SUMMARY
During directed cell migration the movement of the nucleus is coupled to the forward progression of the cell. The microtubule motor cytoplasmic dynein is required for both cell polarization and cell motility. Here, we investigate the mechanism by which dynein contributes to directed migration. Knockdown of dynein slows protrusion of the leading edge and causes defects in nuclear movements. The velocity of nuclear migration was decreased in dynein knockdown cells, and nuclei were mislocalized to the rear of motile cells. In control cells, we observed that wounding the monolayer stimulated a dramatic induction of nuclear rotations at the wound edge, reaching velocities up to 8.5 degrees/min. These nuclear rotations were significantly inhibited in dynein knockdown cells. Surprisingly, centrosomes do not rotate in concert with the nucleus; instead the centrosome remains stably positioned between the nucleus and the leading edge. Together, these results suggest that dynein contributes to migration in two ways: (1) maintaining centrosome centrality by tethering microtubule plus ends at the cortex, and (2) maintaining nuclear centrality by asserting force directly on the nucleus.
INTRODUCTION
Cell migration plays a central role in a variety of biological processes; tissue development, the immune response, and wound healing all require directional migration. Microtubules (MTs) have been shown to contribute to several steps of cell migration, one of the most well characterized being polarity establishment. In migrating fibroblasts, the centrosome reorients between the nucleus and the leading edge, which in turn repositions the Golgi and is thought to establish and maintain cell polarity during migration (Mellor, 2004). Previous work has demonstrated a role for the microtuble motor dynein, its cofactor dynactin, and the dynein regulatory protein Lis1 in reorientation of the centrosome toward the leading edge (Etienne-Manneville and Hall, 2001; Gomes et al., 2005; Palazzo et al., 2001; Yvon et al., 2002). However, inhibition of dynein or dynactin after centrosome reorientation suppresses cell motility, indicating that there is an additional role for dynein in persistent fibroblast motility (Dujardin et al., 2003).
Several studies have suggested a role for dynein and Lis1 in both centrosome and nuclear positioning. During neuronal migration, nascent neurons extend a long leading process from the cell body. The centrosome then migrates into the leading process, and subsequently the nucleus moves forward in a saltatory manner (reviewed in Tsai and Gleeson, 2005). Inhibition of dynein or Lis1 disrupts nuclear migration and causes an increase in nucleus-centrosome distance in neurons (Shu et al., 2004; Tanaka et al., 2004; Tsai et al., 2005), suggesting that dynein and Lis1 couple centrosome and nuclear movements. Studies in lower organisms further support a role for dynein and Lis1 in nucleokinesis. In the filamentous fungus Aspergillus nidulans, nuclei migrate long distances toward the growing tip of the hyphae, a process which requires the fungal homologues of human dynein, dynactin, and Lis1 (reviewed in Tsai and Gleeson, 2005). Studies of pronuclear migration in the one cell stage embryo of Caenorhabditis elegans have also indicated a role for orthologues of dynein heavy chain and Lis1 in nucleokinesis. In this system, the nuclear envelope protein Sun1 and dynein-interacting protein Zyg-12 were suggested to mediate attachment of dynein to the nucleus (Malone et al., 2003).
In contrast to migrating neurons, where there is a loose coupling between nuclei and centrosomes, nuclei and centrosomes in fibroblasts are closely associated. It has been hypothesized that the nucleus and the centrosome are statically tethered in fibroblasts, and that coupling of MTs to the advancing leading edge will pull the nucleus in the direction of migration (Gomes et al., 2005; Neujahr et al., 1998).
In this study, we examine the role of dynein in both centrosome reorientation and nuclear dynamics in migrating fibroblasts. In addition to forward translocation of the nucleus during cell migration, we note a dramatic induction of nuclear rotations upon monolayer wounding. Both the forward translocation and the rotations are dynein-dependent. Further, the rotation of the nucleus is independent of centrosome positioning. These observations suggest a dynamic relationship between the nucleus and the centrosome, and reveal a role for dynein in coupling the nucleus to MTs, therefore maintaining its centrality in migrating cells.
RESULTS
Dynein facilitates lamellar extension in migrating fibroblasts
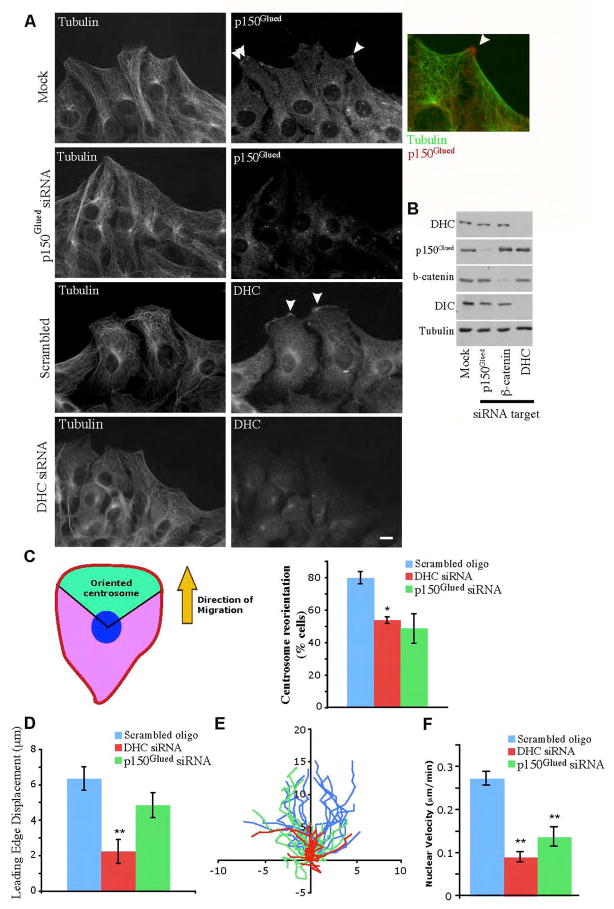
In this study, we used an RNA interference (RNAi) approach to knockdown dynein heavy chain (DHC) and the p150Glued subunit of dynactin, therefore allowing us to analyze the function of dynein and dynactin in migrating cells. NIH/3T3 fibroblasts were grown to confluence and then a wound-healing assay was used to initiate cytoskeletal redistribution and directional migration of cells toward the wound edge (Gundersen and Bulinski, 1988). Cells were mock transfected, transfected with small interfering RNA (siRNA) targeting DHC or p150Glued, or transfected with a scrambled oligonucleotide control; confluent cultures were scratch wounded. Six hours after wounding, the cells were fixed and protein lysates were collected. Western blot analysis revealed that target protein levels were consistently reduced 75–95% in knockdown cells as compared to cells that were mock-treated (Fig. 1B) or transfected with a scrambled oligonucleotide (unpublished data). Observation of fixed cells via immunofluorescence demonstrates that approximately 90% of DHC knockdown cells and 88% of p150Glued knockdown cells show decreased levels of protein targeted by siRNAs. Levels of dynein intermediate chain (DIC) decrease in DHC knockdown cells (Fig. 1B), suggesting that the dynein complex is being destabilized, as has been observed in other cell types (Caviston et al., 2007). However, dynactin is expressed at normal levels in DHC knockdown cells and dynein is expressed at normal levels in p150Glued knockdown cells (Fig. 1B).
