during mitosis is also sensitive to the environment, and exogenous agents that decrease SAM levels, or decrease Dnmt1 levels or enzymatic activity, will result in failure to replicate the patterns, and errors may accumulate over successive rounds of cell division. This can cause aberrant expression of those genes silenced by DNA methylation, and for which the necessary transcription factors are present in the cell. A list of exogenous agents reported to alter DNA methylation is shown in Table 1.
Table 1.
Exogenous agents associated with altered DNA methylation.
| Agent |
|---|
| Folic acid |
| Methionine |
| Choline |
| Hydralazine |
| U0126 |
| PD98059 |
| UV light |
| SP600125 |
| Procainamide |
| 5-azacytidine |
| Genistein |
| Lycopene |
3. Genetics and epigenetics in autoimmunity: lupus
3.1. Genetics of SLE
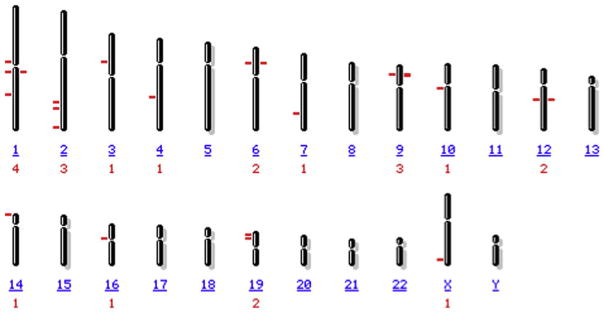
Genetic and epigenetic mechanisms interact to cause lupus-like diseases, and possibly other forms of autoimmunity. A genetic contribution to human lupus is well established. A significant difference in disease concordance between monozygotic twins (25–57%) and dizygotic twins (2–9%) supports a genetic basis [15]. Further evidence for a genetic contribution comes from studies of families in which lupus afflicts members at a rate greater than that of the general population [16]. Recent reviews of lupus genetics [17,18] report more than 20 loci containing lupus associated genes. These are listed in Table 2, and their chromosomal locations shown in Fig. 2. Genes identified include components of the complement activation pathway, IgG Fc receptors, and HLA region genes, supporting earlier work demonstrating important roles for complement, Fc receptors and the HLA genes in lupus autoimmunity. Interestingly, the genome wide searches have also shown a high degree of heterogeneity between populations.
Table 2.
Genes associated with systemic lupus erythematosus.
| Chromosome | Locus | Gene | Ref(s) |
|---|---|---|---|
| 1 | 1p13 | PTPN22 | [84,85] |
| 1q23 | FCGR2A | [20,84,86,87] | |
| 1q23 | FCGR3A | [20,84,88] | |
| 1q32 | IL10 | [22,89,90] | |
| 1p36 | C1Q | [18] | |
| 2 | 2q32 | STAT4 | [38,39,84] |
| 2q33 | CTLA4 | [34,35,91–93] | |
| 2q37 | PDCD1 | [33,94,95] | |
| 3 | 3p14 | PXK | [84] |
| 4 | 4q26 | IL21 | [96] |
| 6 | 6p21 | C2 | [27,28] |
| 6p21 | C4 | [27,29,97] | |
| 6p21 | TNFA | [22,25,98] | |
| 6p21 | TNFB | [26,99,100] | |
| 7 | 7q32 | IRF5 | [101,102] |
| 9 | 9p22 | IFNA | [103] |
| 9p21 | IFNB | [18] | |
| 10 | 10q11 | MBL | [104] |
| 12 | 12q14 | IFNG | [105] |
| 16 | 16p11 | ITGAM | [84] |
| 19 | 19cen-q13 | MAN2B1 | [104,106] |
| 19p13 | C3 | ||
| X | Xq28 | MECP2 | [107] |
Fig. 2.
Chromosomal location of genes predisposing to SLE. The number or letter beneath each chromosome identifies the chromosome, and the number below that shows the number of lupus associated sites on the chromosome. The locations of lupus predisposing genes are indicated by the lines.
Chromosome 1 contains some of the loci most consistently identified in SLE. The linkage interval 1q23 encodes Fcγ receptors FCGR2A and FCGR3A. FCGRs have different affinities for IgG and its subclasses. The Arg variant at amino acid position 131 of FCGR2A (R131) diminishes binding to IgG2. Similarly, a phenylalanine substitution in position 158 (F158) in FCGR3A reduces the IgG1-, IgG3-, and IgG4-binding capacity of the receptor. These variants may result in defective clearance of immune complexes from the circulation, contributing to their deposition in tissues such as the kidney and blood vessels [19,20]. Other disease associated genes on chromosome 1 include PTPN22, IL10, and C1Q. PTPN22 is involved in regulating T cell activation, while a PTPN22 R620W polymorphism is implicated in an increased risk of SLE [21]. IL 10 is an important cytokine with anti-inflammatory and stimulatory activities, and plays a critical role in the regulation of cellular and humoral immune responses. IL-10/TNFα interactions [22] and the IL10 promoter haplotype that produces higher levels of cytokine [23] are all associated with SLE.
The region in chromosome 6 encoding the major histocompatibility complex (MHC) also encodes components of complement pathway (C2, C4) as well as TNFα and TNFβ. TNFα is a multifunctional proinflammatory cytokine involved in regulating a wide spectrum of biological processes including cell proliferation, differentiation, and apoptosis, while TNFβ mediates a variety of inflammatory, immunostimulatory, and antiviral responses. Polymorphisms in these genes have been implicated in SLE susceptibility [24–26]. Deficiencies in complement pathway genes C2 [27,28], C4 [29], C1Q [18] and C3 [30] appear to cause SLE in some people. Polymorphisms in C2, C4A and C4B are in linkage disequilibrium with HLA-B and HLA-DR alleles [31] and may predispose to SLE. In addition, a significant association of the HLA-DRB1 gene with SLE has been established in Latin American populations [32].