Figure 1.
Dynein and dynactin localize to the leading edge of migrating cells and promote centrosome reorientation, protrusion, and nuclear migration. (A) Dynein and dynactin concentrate in cortical patches at the leading edge of migrating cells. NIH/3T3 cells that were mock-transfected or transfected with siRNAs targeting p150Glued or DHC were grown to confluency and the monolayer was wounded. Six hours after wounding, cells were fixed and stained for α-tubulin and either p150Glued (top) or DIC (bottom). Mock-transfected cells show patches of dynein and dynactin at the leading edge of migrating cells (arrowheads). Inset, overlay of cortical patch of p150Glued (red) and projecting MTs (green). Bar, 10 μm. (B) Western blot of protein lysates from cells that were mock-transfected, transfected with siRNAs targeting p150Glued, DHC, or β-catenin, a protein that binds to dynein (Ligon et al., 2001) consistently show knockdown of target protein levels by 75–95%. Levels of DIC decrease in DHC knockdown cells, suggesting the dynein complex is being destabilized. In contrast, knockdown of β-catenin, which binds to DIC but is not part of the dynein motor complex, does not destabilize DIC. (C) Centrosome reorientation is inhibited in dynein and dynactin knockdown cells. Orientation of centrosomes 6 hours after wounding. Centrosome orientation was determined by measuring the position of centrosomes, as determined by γ-tubulin staining, in relation to Hoescht stained-nuclei. Centrosomes in the forward facing 120 degree sector (green zone) were scored as reoriented. (D) Leading edge extension is inhibited in dynein knockdown cells. Displacement of the leading edge over 45 min of wound healing was measured (n=15). (E) Tracks of nuclei centroids during migration of fibroblasts over 45 min. Paths are oriented so cells are migrating towards the top of the graph, each path starts at (0,0). Axis labels are in microns. (F) Rates of nuclear movement is decreased in dynein and dynactin knockdown cells. Nuclear velocity during migration was decreased in DHC and p150Glued siRNA cells, compared to mock transfected cells (n=15). Error bars indicate SEM; * indicates p<0.05.; ** indicates p<0.005.
In mock-treated cells at the wound edge, the MT network is polarized in the direction of the wound (Gundersen and Bulinski, 1988), and dynein and dynactin accumulate in a patch-like pattern (Dujardin et al., 2003) at the tips of protrusions into the wound area (Fig. 1A). These cortical patches of dynein/dynactin may contribute to MT forces that maintain nuclear and centrosomal centrality. Dynactin demonstrates decreased accumulation in patches along the leading edges of dynein knockdown cells (Fig. S 1), indicating that dynein is required for dynactin accumulation at these patches.
Mock-treated and siRNA-treated cells were fixed 6 hours after wounding, a timepoint several hours after centrosome reorientation occurs in these cells (Gundersen and Bulinski, 1988), and stained with an antibody to the centrosomal marker γ-tubulin. In mock-treated cells the majority of centrosomes are perinuclear and positioned in the direction of the wound, whereas cells treated with siRNA against dynein or dynactin have centrosomes that are more evenly distributed around the nucleus (Fig. 1C), indicating they are defective in centrosome reorientation. This defect is consistent with previous observations of reduced centrosome reorientation after dynein or dynactin inhibition (Etienne-Manneville and Hall, 2001; Gomes et al., 2005; Palazzo et al., 2001; Yvon et al., 2002). Although inhibition of dynactin function by overexpression of individual dynactin subunits was previously shown to interfere with MT anchoring at the centrosome (Quintyne et al., 1999), we observe no overt defects in microtubule organization in dynein or dynactin knockdown cells. This may be due to some residual dynactin remaining at the centrosome (Fig. 1A), as RNAi does not cause complete silencing of the target molecule.
In order to investigate a role for dynein and dynactin in persistent cell motility, we observed mock-treated or siRNA-treated cells after monolayer wounding by timelapse microscopy (Video 1). In both control and knockdown cultures, cells at the wound edge show robust lamellar ruffling, consistent with a motile phenotype. However, dynein and dynactin knockdown cells appear to advance into the wound at a slower rate than mock-treated cells. Leading edge displacement in the direction of the wound was measured by marking the change in position of the cell edge over 45 minutes of wound closure. We observed a significant decrease in leading edge extension into the wound in dynein knockdown cells (Fig. 1D). Dynactin knockdown cells showed similar rates of leading edge protrusion compared to control cells, which may indicate some differential dependence for dynein versus dynactin in lamellipodial extension (Fig. 1D).
Dynein and dynactin contribute to nuclear movements in migrating fibroblasts
Cell migration involves both lamellar extension and forward progression of the cell body, including the nucleus. In order to examine the dynamics of nuclei in the knockdown cells, we measured the velocity of nuclear movement by tracking the paths of nuclear centroids by timelapse microscopy (Fig. 1E–F). Nuclei in dynein and dynactin knockdown cells moved at a slower velocity and with less directed movement than nuclei in mock-treated cells (Video 1). These slower velocities were correlated with mislocalization of nuclei in dynein and dynactin knockdown cells. Following either dynein or dynactin knockdown, nuclei were significantly closer to the rear of the cells than nuclei in mock treated cells (0.0098 ± 0.0006 μm−1 in control cells, compared to 0.0048 ± 0.0003 μm−1 in p150Glued knockdown cells and 0.0055 ± 0.0005 μm−1 in DHC knockdown cells when normalized for cell area; n=75; p<0.005 for each). This suggests that dynein and dynactin have a role in coupling nuclear position to cell migration.
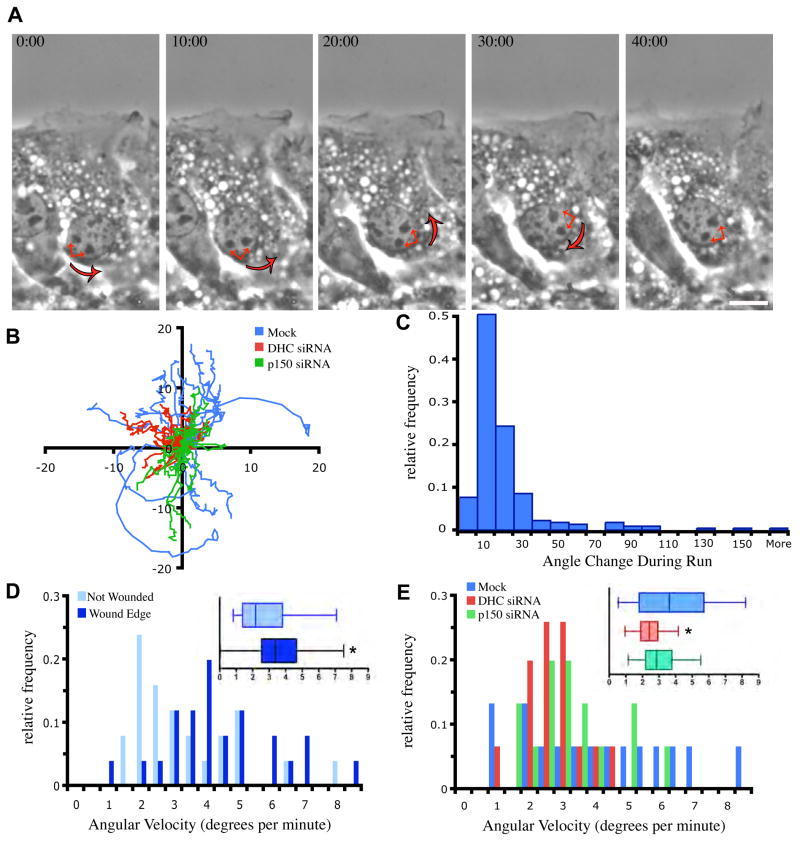
Surprisingly, many control cells demonstrated sustained nuclear rotations during motility (Fig. 2A and S2). These rotations can be monitored by tracking the paths of individual nucleoli, which stay in position relative to each other during interphase (Chubb et al., 2002) (Fig. 2B and S3). We observed rotations in both clockwise and counterclockwise directions in cells at the wounded edge of the monolayer, which reached angular velocities up to 8.5 degrees per min (Fig. 3B–E). Rotations were only observed in the plane of the substratum, as we would expect due to the flat, ellipsoidal shape of fibroblast nuclei (Bolzer et al., 2005). This distinguishes the rotations we observe from rolling cell bodies that have been observed in motile fish keratocytes, an event that appears to be actin-driven (Anderson et al., 1996). Although many rotations result in angle changes of hundreds of degrees, particle tracking at high temporal resolution reveals that most rotations are interrupted by short pauses or directional changes after runs of less than 20 degrees (Fig. 2C). Rotations were observed in cells at, and several rows back from, the wounded edge, but were less frequent in cells in monolayers that had not been wounded (Fig. 2D and Video 2). Nuclear rotations were observed in single migrating NIH/3T3 cells (Levy & Holzbaur, unpublished observations), suggesting that rotations are enhanced specifically in migrating cells, and may reflect increased dynamicity of the cytoskeleton during cell motility.
Figure 2.
Dynein/dynactin-dependent nuclear rotations are enhanced after monolayer wounding. (A) A timeseries of a nucleus that rotates during cell migration. Red arrows indicate 2 nucleoli that stay in position relative to other nucleoli within the nucleus. This nucleus rotates counter-clockwise for the first 30 min, and then switches direction during the final 10 min of this timelapse. Bar, 10 μm. Time is minutes:seconds. (B) Tracks of nucleoli during migration of fibroblasts over 45 min. Large, circular traces seen in mock-transfected cells demonstrate paths of nucleoli in nuclei that are rotating. These are rarely seen in DHC and p150Glued siRNA cells (n=15). Axis labels are in microns. (C) Particle tracking was used to track the duration of rotation of nuclei from cells at the wound edge during migration. A run was defined as rotation in a single direction without pausing (n=225 runs from 25 cells). (D) Angular velocity of nuclei in cells along the wound edge of wounded cultures is higher than that of nuclei in cells that have been grown to confluency, but not wounded (n=25). (E) Angular velocity of nuclei in mock-treated cells along the leading edge is greater than that of DHC and p150Glued siRNA cells, which rarely rotate and thus have a slower angular velocity (n=15). Error bars indicate SEM; * indicates p<0.05.
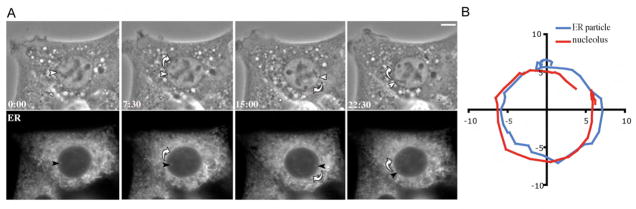
Figure 3.
Nuclear rotation includes ER closely associated with the nuclear membrane. (A) Cells transfected with dsRed2-ER were grown to confluency and wounded. The top panel shows a nucleus rotating clockwise during migration. White arrowhead indicates the position of a nucleolus during rotation. The bottom panel shows the morphology of the ER during the same timeframe. Some small fragments of the ER that are closely opposed to the nucleus rotate with it (black arrowhead, see also Video 3). Peripheral ER shows dynamic remodeling during the rotation. (B) Tracks of the paths of a single nucleolus and an ER particle within the cell shown in part A during nuclear rotation over 45 min. Both follow the same circular path, indicating that they are both rotating in the same manner (see also Video 3). Time is minutes:seconds; bars, 10 microns.
Depolymerization of MTs with high doses (16.6 μM) of nocodozole (NZ) also significantly decreased the velocity of nuclear rotations, demonstrating that nuclear rotations require an intact MT cytoskeleton (Fig. 7 D-E & Video 6). Because dynein and dynactin are known to drive nuclear positioning, we examined the movements of nuclei in cells where DHC or p150Glued had been knocked down. Interestingly, cells in which dynein had been knocked down showed a significant reduction in the velocity of nuclear rotations (Fig. 2E, Fig. S2, and Video 1). It is unlikely that this inhibition in nuclear rotation is secondary to a global disruption in the cellular cytoskeleton, as no overt changes were observed in the distribution of MTs (Fig. 1A), acetylated MTs (unpublished data), vimentin (unpublished data) or actin (Fig. S4) in either dynein or dynactin knockdown cells.
It is possible that the rotations that we observe involve only components within the nucleus, but not the nuclear envelope. Because the outer nuclear membrane is continuous with the ER, we monitored ER dynamics using dsRed2-ER. We observed that segments of the ER that are most closely associated with the nucleus rotate in concert with the nucleus (Fig. 3 and Video 3), suggesting that the entire nucleus, including the nuclear envelope, rotates in migrating cells. However, ER tubules extending farther from the nucleus, as well as most juxtanuclear ER, demonstrated dynamic remodeling over the time period of our observations (tens of minutes). In general the ER did not revolve in concert with the nucleus, nor did ER tubules appear to be trailing behind rotating nuclei, indicating that the majority of the ER network remodels as the nucleus rotates.