Programmed cell death 1 gene (PDCD1), encoded on 2q37, is considered a strong candidate for SLE association. PDCD1 is upregulated in T cells following activation, and inhibits TCR signaling and T/B cell survival. An intronic PDCD SNP alters a binding site for the runt-related transcription factor (RUNX1), suggesting a mechanism through which the SNP may contribute to SLE development [33]. CTLA4, located on 2q33, is a negative costimulatory molecule that inhibits T cell activation, and may help to limit T cell responses under conditions of inflammation. Genetic variability in CTLA4 has been implicated in the development of several autoimmune diseases including SLE [34–36]. Locus 2q32 encodes the STAT4 transcription factor, essential for mediating responses to IL12 in lymphocytes, and regulates T helper cell differentiation. The risk allele of the SNP rs7582694 in STAT4 is associated with severe disease manifestations of systemic lupus erythematosus [37–39].
Thus, loci in chromosome 1, 2 and 6 encode several functionally significant genes associated with SLE pathogenesis. However, as shown in Fig. 2 and Table 2, the genetic predisposition of SLE is not limited to just these regions of the human genome. PTPN2 (protein tyrosine phosphatase, non-receptor type 2) located at 1p13 is considered to be the strongest common genetic risk factor for human autoimmunity outside the MHC. It encodes a lymphoid-specific phosphatase (Lyp), which inhibits T cell receptor signaling through Csk kinase. The X-linked gene methyl-CpG-binding protein 2 (MECP2), located on Xq28 and encoding a protein that represses transcription from methylated promoters, has also been associated with lupus. Polymorphisms in MECP2 may have relevance to the epigenetic DNA methylation changes found in lupus and discussed below.
3.2. Epigenetics of SLE
While genetic factors clearly influence lupus susceptibility, incomplete disease concordance between identical twins suggests a requirement for non-genetic factors, presumably from the environment [15,40]. Convincing evidence indicates that epigenetic mechanisms, and in particular impaired T cell DNA methylation, provide this additional factor.
Early work revealed that normal, antigen specific CD4 + T cells treated with DNA methylation inhibitors like 5-azacytidine will lose restriction for nominal antigen and respond to self class II MHC molecules presenting inappropriate antigens [41]. These epigenetically modified, autoreactive cells resemble those causing chronic graft-vs-host disease, which presents primarily as lupus in murine models [42]. Injecting demethylated murine CD4 + T cells into syngeneic hosts causes a lupus-like disease [43,44] supporting the association between autoreactive responsiveness to self class II MHC molecules and lupus-like autoimmunity.
The observation that a DNA demethylating drug can cause a lupus-like disease suggests that drugs which cause a lupus-like disease may inhibit DNA methylation. Procainamide and hydralazine, which cause ANA’s in most people and a lupus-like disease in a genetically predisposed subset [45], were subsequently found to inhibit DNA methylation [46], and murine T cells treated with these drugs caused lupus-like autoimmunity in syngeneic mice identical to that caused by 5-azacytidine treated T cells [7]. Procainamide is a competitive inhibitor of Dnmt1, the enzyme responsible for maintaining DNA methylation patterns in differentiated cells [47,48], while hydralazine prevents Dnmt1 upregulation during mitosis by blocking ERK pathway signaling at PKCδ [49]. These reports thus raise the possibility that other agents encountered in the environment may also cause a lupus-like disease in genetically predisposed individuals, either by inhibiting Dnmt1 activity or by decreasing Dnmt1 levels.
Patients with idiopathic lupus have changes in T cell signaling identical to those caused by hydralazine. Initial studies demonstrated that T cells from patients with active lupus have hypomethylated DNA, due to decreased Dnmt1 levels and activity [50,51]. Interestingly, the decrease in Dnmt1 levels is due to impaired ERK pathway signaling [50] caused by a block at PKCδ, also inhibited by hydralazine [49]. Inducing an ERK pathway defect selectively in T cells of adult transgenic mice is sufficient to decrease Dnmt1, demethylate DNA and cause anti-DNA antibodies, persuasively demonstrating that acquired T cell ERK pathway defects are sufficient to cause lupus-like autoimmunity, likely through DNA demethylation [52].
Persuasive evidence also demonstrates that CD4 + T cells from patients with active lupus demethylate the same DNA sequences and overexpress the same genes demethylated by 5-azacytidine in vitro, and that overexpression of the demethylated genes contributes to the development of lupus-like autoimmunity. 5-azacytidine causes T cell autoreactivity by demethylating sequences just upstream of the ITGAL promoter, encoding CD11a, leading to overexpression of LFA-1 (CD11a/CD18) [53]. During T cell activation by antigen presenting cells, LFA-1 surrounds the T cell receptor-MHC complex, providing stabilization to the interaction as well as costimulatory signals [54]. Overexpressing LFA-1 may overstabilize the lower affinity interaction between the TCR and self class II MHC molecules as well as increasing costimulatory signals, permitting activation of T cells by the class II MHC molecules without the appropriate antigen. Increasing T cell LFA-1 expression by transfection causes an identical MHC-specific autoreactivity in vitro, and injecting LFA-1 transfected CD4 + T cells into syngeneic mice causes lupus-like autoimmunity [55], demonstrating a causative role for LFA-1 overexpression in T cell autoreactivity and in lupus pathogenesis. Importantly, CD4 + T cells from lupus patients demethylate the same ITGAL regulatory sequences and overexpress LFA-1 on an autoreactive T cell subset [56], indicating that the same mechanism contributes to idiopathic lupus and the demethylation lupus model.