Nuclear rotations are not accompanied by centrosome movements
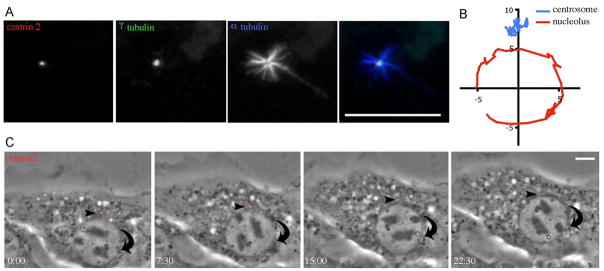
Previous studies have demonstrated that the centrosome precedes the nucleus in forward movement during cell migration, leading to the hypothesis that protein-protein interactions tether the centrosome to the nucleus, and that MT interactions with the cortex maintain centrosome and nuclear centrality (Gomes et al., 2005; Morris, 2003; Tanaka et al., 2004; Tsai and Gleeson, 2005). If this hypothesis were true, then we would expect the centrosome to move in concert with a rotating nucleus. Therefore, we transfected cells with 3XGFP-EMTB, a construct that brightly labels MTs without perturbing MT dynamics (Faire et al., 1999). Live cell imaging of both MTs and the nucleus, as visible by phase, demonstrates that the MT network does not follow the circular movements of rotating nuclei (Video 4). To look specifically at centrosome dynamics, we transfected cells with dsRed1-Centrin2 (Tanaka et al., 2004). The majority of transfected cells showed a single or two adjacent centrin-positive puncta close to the nucleus that are associated with the centrosome marker gamma-tubulin (Fig. 4A). Visualization of MT regrowth after nocodazole washout shows that these puncta are capable of MT nucleation (Fig. 4A), corroborating that Centrin2 is an accurate centrosome marker. Live cell imaging of dsRed1-Centrin2 and the nucleus, as visible by phase, demonstrates that the centrosome maintains its orientation between the nucleus and the leading edge, even in cells where the nucleus rotates (Fig. 4B-C and Video 5). Therefore, the centrosome is not in a static interaction with the nucleus as it rotates.
Figure 4.
Nuclear rotations are not coupled to centrosome rotation. (A) Exogenous centrin2 closely associates with endogenous γ tubulin in puncta capable of MT nucleation, supporting its functional role as a centrosome marker. Cells transfected with dsRed1-Centrin2 (red) were subjected to nocodazole treatment. After a 5 min wash, cells were fixed and stained for γ tubulin (green) and α tubulin (blue). (B) Tracks of a single nucleolus (red) and a centrosome (blue) within the cell shown in part D during nuclear rotation over 45 min. The cell is migrating toward the top of the graph and the nucleus rotates while the centrosome remains between the nucleus and the leading edge. (C) Cells transfected with dsRed1-Centrin2 (red), a centrosomal marker, were grown to confluency and wounded. The arrowhead indicates a centrosome that stays positioned between the nucleus and the leading edge while the nucleus rotates clockwise (also see Video 5). Time is minutes:seconds; bars, 10 microns.
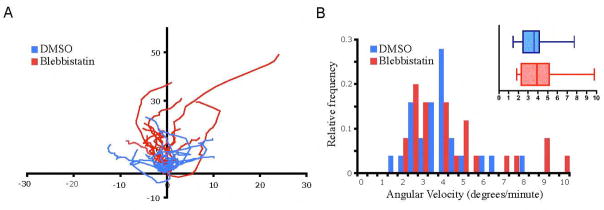
Previous work suggests that retrograde actin flow and myosin II contribute to nuclear positioning by pushing the nucleus rearward during centrosome reorientation in fibroblasts (Gomes et al., 2005). In order to examine if this force contributes to nuclear rotation during forward migration, we observed nuclear dynamics in cells treated with blebbistatin (BB), a myosin II inhibitor that has been shown to block actin retrograde flow (Ponti et al., 2004; Straight et al., 2003). As seen in fibroblasts with reduced levels of myosin II A, cells treated with BB demonstrate exaggerated membrane ruffling, enhanced migration, and reduced cell body retraction (Even-Ram et al., 2007; Vicente-Manzanares et al., 2007). Cell treated with BB showed no inhibition of nuclear rotations (Fig. 5 and Video 6), indicating that myosin II and retrograde actin flow do not drive these movements. In fact, some cells treated with BB reached angular velocities above those observed in DMSO-treated cells, up to 9.8 μm/min (Fig. 5B). This suggests that actomyosin may be dampening nuclear rotations in control cells, perhaps because actin/myosin II stress fibers act as a physical hindrance to dynamic nuclear movements.
Figure 5.
Nuclear rotations are independent of myosin II activity. (A) Tracks of nucleoli during migration of DMSO or blebbistatin-treated fibroblasts over 45 min (n=15). Axis labels are in microns. (B) Angular velocity of nuclei in the presence of myosin II inhibitor blebbistatin (n=25).
Nuclear rotations do not depend on dynactin localized to either the leading edge or the Golgi complex of migrating cells
There are two locations in the cell that dynein may be actively contributing to migration: at the cell cortex or at the nucleus. Dynein and dynactin accumulate in patches (Dujardin et al., 2003) at the leading edges of migrating fibroblasts (Fig. 1A), where they may act to influence MT dynamics or exert force on MTs that project towards the cortex in order to polarize the centrosome during migration. Alternatively, dynein/dynactin may localize near the nucleus to contribute to migration: either interacting directly with components of the nuclear envelope (Malone et al., 2003; Salina et al., 2002) or accumulating at the perinuclear Golgi (Fath et al., 1994).
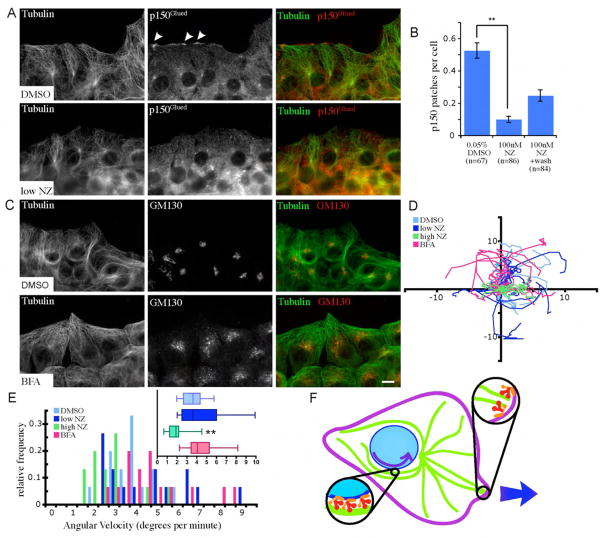
In order to specifically inhibit dynein/dynactin accumulation at the leading edge, we treated NIH/3T3 cells with a low dose (100 nM) of NZ. Although high doses of NZ are known to depolymerize MTs, concentrations in the nanomolar range have been shown to inhibit MT dynamics without causing disassembly (Vasquez et al., 1997). After treatment with low-dose NZ for 30 min, cells have normal MT distribution but accumulation of dynactin patches at the leading edge is inhibited (Fig. 6A-B), suggesting that the formation and/or maintenance of these patches are dependent on dynamic MTs. Centrosome reorientation is also inhibited in the presence of low-dose NZ (our unpublished results and see Gomes et al., 2005; Magdalena et al., 2003; Yvon et al., 2002), which may indicate a role for dynactin patches at the leading edge in coupling MT plus-ends to the cortex. Dynamic MT plus ends may act to deliver molecules to the cortex that act to maintain dynactin patches. One possible candidate is dynein, which is necessary for the formation of dynactin patches at the leading edge (Fig. S 1). This system would parallel observations in both budding yeast and Aspergillus nidulans: dynein is off-loaded from MT plus ends to the cortex, where it exerts pulling forces on MTs (Lee et al., 2003; Sheeman et al., 2003; Xiang and Fischer, 2004).
Figure 6.
Nuclear rotations are independent of leading-edge- and Golgi-localized dynein. (A) Cells treated with low doses of nocodazole (NZ) 5.5 hours after wounding maintain the MT network morphology (green), but lose patches of p150Glued (red) at the leading edge. (B) Dynactin patch accumulation is inhibited in cells treated with low-doses of NZ. Patches are partially rescued 30 min after NZ washout. (C) Cells treated with BFA 5.5 hours after wounding maintain MT network morphology (green) but have fragmented Golgi (red, Golgi marker GM130). Bar, 10 microns. (D) Tracks of nucleoli treated with DMSO, BFA, or NZ during migration of fibroblasts over 45 min. Axis labels are in microns. (E) Angular velocity nuclei of cells treated with DMSO (control), NZ, or BFA (n=15). (F) Scheme for two roles for dynein/dynactin in cell motility: (1) Dynein and dynactin accumulate in cortical patches at the leading edge, where they interact with MTs and mediate centrosome and MT orientation during motility, and (2) dynein and dynactin interact with the nuclear envelope and transport the nucleus along MTs on the sides of the nucleus. Error bars indicate SEM; ** indicates p<0.005.
We used low doses of NZ to assess nuclear rotations in the absence of dynein/dynactin accumulation at the leading edge. Cells monolayers were wounded and allowed to recover for a minimum of 4 hours to allow for centrosome reorientation toward the wound edge prior to addition of 100nM NZ. Timelapse microscopy demonstrates that nuclear rotations occurred even in absence of dynactin patches at the leading edge at angular velocities similar to those observed in control, DMSO-treated cells (Fig. 6D-E, Video 6). It should be noted that although nuclear rotations were not disrupted, the cells did not progress forward after low doses of NZ were applied, as has been seen previously (Liao et al., 1995). This may be due directly to the reduction in MT dynamics, or indirectly due to less dynamic MTs not reaching cortical patches of dynein/dynactin.
Dynein and dynactin are known to localize to the Golgi membrane (Fath et al., 1994) and to maintain the perinuclear localization of the Golgi complex (Corthesy-Theulaz et al., 1992). It is possible that the enrichment of dynein/dynactin at the Golgi could drive nuclear rotations. To investigate this we treated cells with Brefeldin A (BFA) to disrupt the Golgi complex. In cells that recovered from wounding in the presence of BFA fragments of Golgi were dispersed throughout the cytoplasm with no perinuclear stacks remaining (Fig. 6C). Centrosomes were polarized normally, and dynactin patches accumulated at the leading edge (unpublished data). After monolayer wounding, cells demonstrated nuclear rotations that were robust, even in the presence of BFA (Fig. 6D, Video 6). The angular velocities of these rotations were comparable to rotations observed in control, DMSO-treated cells (Fig. 6E).
DISCUSSION
Dynein and dynactin have been shown to contribute to centrosome reorientation and polarity establishment during both directed cell migration and immunological synapse formation (Combs et al., 2006; Etienne-Manneville and Hall, 2001; Palazzo et al., 2001; Yvon et al., 2002). However, microinjection of single cells at an in vitro wound edge with inhibitory antibodies to dynein after centrosome reorientation will cause the injected cell to fall behind in the field of migrating cells (Dujardin et al., 2003), suggesting that dynein has an additional role in directed cell motility. Here, we identify a role for dynein in driving nuclear movement during fibroblast migration.
Wounded cell monolayer cultures provide a system in which the kinetics and direction of cell movement can be synchronized throughout a population of cells. An early step in wound healing involves reorientation of the centrosome to a position between the nucleus and the leading edge (Gundersen and Bulinski, 1988), a process that is dynein-dependent (Etienne-Manneville and Hall, 2001; Palazzo et al., 2001; Yvon et al., 2002) and cell-type specific, as epithelial cells do not demonstrate centrosome reorientation (Danowski et al., 2001; Yvon et al., 2002). The predominant model for centrosome reorientation involves patches of cortical dynein tethering MT plus ends to the cortex, which maintains the centrosome in the center of the cell. Concurrently, actin retrograde flow moves the nucleus towards the rear, so that it is between the centrosome and the trailing edge of the cell (Gomes et al., 2005). The involvement of cortical dynein in this process is supported by experiments that show that low doses of nocodazole, which block accumulation of cortical dynein, inhibit centrosome reorientation (Fig. 6A & B and Gomes et al., 2005; Magdalena et al., 2003; Yvon et al., 2002).
Studies of neuronal motility suggest that dynein has roles in both centrosome and nuclear positioning. Inhibition of dynein and Lis1 increases the nuclear-centrosomal distance, suggesting impaired nucleokinesis during neuronal migration (Tanaka et al., 2004). Unlike the saltatory movement of nuclei in neurons, migration of fibroblast nuclei are temporally coupled to both leading edge extension and centrosome movement. Additionally, imaging of GFP-tagged tubulin in migrating NIH/3T3 cells demonstrates that the centrosome and the nucleus are physically close to one another (Gomes and Gundersen, 2006). Such observations have led to the hypothesis that in fibroblasts, the interaction of MTs with the leading edge couples centrosome movement to leading edge protrusion, and that a physical interaction of the centrosome with the nucleus passively pulls the nucleus forward.