The 5-azacytidine T cell DNA demethylation model also predicted other T cell DNA methylation alterations that were subsequently found in lupus T cells. 5-azacytidine was shown to demethylate and cause overexpression of the cytotoxic molecule perforin and the B cell costimulatory molecule CD70 in CD4 + T cells, and CD4 + T cells from lupus patients were found to demethylate the same regulatory sequences and overexpress the same molecules that were affected by 5-azacytidine [57,58]. Perforin contributes to autoreactive macrophage killing by lupus T cells, generating a source of antigenic apoptotic nucleosomes [58], while CD70 overexpression contributes to B cell overstimulation and antibody overproduction [59]. Thus the DNA demethylation model can predict changes in T cell gene expression that contribute to lupus pathogenesis. These studies also demonstrate that demethylated CD4 + T cells are sufficient to cause autoimmunity, indicating that demethylated, autoreactive T cells are the cells responsible for breaking tolerance and activating lupus in genetically predisposed people.
3.3. Epigenetics and X chromosome reactivation in women with lupus
CD40L is another B cell costimulatory molecule overexpressed on lupus T cells and contributing to autoantibody production [60]. Interestingly though, CD40LG is on the X chromosome, so men have only one copy while women have 2, and the copy on the inactive X in women is silenced by mechanisms including DNA methylation. Studies similar to those described above demonstrated that 5-azacytidine caused overexpression of CD40L on CD4 + T cells from women but not men, and that the overexpression was associated with demethylation of CD40LG on the inactive X [61]. Further, women with active lupus demethylated and overexpressed CD40L, while men with active lupus did not overexpress CD40L [61]. Since CD40L overexpression contributes to the pathogenesis of lupus-like autoimmunity [62], demethylation of genes on the inactive X may predispose women to SLE.
3.4. Genetics and epigenetics in drug induced lupus
Drug induced lupus currently provides the best example of how an environmental agent that affects DNA methylation can cause autoimmunity in a genetically predisposed host. Studies summarized above demonstrate that the lupus-inducing drugs procainamide and hydralazine are DNA methylation inhibitors [46], that human CD4 + T cells treated with these drugs or the DNA methyltransferase inhibitor 5-azacytidine overexpress methylation sensitive genes and become autoreactive, and that murine CD4 + T cells treated with procainamide, hydralazine or 5-azacytidine cause a lupus-like disease in syngeneic mice [7]. These reports suggest that procainamide and hydralazine may cause a lupus-like disease in humans through similar epigenetic mechanisms.
Interestingly, while a majority of individuals will develop a positive ANA if they receive procainamide or hydralazine long enough, people those genetically metabolize hydralazine or procainamide quickly (rapid acetylators) take longer to develop ANA’s than those that metabolize these drugs slowly [45], demonstrating an epigenetic link between “environment” and genetics in the autoantibodies induced by these drugs. Similarly, HLA-DR4 and female sex are reported to predispose individuals to the development of symptomatic hydralazine-induced lupus [63]. It is reasonable to propose that genetic elements will also promote the development of lupus-like autoimmunity in people with hypomethylated DNA caused by other environmental agents affecting DNA methylation.
4. Genetics and epigenetics of rheumatoid arthritis
4.1. Genetics of RA
Rheumatoid arthritis (RA) is a systemic autoimmune disease affecting approximately 3% of the population worldwide, and characterized by chronic inflammation and destruction of the synovial joints leading to progressive joint damage and disability. Familial and twin studies suggest a >50% genetic contribution to RA, and a high concordance rate in monozygotic twins (12–30%). RA is also more prevalent in first-degree relatives [64]. A lower prevalence of RA in rural Africans than their counterparts who migrate to the towns suggests the importance of environmental factors in RA [65]. Genes known to be associated with RA are shown in Table 3. The contribution of human leukocyte antigen (HLA) genes at 6p21 shows the strongest linkage to RA. HLA-DRB1 allele variants are well characterized and their predisposition to RA is clear [66]. However, familial risk due to the HLA genes has been estimated to be only ~30%, suggesting that non-HLA genes may play a significant role in RA susceptibility. The PTPN2 1858T variant was found to be associated with RA, T1DM and SLE [67]. However, inconsistent results have been obtained in different populations for the association of CTLA + 49/G SNP to RA [68]. An SNP haplotype marked by rs7574865 of STAT4 is also associated with susceptibility to both RA and SLE.
Table 3.
Genes associated rheumatoid arthritis.
4.2. Epigenetics of RA
In contrast to lupus where T cell DNA demethylation can be shown to cause autoimmunity, less is known about epigenetics in RA, and the abnormal cell(s) responsible for initiating RA are uncertain. Current models propose that the initiating event is a T cell response to an agent acquired from the environment, but the nature of the environmental trigger is uncertain [69]. Aberrant expression of genes in RA synovial fibroblasts (RASF) lacking specific genetic mutations suggests that epigenetic mechanisms may be involved. Activated RASF demonstrate DNA demethylation and increased expression of normally methylated repetitive DNA elements such as the retrotransposon LINE-1, hypermethylation of regulatory and death-associated proteins, and histone deacetylase induced changes in the posttranslational activation of transcriptional activators and nuclear proteins [70]. T cell DNA is demethylated in RA [51], and may result in the generation of autoreactive T and/or B cell clones in RA as it does in lupus. The CD21 promoter is also demethylated in RA PBMC and synovial fluid cells [71]. Further evidence for an interaction of environmental and genetic factors in RA pathogenesis comes from studies examining cigarette smoking and susceptibility genes in RA patients. The presence of risk factor PTPN22 R620W interacts with heavy cigarette smoking in a multiplicative manner in RA [72]. How and if smoking influences epigenetic mechanisms is unknown.
5. Genetics and epigenetics of multiple sclerosis
5.1. Genetics of MS
Multiple sclerosis is a chronic inflammatory neurodegenerative autoimmune disease also believed to be caused by genetic and environmental factors. A concordance rate of 30% in monozygotic twins compared to 5% for dizygotic indicates a significant genetic component in disease susceptibility. The HLA gene cluster at chromosome 6p21.3 has been established by both candidate gene association and whole genome linkage analysis [73]. HLA-DR2 phenotype, HLA-DRB1*1501, DQAl*0102, and DQB1*0602, are strongly associated with MS susceptibility. HLA-DRB1*1501 is most consistently associated with MS in genome wide linkage studies. T cells recognize antigen presented by molecules of the MHC, and polymorphic HLA-DRB1 residues can affect the shape and charge at the peptide-binding site of the HLA molecule. HLA factors account for 20–60% of the MS genetic risk. Non-MHC loci also associated with MS (Table 4) including cytokines and their receptors which may drive the inflammatory process in an MS plaque. Increased levels of the proinflammatory cytokines IL1, IL4, IL6 and TNFα, and decreased levels of anti-inflammatory cytokines IL10 and IL4 have been reported to correlate with disease progression [74].
Table 4.
Non-MHC genes associated multiple sclerosis.
5.2. Epigenetics of MS
The discordance rate of 70% among monozygotic twins highlights the importance of environmental factors in disease pathogenesis. Studies of individuals with similar genetic backgrounds but living in different parts of the world have revealed significant differences in disease prevalence [75]. Sunlight exposure appears to have a protective role in MS, possibly mediated by vitamin D3 [76]. Epstein–Barr virus (EBV) infection has also been implicated in the inflammatory response in MS [73]. However, less is known about the epigenetics of MS. Myelin basic protein (MBP) citrullination can result in a loss of myelin stability in MS brains, and the enzyme peptide arginine deiminase 2 (PAD2) catalyzes the MBP citrullination. Increased citrullinated MBP in MS white matter may be due to hypomethylation of the PAD2 promoter and subsequent PAD2 overexpression [77].
6. Genetics and epigenetics of type 1 diabetes mellitus
6.1. Genetics of T1DM
T1DM is another example of an autoimmune disease with genetic and environmental components. A concordance rate of 30–50% in monozygotic compared to 5% in dizygotic twins demonstrates a significant genetic component in disease susceptibility. Common genetic elements appear to contribute to a strikingly higher prevalence of T1DM and MS in the population of Sardinia. Whole genome linkage scans have identified chromosome regions 6q26, 10q21.2, 20p12.3 and 22q11.22 as common to both autoimmune diseases in this population [78]. A comprehensive review reveals several other genetic susceptibility loci [79]. The major locus determining T1DM familial aggregation is the HLA region on chromosome 6p21, and accounts for about 50% of the familial clustering. HLA haplotypes DR4-DQ8, DR3-DQ2 are present in 90% of T1DM patients. DR15- DR6 has a protective association with T1DM [79]. Table 5 shows non-HLA loci associated with T1DM. The insulin gene (INS) locus at 11p15 is the second most important susceptibility locus. A region containing a variable number of tandem repeats (VNTR), located upstream of the insulin gene, is associated with T1DM. Variation in the size of the VNTR correlates with the thymic expression of insulin mRNA. The short form of the insulin VNTR is associated with type 1 diabetes whereas the long form is associated with decreased risk [80].
Table 5.
Non-MHC genes associated with type 1 diabetes.
6.2. Epigenetics of T1DM
Non-genetic factors appear to enhance the autoimmune process causing destruction of islet beta cells in T1DM. Environmental factors proposed include nutrition and viruses [81]. The most persuasive evidence for a viral etiology is provided by the congenital rubella syndrome [82]. However, no direct evidence for epigenetic changes associated with T1DM is available. Since nutrition provides the methyl donors (methionine, choline) and cofactors (folic acid, vitamin B12 and pyridoxal phosphate) essential for DNA and histone methylation [83], it is possible that epigenetic mechanisms contribute to T1DM as well.
7. Summary
Many of the poorly understood autoimmune diseases have a genetic and an environmental component. Understanding of autoimmunity genetics is developing rapidly, but how the environmental contributes to disease development is still largely unknown. Convincing evidence indicates that the environment modifies the immune system through epigenetic mechanisms to cause SLE, raising the possibility that epigenetics may contribute to the development of other forms of autoimmunity. Further study of epigenetic abnormalities in other diseases, such as RA, MS and T1DM, may clarify how the environment alters the immune system in these diseases, and suggest ways to ameliorate these conditions.
Acknowledgments
This work was supported by PHS grants AG25877, ES015214 and AR056370, and a Merit grant from the Dept. of Veterans Affairs. The authors thank Ms. Cindy Bourke for her expert secretarial assistance.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit:
References
- 1.Wandstrat A, Wakeland E. The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat Immunol. 2001;2:802–9. doi: 10.1038/ni0901-802. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon-Riquelme ME. Recent advances in the genetics of autoimmune diseases. Ann N Y Acad Sci. 2007;1110:1–9. doi: 10.1196/annals.1423.001. [DOI] [PubMed] [Google Scholar]
- 3.Strickland FM, Richardson BC. Epigenetics in human autoimmunity. Epigenetics in autoimmunity - DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity. 2008;41:278–86. doi: 10.1080/08916930802024616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed SA, Hissong BD, Verthelyi D, Donner K, Becker K, Karpuzoglu-Sahin E. Gender and risk of autoimmune diseases: possible role of estrogenic compounds. Environ Health Perspect. 1999;107(Suppl 5):681–6. doi: 10.1289/ehp.99107s5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rider V, Foster RT, Evans M, Suenaga R, Abdou NI. Gender differences in autoimmune diseases: estrogen increases calcineurin expression in systemic lupus erythematosus. Clin Immunol Immunopathol. 1998;89:171–80. doi: 10.1006/clin.1998.4604. [DOI] [PubMed] [Google Scholar]