We now propose a role for dynein in driving nuclear movement along perinuclear MTs in migrating fibroblasts, similar to the role for dynein and Lis1 in neuronal nucleokinesis. Knockdown of dynein in fibroblasts inhibits centrosome reorientation at the initiation of fibroblast motility, but defects in nuclear migration during later phases of migration were also observed: nuclear displacement in the direction of migration (Fig. 1F), as well as the velocity of nuclear rotations that are induced upon wounding, were reduced in dynein knockdown cells (Fig. 2E). Imaging of the centrosome during migration revealed that although the nucleus and centrosome are physically close to one another, they are not statically associated, as nuclear rotations are not accompanied by centrosome revolutions (Fig. 4). These data support previous observations of GFP-γ-tubulin in wound edge cells, which demonstrated that motion of the nucleus and the centrosome were uncoordinated (Yvon et al., 2002). While centrosome positioning appears to be mediated by cortical dynein (Morris, 2003), selective disruption of leading edge dynein patches did not inhibit nuclear rotations (Fig. 6D & E), suggesting that nuclear rotations are driven by dynein at the nuclear membrane. Such a mechanism could dynamically couple the nucleus to perinuclear MTs, therefore maintaining its centrality during cell migration.
Previous studies have demonstrated an interaction of dynein with isolated nuclei from cerebellar granule neurons (Tanaka et al., 2004) and with the nuclear envelope of NRK cells (Salina et al., 2002) and C. elegans embryonic and gonadal cells (Malone et al., 2003). The p62 subunit of dynactin facilitates the association of dynein with the nuclear envelope (Salina et al., 2002); the nuclear envelope component that facilitates this interaction in vertebrate cells is unknown, but may include SUN-domain proteins (Worman and Gundersen, 2006).
Do nuclear rotations promote migration? The spatial positioning of chromatin is proposed to play an important role in gene regulation, but it is generally believed to be the position of the gene with respect to the nucleus, rather than the entire cell, that influences transcription (Meaburn and Misteli, 2007). Additionally, nuclear rotations are often sustained for more than 360 degrees, making it unlikely that they act to redirect a certain domain of the nucleus to a specific position within the cell. Only a subset of cells at the wound edge show sustained rotations in a single direction while migrating, but these cells do not migrate faster or with more directional persistence than cells without rotating nuclei. It is therefore likely that the rotations themselves do not contribute to migration, but that the rotations reveal the mechanism by which dynein maintains nuclear centrality in migrating cells. Observation of wound-edge cells with nuclei that are not rotating in a sustained manner reveals many stochastic nuclear movements that appear to be very short rotations that frequently switch direction. This “jostling” seems to center nuclei in cells as they move forward. In support of this idea, knockdown of dynein is accompanied by a rearward shift in nuclei position, so that they become situated in the back of migrating cells. This may be a passive shift in position, due to the forward movement of the cell body when uncoupled to the nucleus. Alternatively, retrograde actin flow may be actively pushing the nucleus rearward, as has been shown to occur during centrosome reorientation (Gomes et al., 2005). In support of this idea, rotations were robust in cells treated with BB (Fig. 5).
Nuclear rotations have been observed previously in cell culture (Englander and Rubin, 1987; Paddock and Albrecht-Buehler, 1986; Paddock and Albrecht-Buehler, 1988; Pomerat, 1953) and in vivo during the early stages of brain development in zebrafish embryos (Herbomel, 1999). Rotations involve both the interior and exterior of the nucleus, and require an intact MT cytoskeleton (Ji et al., 2007). Here we show nuclear rotations are induced upon stimulation of cell migration. We also observe a large reduction in rotations in the presence of nocodazole (Fig. 6D and 6 E). Furthermore, dynein was identified as the driving force for rotations in migrating NIH/3T3 fibroblasts. The repolarization that occurs with onset of migration may induce rotations by causing a redistribution of dynein associated with the nucleus. An asymmetric distribution will result in an imbalance of force leading to sustained rotation in a single direction. Stochastic redistribution of dynein motors during motility may lead to the pauses and directional reversals we also observed. The observation that some rotations persist beyond 360 degrees also suggest that multiple dynein molecules act to drive rotations, so that as one dynein may lose its attachment to an MT, other molecules may maintain nuclear-MT attachment and minus-end directed movement.
Several studies have observed nuclear rotation in response to perturbation of intermediate filaments, including vimentin and lamin B1 (Hay and De Boni, 1991; Ji et al., 2007). The results presented here, in which induction of rotations was observed in response to stimulation of cell migration, might suggest that certain nuclear/cytoskeletal connections are relaxed when cells transition from a sedentary to migratory state. Intriguingly, disruption of actin stress fibers and myosin II activity with blebbistatin stimulated rotations that reached higher velocities than those observed in control cells (Fig. 5), although the differences were not statistically significant. Nonetheless, these data raise the possibility that actin/nuclear connections or actin stress fibers dampen dynein-driven rotations. Alternatively, rearward nuclear movement driven by actin retrograde flow (Gomes et al., 2005) may act as an opposing force to forward, dynein-driven nuclear movement. The balance of these forces may act to maintain the nucleus in the center of migrating cells.
In conclusion, we propose a model wherein dynein has two distinct functions in migrating fibroblasts (Fig. 6F). First, cortical patches of dynein interact with MT plus ends to facilitate centrosome reorientation. Second, nuclear envelope-associated dynein interacts with the nucleus and walks along perinuclear MTs to maintain nuclear centrality as the cell translocates. These two roles for dynein make it a central component in both polarity establishment and cell body translocation during fibroblast migration.
MATERIALS AND METHODS
Cell Culture, Transfection, and Wounding
GFPTub3T3 cells (a kind gift from G. Gundersen, (Gomes et al., 2005) were maintained in DME containing 10% newborn calf serum (NCS). For siRNA transfections, 60 nM RNA oligonucleotides (Dharmacon) were transfected using Lipofectamine RNAiMAX (Invitrogen). The following target sequences were used for DHC: DHC #1: GAAAUCAACUUGCCCGAUA and DHC #2: CCAAAUACCUACAUUACUU); p150Glued: p150 #1: CGAGCUCACCACAGACCUG and p150 #2: CCUACGCAAUCCGACCGAG; β-cateninn, CAGGGUGCUAUUCCACGACUA (alternate: CAGAUAGAAAUGGUCCGA); scrambled against p150 #1, CCUACGCAAUCCGACCGAG. For DNA transfections, Amaxa Nucleofector program U-30 was used to transfect 5 μg of DNA into ~1x106 cells using Nucleofector Kit R. pDsRed1-Cent2 was a kind gift from J. Gleeson (Tanaka et al., 2004); pDsRed2-ER (Clontech) was a kind gift from A. Akhmanova; 3XGFP-EMTB was a kind gift from J.C. Bulinski (Faire et al., 1999). For drug treatments, cells were incubated with 0.05% DMSO, 16.6 μM (high dose) or 100 nM (low dose) NZ (Sigma), 2.5 μg/ml BFA (Sigma), or 50 μM BB (Tocris).