- 6.Sinha S, Kaler LJ, Proctor TM, Teuscher C, Vandenbark AA, Offner H. IL-13-mediated gender difference in susceptibility to autoimmune encephalomyelitis. J Immunol. 2008;180:2679–85. doi: 10.4049/jimmunol.180.4.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yung R, Chang S, Hemati N, Johnson K, Richardson B. Mechanisms of drug-induced lupus. IV. Comparison of procainamide and hydralazine with analogs in vitro and in vivo. Arthritis Rheum. 1997;40:1436–43. doi: 10.1002/art.1780400811. [DOI] [PubMed] [Google Scholar]
- 8.Herrera BM, Cader MZ, Dyment DA, Bell JT, Deluca GC, Willer CJ, et al. Multiple sclerosis susceptibility and the X chromosome. Mult Scler. 2007;13:856–64. doi: 10.1177/1352458507076961. [DOI] [PubMed] [Google Scholar]
- 9.Yin X, Latif R, Tomer Y, Davies TF. Thyroid epigenetics: X chromosome inactivation in patients with autoimmune thyroid disease. Ann N Y Acad Sci. 2007;1110:193–200. doi: 10.1196/annals.1423.021. [DOI] [PubMed] [Google Scholar]
- 10.Bird AP. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 11.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 12.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–9. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 15.Jarvinen P, Aho K. Twin studies in rheumatic diseases. Semin Arthritis Rheum. 1994;24:19–28. doi: 10.1016/0049-0172(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 16.Sestak AL, Shaver TS, Moser KL, Neas BR, Harley JB. Familial aggregation of lupus and autoimmunity in an unusual multiplex pedigree. J Rheumatol. 1999;26:1495–9. [PubMed] [Google Scholar]
- 17.Sestak A, O’Neil KM. Familial lupus and antiphospholipid syndrome. Lupus. 2007;16:556–63. doi: 10.1177/0961203307078071. [DOI] [PubMed] [Google Scholar]
- 18.Sestak AL, Nath SK, Sawalha AH, Harley JB. Current status of lupus genetics. Arthritis Res Ther. 2007;9:210. doi: 10.1186/ar2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonsen A, Bengtsson AA, Sturfelt G, Truedsson L. Analysis of HLA DR, HLA DQ, C4A, FcgammaRIIa, FcgammaRIIIa, MBL, and IL-1Ra allelic variants in Caucasian systemic lupus erythematosus patients suggests an effect of the combined FcgammaRIIa R/R and IL-1Ra 2/2 genotypes on disease susceptibility. Arthritis Res Ther. 2004;6:R557–62. doi: 10.1186/ar1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karassa FB, Trikalinos TA, Ioannidis JP. The role of FcgammaRIIA and IIIA polymorphisms in autoimmune diseases. Biomed Pharmacother. 2004;58:286–91. doi: 10.1016/j.biopha.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75:504–7. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suarez A, Lopez P, Mozo L, Gutierrez C. Differential effect of IL10 and TNF{alpha} genotypes on determining susceptibility to discoid and systemic lupus erythematosus. Ann Rheum Dis. 2005;64:1605–10. doi: 10.1136/ard.2004.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosado S, Rua-Figueroa I, Vargas JA, Garcia-Laorden MI, Losada-Fernandez I, Martin-Donaire T, et al. Interleukin-10 promoter polymorphisms in patients with systemic lupus erythematosus from the Canary Islands. Int J Immunogenet. 2008;35:235–42. doi: 10.1111/j.1744-313X.2008.00762.x. [DOI] [PubMed] [Google Scholar]
- 24.Tarassi K, Carthy D, Papasteriades C, Boki K, Nikolopoulou N, Carcassi C, et al. HLA-TNF haplotype heterogeneity in Greek SLE patients. Clin Exp Rheumatol. 1998;16:66–8. [PubMed] [Google Scholar]
- 25.Sullivan KE, Suriano A, Dietzmann K, Lin J, Goldman D, Petri MA. The TNFalpha locus is altered in monocytes from patients with systemic lupus erythematosus. Clin Immunol. 2007;123:74–81. doi: 10.1016/j.clim.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HY, Lee SH, Yang HI, Park SH, Cho CS, Kim TG, et al. TNFB gene polymorphism in patients with systemic lupus erythematosus in Korean. Korean J Intern Med. 1995;10:130–6. doi: 10.3904/kjim.1995.10.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan KE, Jawad AF, Piliero LM, Kim N, Luan X, Goldman D, et al. Analysis of polymorphisms affecting immune complex handling in systemic lupus erythematosus. Rheumatology (Oxford) 2003;42:446–52. doi: 10.1093/rheumatology/keg157. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan KE, Petri MA, Schmeckpeper BJ, McLean RH, Winkelstein JA. Prevalence of a mutation causing C2 deficiency in systemic lupus erythematosus. J Rheumatol. 1994;21:1128–33. [PubMed] [Google Scholar]
- 29.Sullivan KE, Kim NA, Goldman D, Petri MA. C4A deficiency due to a 2 bp insertion is increased in patients with systemic lupus erythematosus. J Rheumatol. 1999;26:2144–7. [PubMed] [Google Scholar]
- 30.Miyagawa H, Yamai M, Sakaguchi D, Kiyohara C, Tsukamoto H, Kimoto Y, et al. Association of polymorphisms in complement component C3 gene with susceptibility to systemic lupus erythematosus. Rheumatology (Oxford) 2008;47:158–64. doi: 10.1093/rheumatology/kem321. [DOI] [PubMed] [Google Scholar]
- 31.Speirs C, Fielder AH, Chapel H, Davey NJ, Batchelor JR. Complement system protein C4 and susceptibility to hydralazine-induced systemic lupus erythematosus. Lancet. 1989;1:922–4. doi: 10.1016/s0140-6736(89)92506-3. [DOI] [PubMed] [Google Scholar]
- 32.Castano-Rodriguez N, Diaz-Gallo LM, Pineda-Tamayo R, Rojas-Villarraga A, Anaya JM. Meta-analysis of HLA-DRB1 and HLA-DQB1 polymorphisms in Latin American patients with systemic lupus erythematosus. Autoimmun Rev. 2008;7:322–30. doi: 10.1016/j.autrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet. 2002;32:666–9. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 34.Graham DS, Wong AK, McHugh NJ, Whittaker JC, Vyse TJ. Evidence for unique association signals in SLE at the CD28-CTLA4-ICOS locus in a family-based study. Hum Mol Genet. 2006;15:3195–205. doi: 10.1093/hmg/ddl395. [DOI] [PubMed] [Google Scholar]
- 35.Parks CG, Hudson LL, Cooper GS, Dooley MA, Treadwell EL, St Clair EW, et al. CTLA-4 gene polymorphisms and systemic lupus erythematosus in a population-based study of whites and African-Americans in the southeastern United States. Lupus. 2004;13:784–91. doi: 10.1191/0961203304lu1085oa. [DOI] [PubMed] [Google Scholar]
- 36.Barreto M, Santos E, Ferreira R, Fesel C, Fontes MF, Pereira C, et al. Evidence for CTLA4 as a susceptibility gene for systemic lupus erythematosus. Eur J Hum Genet. 2004;12:620–6. doi: 10.1038/sj.ejhg.5201214. [DOI] [PubMed] [Google Scholar]
- 37.Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet. 2008;4:e1000084. doi: 10.1371/journal.pgen.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigurdsson S, Nordmark G, Garnier S, Grundberg E, Kwan T, Nilsson O, et al. A common STAT4 risk haplotype for Systemic Lupus Erythematosus is over-expressed, correlates with anti-dsDNA production and shows additive effects with two IRF5 risk alleles. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi S, Ikari K, Kaneko H, Kochi Y, Yamamoto K, Shimane K, et al. Association of STAT4 with susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Japanese population. Arthritis Rheum. 2008;58:1940–6. doi: 10.1002/art.23494. [DOI] [PubMed] [Google Scholar]
- 40.Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–8. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 41.Richardson B. Effect of an inhibitor of DNA methylation on T cells. II.5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum Immunol. 1986;17:456–70. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- 42.Rolink AG, Gleichmann E. Allosuppressor- and allohelper-T cells in acute and chronic graft-vs.-host (GVH) disease. III. Different Lyt subsets of donor T cells induce different pathological syndromes. J Exp Med. 1983;158:546–58. doi: 10.1084/jem.158.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, et al. Treating activated CD4 + T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92:38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–35. [PubMed] [Google Scholar]
- 45.Yung RL, Richardson BC. Drug-induced lupus. Rheum Dis Clin North Am. 1994;20:61–86. [PubMed] [Google Scholar]
- 46.Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–200. [PubMed] [Google Scholar]
- 47.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280:40749–56. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheinbart LS, Johnson MA, Gross LA, Edelstein SR, Richardson BC. Procainamide inhibits DNA methyltransferase in a human T cell line. J Rheumatol. 1991;18:530–4. [PubMed] [Google Scholar]
- 49.Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–63. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 50.Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, McCune WJ, et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 2001;44:397–407. doi: 10.1002/1529-0131(200102)44:2<397::AID-ANR59>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 51.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–73. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 52.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and over-expression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–78. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alcolado JC, Laji K, Gill-Randall R. Maternal transmission of diabetes. Diabet Med. 2002;19:89–98. doi: 10.1046/j.1464-5491.2002.00675.x. [DOI] [PubMed] [Google Scholar]
- 54.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–9. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 55.Yung R, Powers D, Johnson K, Amento E, Carr D, Laing T, et al. Mechanisms of drug-induced lupus. II. T cells overexpressing lymphocyte function-associated antigen 1 become autoreactive and cause a lupuslike disease in syngeneic mice. J Clin Invest. 1996;97:2866–71. doi: 10.1172/JCI118743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1282–91. doi: 10.1002/art.10234. [DOI] [PubMed] [Google Scholar]
- 57.Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–9. doi: 10.4049/jimmunol.174.10.6212. [DOI] [PubMed] [Google Scholar]
- 58.Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172:3652–61. doi: 10.4049/jimmunol.172.6.3652. [DOI] [PubMed] [Google Scholar]
- 59.Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–60. doi: 10.1002/art.20255. [DOI] [PubMed] [Google Scholar]
- 60.Koshy M, Berger D, Crow MK. Increased expression of CD40 ligand on systemic lupus erythematosus lymphocytes. J Clin Invest. 1996;98:826–37. doi: 10.1172/JCI118855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–8. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 62.Crow MK, Kirou KA. Regulation of CD40 ligand expression in systemic lupus erythematosus. Curr Opin Rheumatol. 2001;13:361–9. doi: 10.1097/00002281-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 63.Batchelor JR, Welsh KI, Tinoco RM, Dollery CT, Hughes GR, Bernstein R, et al. Hydralazine-induced systemic lupus erythematosus: influence of HLA-DR and sex on susceptibility. Lancet. 1980;1:1107–9. doi: 10.1016/s0140-6736(80)91554-8. [DOI] [PubMed] [Google Scholar]