Cells were grown to confluency and monolayers were wounded using a ~25 μg glass scribe or p200 micropipette tip. Cells were fixed in methanol with 1mM EGTA for 10 min at −20°C, air-dried, and blocked with PBS containing 1% BSA and 5% normal goat serum; coverslips were incubated with mouse primary antibodies to α-tubulin (clone DM1A, Sigma), p150Glued (BD Biosciences), DIC (Chemicon), β-catenin (BD Biosciences), GM130 (BD Biosciences), acetylated tubulin (Sigma) and vimentin (Sigma) or rabbit primary antibodies to DHC (Santa Cruz), γ-tubulin (Covance), and p150Glued (Tokito et al., 1996). Coverslips were incubated with alexa-conjugated secondary antibodies from Molecular Probes and mounted with ProLong Gold (Molecular Probes). Alternatively, cells were fixed in 4% paraformadahyde, permeabilized in 0.25% TritonX 100, and stained with TRITC-conjugated phalloidin (Sigma) for visualization of actin.
Cells were lysed in 10 mM Tris with 25 mM NaCl, 0.25% IGEPAL, and 5 mM EDTA, denatured in gel sample buffer, and western blotted using SDS-PAGE and the above antibodies, followed by chemiluminescence detection (Western Lightening, Perkins Elmer).
Image Acquisition
Images were acquired using a Leica DMIRBE microscope with 40X/0.70NA, 63X/1.32NA, and 100X/1.4NA objectives and an Orca ER CCD camera (Hamamatsu) with OpenLab software (Improvision). For time-lapse imaging, cells were seeded on glass bottom dishes (World Precision Instruments), imaged in phenol red-free DMEM containing 10% NCS, 25 mM HEPES, 1% OxyFluor (Oxyrase), and sealed with mineral oil (Sigma). The microscope stage was maintained at 37°C. Time-lapse recordings were written with OpenLab software. Figures were prepared using Adobe Photoshop.
Data Analysis
All data analysis was done with ImageJ software. p150Glued leading edge patches were defined as areas of fluorescence intensity at the leading edge that were 1.5-fold greater than the fluorescence intensity of the adjacent area to the patch away from the wound edge after linearly adjusting the contrast of the image to 0.5% saturated pixels. Centrosome orientation in fixed cells was measured by calculating the angle of the centrosome, as determined by anti-γ-tubulin antibody staining, relative to a line bisecting the nucleus and parallel to the wound. Leading edge displacement was determined by measuring the distance the lamella moved perpendicular to the direction of the wound over the course of a 45 min movie. Leading edge displacement was measured at 5 randomly selected points along the wound edge per time-lapse. The rates of forward movement of the entire nucleus were analyzed by tracking the movements of the nucleus centroid over time. Measurement of nuclear position within the cell was analyzed by drawing a line perpedicular to the direction of the wound, starting at the rearmost point of the nucleus and ending at the trailing edge of the cell. Nucleoli and ER particle tracking was performed with the “Manual Tracking” plug-in for ImageJ submitted to http://rsb.info.nih.gov/ij/ by Fabrice P. Cordelieres. Angular velocity was calculated by measuring the distance traveled by a particle relative to the centroid of the nucleus over time.
Supplementary Material
Figure S1. DHC knockdown decreases dynactin patches at leading edge. NIH/3T3 cells were transfected with mock siRNA or with siRNA targeting DHC and were grown to confluency. The monolayer was wounded. Six hours after wounding, cells were fixed and stained for tubulin (green) and p150Glued (red). Bar, 10 microns. DHC knockdown cells had less p150Glued leading edge patches (arrowheads) than mock-transfected cells. Error bars indicate SEM; * indicates p<0.05.
Figure S2. Multiple independent siRNA oligos induce similar phenotypes. Cells that are mock transfected or transfected with scrambled siRNA (scrambled siRNA corresponds to oligo p150 #1) show no difference in the velocity of nuclear rotations. Transfection of cells with alternate oligos against DHC and p150Glued show no difference when compared to one another, and all induce a significant reduction in angular velocity of rotations compared to mock-treated cells. siRNAs DHC #1, DHC #2, p150 #1, and p150 #2 all show similar inhibition of centrosome reorientation compared to either mock-treated or scrambled siRNA-transfected samples (data not shown). Error bars indicate SEM; ** indicates p<0.005; n.d. indicated no difference; n=10.
Figure S3. Nucleoli remain in position relative to one another in migrating cells. (A) Tracks of the paths of 4 nucleoli in a single nucleus that is not rotating. Each path was normalized to the center of the nucleus to correct for migration of the nucleus towards the wound. Each nucleolus stays stationary relative to its position in the nucleus. Axis labels are in microns, time is in minutes:seconds. (B) Tracks of the paths of 4 nucleoli in a single nucleus that is rotating while migrating into the wound. Each nucleolus follows a similar circular path in relation to the center of the nucleus, but all nucleoli do not change position relative to one another. Axis labels are in microns, time is in minutes:seconds.
Figure S4. Knockdown of dynein or dynactin does not alter actin distribution. Immunofluorescence of cells that were fixed 6 hours after monolayer wounding and stained with an antibody to α-tubulin (green) and TRITC-conjugated phalloidin (red). Bar, 10 microns.
Acknowledgments
This work was supported by NIH grant GM068591 to ELFH and NIH/NIA predoctoral training grant T32 AG0025 to JRL.