- 64.Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH, Caeiro F, Massardo L, Villa AR, et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–47. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 65.Wordsworth P, Bell J. Polygenic susceptibility in rheumatoid arthritis. Ann Rheum Dis. 1991;50:343–6. doi: 10.1136/ard.50.6.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orozco G, Rueda B, Martin J. Genetic basis of rheumatoid arthritis. Biomed Pharmacother. 2006;60:656–62. doi: 10.1016/j.biopha.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 67.Orozco G, Pascual-Salcedo D, Lopez-Nevot MA, Cobo T, Cabezon A, Martin-Mola E, et al. Auto-antibodies, HLA and PTPN22: susceptibility markers for rheumatoid arthritis. Rheumatology (Oxford) 2008;47:138–41. doi: 10.1093/rheumatology/kem343. [DOI] [PubMed] [Google Scholar]
- 68.Zhernakova A, Eerligh P, Barrera P, Wesoly JZ, Huizinga TW, Roep BO, et al. CTLA4 is differentially associated with autoimmune diseases in the Dutch population. Hum Genet. 2005;118:58–66. doi: 10.1007/s00439-005-0006-z. [DOI] [PubMed] [Google Scholar]
- 69.Firestein G. Rheumatoid synovitis and pannus. In: SA, Hochberg M, Smolen J, Weinblatt M, Weisman M, editors. Rheumatology. New York: Mosby; 2003. pp. 876–879. [Google Scholar]
- 70.Sanchez-Pernaute O, Ospelt C, Neidhart M, Gay S. Epigenetic clues to rheumatoid arthritis. J Autoimmun. 2008;30:12–20. doi: 10.1016/j.jaut.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 71.Schwab J, Illges H. Silencing of CD21 expression in synovial lymphocytes is independent of methylation of the CD21 promoter CpG island. Rheumatol Int. 2001;20:133–7. doi: 10.1007/s002960000090. [DOI] [PubMed] [Google Scholar]
- 72.Costenbader KH, Chang SC, De Vivo I, Plenge R, Karlson EW. Genetic polymorphisms in PTPN22, PADI-4, and CTLA-4 and risk for rheumatoid arthritis in two longitudinal cohort studies: evidence of gene-environment interactions with heavy cigarette smoking. Arthritis Res Ther. 2008;10:R52. doi: 10.1186/ar2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. 2008;9:516–26. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 74.Ramagopalan SV, Deluca GC, Degenhardt A, Ebers GC. The genetics of clinical outcome in multiple sclerosis. J Neuroimmunol. 2008 doi: 10.1016/j.jneuroim.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 75.Alonso A, Hernan MA. Temporal trends in the incidence of multiple sclerosis: a systematic review. Neurology. 2008;71:129–35. doi: 10.1212/01.wnl.0000316802.35974.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kragt JJ, Van Amerongen BM, Killestein J, Dijkstra CD, Uitdehaag BM, Polman CH, et al. Higher levels of 25-hydroxyvitamin D are associated with a lower incidence of multiple sclerosis only in women. Mult Scler. 2008 doi: 10.1177/1352458508095920. [DOI] [PubMed] [Google Scholar]
- 77.Moscarello MA, Mastronardi FG, Wood DD. The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochem Res. 2007;32:251–6. doi: 10.1007/s11064-006-9144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pitzalis M, Zavattari P, Murru R, Deidda E, Zoledziewska M, Murru D, et al. Genetic loci linked to type 1 diabetes and multiple sclerosis families in Sardinia. BMC Med Genet. 2008;9:3. doi: 10.1186/1471-2350-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ounissi-Benkalha H, Polychronakos C. The molecular genetics of type 1 diabetes: new genes and emerging mechanisms. Trends Mol Med. 2008;14:268–75. doi: 10.1016/j.molmed.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 80.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng H, Hagopian W. Environmental factors in the development of Type 1 diabetes. Rev Endocr Metab Disord. 2006;7:149–62. doi: 10.1007/s11154-006-9024-y. [DOI] [PubMed] [Google Scholar]
- 82.Robles DT, Eisenbarth GS. Type 1A diabetes induced by infection and immunization. J Autoimmun. 2001;16:355–62. doi: 10.1006/jaut.2000.0483. [DOI] [PubMed] [Google Scholar]
- 83.Poirier LA. The effects of diet, genetics and chemicals on toxicity and aberrant DNA methylation: an introduction. J Nutr. 2002;132:2336S–9. doi: 10.1093/jn/132.8.2336S. [DOI] [PubMed] [Google Scholar]
- 84.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sullivan KE. Genetics of systemic lupus erythematosus. Clinical implications. Rheum Dis Clin North Am. 2000;26:229–56. v–vi. doi: 10.1016/s0889-857x(05)70137-x. [DOI] [PubMed] [Google Scholar]
- 86.Karassa FB, Bijl M, Davies KA, Kallenberg CG, Khamashta MA, Manger K, et al. Role of the Fcgamma receptor IIA polymorphism in the antiphospholipid syndrome: an international meta-analysis. Arthritis Rheum. 2003;48:1930–8. doi: 10.1002/art.11059. [DOI] [PubMed] [Google Scholar]
- 87.Karassa FB, Trikalinos TA, Ioannidis JP. Role of the Fcgamma receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum. 2002;46:1563–71. doi: 10.1002/art.10306. [DOI] [PubMed] [Google Scholar]
- 88.Karassa FB, Trikalinos TA, Ioannidis JP. The Fc gamma RIIIA-F158 allele is a risk factor for the development of lupus nephritis: a meta-analysis. Kidney Int. 2003;63:1475–82. doi: 10.1046/j.1523-1755.2003.00873.x. [DOI] [PubMed] [Google Scholar]
- 89.DAS, Giordano M, Mellai M, Lanceni M, Barizzone N, Marchini M, et al. Association tests with systemic lupus erythematosus (SLE) of IL10 markers indicate a direct involvement of a CA repeat in the 5′ regulatory region. Genes Immun. 2002;3:454–63. doi: 10.1038/sj.gene.6363928. [DOI] [PubMed] [Google Scholar]
- 90.Alarcon-Riquelme ME, Lindqvist AK, Jonasson I, Johanneson B, Sandino S, Alcocer-Varela J, et al. Genetic analysis of the contribution of IL10 to systemic lupus erythematosus. J Rheumatol. 1999;26:2148–52. [PubMed] [Google Scholar]