Abbreviations
- BB
Blebbistatin
- BFA
Brefeldin A
- DHC
Dynein heavy chain
- MT
Microtubule
- NZ
Nocodazole
References
- Anderson KI, Wang YL, Small JV. Coordination of protrusion and translocation of the keratocyte involves rolling of the cell body. J Cell Biol. 1996;134:1209–18. doi: 10.1083/jcb.134.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur EL. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007;104:10045–50. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–45. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Combs J, Kim SJ, Tan S, Ligon LA, Holzbaur EL, Kuhn J, Poenie M. Recruitment of dynein to the Jurkat immunological synapse. Proc Natl Acad Sci U S A. 2006;103:14883–8. doi: 10.1073/pnas.0600914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthesy-Theulaz I, Pauloin A, Pfeffer SR. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol. 1992;118:1333–45. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danowski BA, Khodjakov A, Wadsworth P. Centrosome behavior in motile HGF-treated PtK2 cells expressing GFP-gamma tubulin. Cell Motil Cytoskeleton. 2001;50:59–68. doi: 10.1002/cm.1041. [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–11. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander LL, Rubin LL. Acetylcholine receptor clustering and nuclear movement in muscle fibers in culture. J Cell Biol. 1987;104:87–95. doi: 10.1083/jcb.104.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–98. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, Bulinski JC. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci. 1999;112 (Pt 23):4243–55. doi: 10.1242/jcs.112.23.4243. [DOI] [PubMed] [Google Scholar]
- Fath KR, Trimbur GM, Burgess DR. Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J Cell Biol. 1994;126:661–75. doi: 10.1083/jcb.126.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ER, Gundersen GG. Real-time centrosome reorientation during fibroblast migration. Methods Enzymol. 2006;406:579–92. doi: 10.1016/S0076-6879(06)06045-9. [DOI] [PubMed] [Google Scholar]
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–63. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci U S A. 1988;85:5946–50. doi: 10.1073/pnas.85.16.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay M, De Boni U. Chromatin motion in neuronal interphase nuclei: changes induced by disruption of intermediate filaments. Cell Motil Cytoskeleton. 1991;18:63–75. doi: 10.1002/cm.970180107. [DOI] [PubMed] [Google Scholar]
- Herbomel P. Spinning nuclei in the brain of the zebrafish embryo. Curr Biol. 1999;9:R627–8. doi: 10.1016/s0960-9822(99)80407-2. [DOI] [PubMed] [Google Scholar]
- Ji JY, Lee RT, Vergnes L, Fong LG, Stewart CL, Reue K, Young SG, Zhang Q, Shanahan CM, Lammerding J. Cell nuclei spin in the absence of lamin b1. J Biol Chem. 2007;282:20015–26. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–64. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Nagasaki T, Gundersen GG. Low concentrations of nocodazole interfere with fibroblast locomotion without significantly affecting microtubule level: implications for the role of dynamic microtubules in cell locomotion. J Cell Sci. 1995;108 (Pt 11):3473–83. doi: 10.1242/jcs.108.11.3473. [DOI] [PubMed] [Google Scholar]
- Ligon LA, Karki S, Tokito M, Holzbaur EL. Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat Cell Biol. 2001;3:913–7. doi: 10.1038/ncb1001-913. [DOI] [PubMed] [Google Scholar]
- Magdalena J, Millard TH, Machesky LM. Microtubule involvement in NIH 3T3 Golgi and MTOC polarity establishment. J Cell Sci. 2003;116:743–56. doi: 10.1242/jcs.00288. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–36. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Mellor H. Cell motility: Golgi signalling shapes up to ship out. Curr Biol. 2004;14:R434–5. doi: 10.1016/j.cub.2004.05.038. [DOI] [PubMed] [Google Scholar]
- Morris NR. Nuclear positioning: the means is at the ends. Curr Opin Cell Biol. 2003;15:54–9. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Neujahr R, Albrecht R, Kohler J, Matzner M, Schwartz JM, Westphal M, Gerisch G. Microtubule-mediated centrosome motility and the positioning of cleavage furrows in multinucleate myosin II-null cells. J Cell Sci. 1998;111 (Pt 9):1227–40. doi: 10.1242/jcs.111.9.1227. [DOI] [PubMed] [Google Scholar]
- Paddock SW, Albrecht-Buehler G. The degree of coupling of nuclear rotation in binucleate 3T3 cells. Exp Cell Res. 1986;166:113–26. doi: 10.1016/0014-4827(86)90512-4. [DOI] [PubMed] [Google Scholar]
- Paddock SW, Albrecht-Buehler G. Rigidity of the nucleus during nuclear rotation in 3T3 cells. Exp Cell Res. 1988;175:409–13. doi: 10.1016/0014-4827(88)90205-4. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Joseph HL, Chen YJ, Dujardin DL, Alberts AS, Pfister KK, Vallee RB, Gundersen GG. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001;11:1536–41. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- Pomerat CM. Rotating nuclei in tissue cultures of adult human nasal mucosa. Exp Cell Res. 1953;5:191–6. doi: 10.1016/0014-4827(53)90104-9. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–6. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–34. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–72. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Shu T, Ayala R, Nguyen MD, Xie Z, Gleeson JG, Tsai LH. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 2004;44:263–77. doi: 10.1016/j.neuron.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–7. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–21. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokito MK, Howland DS, Lee VM, Holzbaur EL. Functionally distinct isoforms of dynactin are expressed in human neurons. Mol Biol Cell. 1996;7:1167–80. doi: 10.1091/mbc.7.8.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–45. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–8. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol Biol Cell. 1997;8:973–85. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–80. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Gundersen GG. Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 2006;16:67–9. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Xiang X, Fischer R. Nuclear migration and positioning in filamentous fungi. Fungal Genet Biol. 2004;41:411–9. doi: 10.1016/j.fgb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Yvon AM, Walker JW, Danowski B, Fagerstrom C, Khodjakov A, Wadsworth P. Centrosome reorientation in wound-edge cells is cell type specific. Mol Biol Cell. 2002;13:1871–80. doi: 10.1091/mbc.01-11-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. DHC knockdown decreases dynactin patches at leading edge. NIH/3T3 cells were transfected with mock siRNA or with siRNA targeting DHC and were grown to confluency. The monolayer was wounded. Six hours after wounding, cells were fixed and stained for tubulin (green) and p150Glued (red). Bar, 10 microns. DHC knockdown cells had less p150Glued leading edge patches (arrowheads) than mock-transfected cells. Error bars indicate SEM; * indicates p<0.05.
Figure S2. Multiple independent siRNA oligos induce similar phenotypes. Cells that are mock transfected or transfected with scrambled siRNA (scrambled siRNA corresponds to oligo p150 #1) show no difference in the velocity of nuclear rotations. Transfection of cells with alternate oligos against DHC and p150Glued show no difference when compared to one another, and all induce a significant reduction in angular velocity of rotations compared to mock-treated cells. siRNAs DHC #1, DHC #2, p150 #1, and p150 #2 all show similar inhibition of centrosome reorientation compared to either mock-treated or scrambled siRNA-transfected samples (data not shown). Error bars indicate SEM; ** indicates p<0.005; n.d. indicated no difference; n=10.
Figure S3. Nucleoli remain in position relative to one another in migrating cells. (A) Tracks of the paths of 4 nucleoli in a single nucleus that is not rotating. Each path was normalized to the center of the nucleus to correct for migration of the nucleus towards the wound. Each nucleolus stays stationary relative to its position in the nucleus. Axis labels are in microns, time is in minutes:seconds. (B) Tracks of the paths of 4 nucleoli in a single nucleus that is rotating while migrating into the wound. Each nucleolus follows a similar circular path in relation to the center of the nucleus, but all nucleoli do not change position relative to one another. Axis labels are in microns, time is in minutes:seconds.
Figure S4. Knockdown of dynein or dynactin does not alter actin distribution. Immunofluorescence of cells that were fixed 6 hours after monolayer wounding and stained with an antibody to α-tubulin (green) and TRITC-conjugated phalloidin (red). Bar, 10 microns.