- 91.Torres B, Aguilar F, Franco E, Sanchez E, Sanchez-Roman J, Jimenez Alonso J, et al. Association of the CT60 marker of the CTLA4 gene with systemic lupus erythematosus. Arthritis Rheum. 2004;50:2211–5. doi: 10.1002/art.20347. [DOI] [PubMed] [Google Scholar]
- 92.Aguilar F, Torres B, Sanchez-Roman J, Nunez-Roldan A, Gonzalez-Escribano MF. CTLA4 polymorphism in Spanish patients with systemic lupus erythematosus. Hum Immunol. 2003;64:936–40. doi: 10.1016/s0198-8859(03)00171-x. [DOI] [PubMed] [Google Scholar]
- 93.Kristiansen OP, Larsen ZM, Pociot F. CTLA-4 in autoimmune diseases–a general susceptibility gene to autoimmunity? Genes Immun. 2000;1:170–84. doi: 10.1038/sj.gene.6363655. [DOI] [PubMed] [Google Scholar]
- 94.Prokunina L, Gunnarsson I, Sturfelt G, Truedsson L, Seligman VA, Olson JL, et al. The systemic lupus erythematosus-associated PDCD1 polymorphism PD1.3A in lupus nephritis. Arthritis Rheum. 2004;50:327–8. doi: 10.1002/art.11442. [DOI] [PubMed] [Google Scholar]
- 95.Prokunina L, Padyukov L, Bennet A, de Faire U, Wiman B, Prince J, et al. Association of the PD-1.3A allele of the PDCD1 gene in patients with rheumatoid arthritis negative for rheumatoid factor and the shared epitope. Arthritis Rheum. 2004;50:1770–3. doi: 10.1002/art.20280. [DOI] [PubMed] [Google Scholar]
- 96.Sawalha AH, Kaufman KM, Kelly JA, Adler AJ, Aberle T, Kilpatrick J, et al. Genetic association of interleukin-21 polymorphisms with systemic lupus erythematosus. Ann Rheum Dis. 2008;67:458–61. doi: 10.1136/ard.2007.075424. [DOI] [PubMed] [Google Scholar]
- 97.Mascart-Lemone F, Hauptmann G, Goetz J, Duchateau J, Delespesse G, Vray B, et al. Genetic deficiency of C4 presenting with recurrent infections and a SLE-like disease. Genetic and immunologic studies. Am J Med. 1983;75:295–304. doi: 10.1016/0002-9343(83)91208-1. [DOI] [PubMed] [Google Scholar]
- 98.Sullivan KE, Wooten C, Schmeckpeper BJ, Goldman D, Petri MA. A promoter polymorphism of tumor necrosis factor alpha associated with systemic lupus erythematosus in African-Americans. Arthritis Rheum. 1997;40:2207–11. doi: 10.1002/art.1780401215. [DOI] [PubMed] [Google Scholar]
- 99.Takeuchi F, Nakano K, Nabeta H, Hong GH, Kawasugi K, Mori M, et al. Genetic contribution of the tumour necrosis factor (TNF) B + 252*2/2 genotype, but not the TNFa,b microsatellite alleles, to systemic lupus erythematosus in Japanese patients. Int J Immunogenet. 2005;32:173–8. doi: 10.1111/j.1744-313X.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- 100.Parks CG, Pandey JP, Dooley MA, Treadwell EL, St Clair EW, Gilkeson GS, et al. Genetic polymorphisms in tumor necrosis factor (TNF)-alpha and TNF-beta in a population-based study of systemic lupus erythematosus: associations and interaction with the interleukin-1alpha-889 C/T polymorphism. Hum Immunol. 2004;65:622–31. doi: 10.1016/j.humimm.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 101.Cunninghame Graham, Manku DSH, Wagner S, Reid J, Timms K. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum Mol Genet. 2007;16:579–91. doi: 10.1093/hmg/ddl469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ferreiro-Neira I, Calaza M, Alonso-Perez E, Marchini M, Scorza R, Sebastiani GD, et al. Opposed independent effects and epistasis in the complex association of IRF5 to SLE. Genes Immun. 2007;8:429–38. doi: 10.1038/sj.gene.6364407. [DOI] [PubMed] [Google Scholar]
- 103.Nakashima H, Matsuno S, Akahoshi M, Miyake K, Inoue Y, Tanaka Y, et al. Association between IFNA genotype and the risk of systemic lupus erythematosus. Clin Rheumatol. 2005;24:38–40. doi: 10.1007/s10067-004-0966-8. [DOI] [PubMed] [Google Scholar]
- 104.Lee YH, Witte T, Momot T, Schmidt RE, Kaufman KM, Harley JB, et al. The mannose-binding lectin gene polymorphisms and systemic lupus erythematosus: two case-control studies and a meta-analysis. Arthritis Rheum. 2005;52:3966–74. doi: 10.1002/art.21484. [DOI] [PubMed] [Google Scholar]
- 105.Tangwattanachuleeporn M, Sodsai P, Avihingsanon Y, Wongpiyabovorn J, Wongchinsri J, Hirankarn N. Association of interferon-gamma gene polymorphism (+874A) with arthritis manifestation in SLE. Clin Rheumatol. 2007;26:1921–4. doi: 10.1007/s10067-007-0699-6. [DOI] [PubMed] [Google Scholar]
- 106.Sullivan KE, Wooten C, Goldman D, Petri M. Mannose-binding protein genetic polymorphisms in black patients with systemic lupus erythematosus. Arthritis Rheum. 1996;39:2046–51. doi: 10.1002/art.1780391214. [DOI] [PubMed] [Google Scholar]
- 107.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS ONE. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rooryck C, Barnetche T, Richez C, Laleye A, Arveiler B, Schaeverbeke T. Influence of FCGR3A-V212Fand TNFRSF1B-M196R genotypes inpatients with rheumatoid arthritis treated with infliximab therapy. Clin Exp Rheumatol. 2008;26:340–2. [PubMed] [Google Scholar]
- 109.Orozco G, Sanchez E, Gonzalez-Gay MA, Lopez-Nevot MA, Torres B, Pascual-Salcedo D, et al. SLC22A4, RUNX1, and SUMO4 polymorphisms are not associated with rheumatoid arthritis: a case-control study in a Spanish population. J Rheumatol. 2006;33:1235–9. [PubMed] [Google Scholar]
- 110.de Vries N, Tijssen H, Jarvinen P, Aho K, van de Putte LB. HLA-DRB1 in eight Finnish monozygotic twin pairs concordant for rheumatoid arthritis. Tissue Antigens. 1997;49:277–9. doi: 10.1111/j.1399-0039.1997.tb02752.x. [DOI] [PubMed] [Google Scholar]