Abstract
Various environmental and internal cues play essential roles in regulating diverse aspects of plant growth and development. Phytohormones usually coordinate multiple stimuli to directly regulate multiple developmental programs. Recent studies have provided progresses into the complexity of their cross talk. Particularly, the signaling pathways of various phytohormones have been revealed, leading to the discovery of the mechanisms of the interplay among different hormone signaling pathways. This review focuses on the recent advances of the signaling cross-talk between brassinosteroids and other hormones, including abscisic acid, auxin, gibberellins, ethylene and jasmonate.
Key words: brassinosteroids, plant hormone, abscisic acid, auxin, cross talk, signaling
Introduction
Different from animals, plants need to constantly regulate their developmental and physiological processes to respond to various external and internal stimuli in their sessile lifestyle. Studies have revealed that multiple networks among different hormones and their signaling pathways exist, and play key roles in establishing the developmental program in plants.1,2 In addition, a large set of microarray data demonstrated that many genes are co-regulated by different sets of hormones, suggesting the importance of hormone signaling in coordinately regulating the same biological process in plants.3,4 With the hormonal biosynthesis and signaling pathways established, recent studies have discovered many physiological and molecular mechanisms of hormonal cross-talk.
Brassinosteroids (BRs) distribute in almost all tissues of plants as a unique class of plant polyhydroxysteroids that are structurally analogous to the well-studied animal and insect steroids. Physiological studies, as well as the characterization of BR biosynthetic and signaling mutants, have demonstrated that BRs inhibit root elongation, and promote stem elongation, pollen tube growth, leaf bending, epinasty, early flowering and xylem differentiation, through regulating cell expansion, cell division, and cell differentiation.5,6
In the last decade, many key components of the BR signaling pathway have been isolated and characterized, although there is still a significant gap present in the pathway. The BR’s receptor BRI1 was discovered as a plasma membrane-localized leucine-rich-repeat receptor kinase.7 Direct binding of BRs to the island domain of BRI1,7–10 triggers the phosphorylation of BRI1 on multiple sites, which leads to the conformational change of the preformed homodimer of BRI1, dissociation of a negative regulator of BKI1 from plasma membrane, and association of BAK1 with BRI1,11,12 and further promote the activity of receptor complex.13,14 Two downstream components, a GSK3-like protein kinase BIN215,16 and a protein phosphatase BSU1,17 control the phosphorylation states of a family of plant-specific transcription factors, including BES1 and BZR1.18,19 Phosphorylated BES1 and BZR1 are less stable,18,20 are more likely to be retained to the cytoplasm by 14-3-3 proteins,21 and have less DNA binding affinity,22 than the dephosphorylated forms, which can directly bind to the promoter regions of BR-responsive genes and regulate BR-related biological processes. Recently, proteomic study identified other substrates of BRI1 kinase, three homologous BR-signaling kinases BSKs, that may activate downstream BR signal transduction.23 However, the links between the BRI1 receptor and BIN2 are still missing.
Recently, it is found that BRs can cross talk with numerous other hormones in regulating many developmental processes in plants. With many components of signaling pathways of BRs and other hormones found and characterized, it becomes possible to explore the mechanisms underlying the cross talk between BRs and other hormones. In this review, we will describe the recent advances in addressing how BRs interact with other plant hormones, including abscisic acid (ABA), auxin, gibberellins (GAs), ethylene, cytokinin and jasmonate (JA).
Brassinosteroids and Abscisic Acid
To date, many physiological and genetic studies have demonstrated that BRs and ABA can co-regulate many developmental processes.24–27 It is well known that ABA is required to establish seed dormancy during embryo maturation and to inhibit seed germination,24 whereas BRs promote seed germination, possibly through enhancing the embryo growth potential to antagonize ABA’s effect.24,27,28 It is reported that the BR related mutants also have altered response to ABA. In Arabidopsis, the BR biosynthetic mutant det2-1, the BR responsive mutant bri1, and the BKI1 overexpression line (BKI1-OX) are more sensitive to ABA on seed germination than wild types, although these mutants germinate well under normal growth condition;27,29 by contrast, overexpression of BRI1 leads to an enhanced resistance to ABA on seed germination.29 Moreover, studies also have revealed that mutants of possible components of ABA signaling pathway, gpa1, agb1 and gcr1 are more sensitive to ABA on seed germination, and also show altered sensitivities to BRs.30–32
Microarray data showed that BRs and ABA can co-regulate the expression of hundreds of genes.4 Specifically, more than thirty-five percent of BR-regulated genes are also regulated by ABA, indicating that ABA may regulate BR signaling.3,4 Early responsive genes of ABA, RD29A and RD22 are also regulated by BRs in Arabidopsis,33 further indicating the possible cross-talk between BR and ABA. However, whether the interaction between of BR and ABA is through their primary signaling cascades, parallel pathways, or secondary effect, is largely unknown.
Using genetic and biochemical approaches, Zhang et al.29 proposed a model to describe the interplay between BR and ABA primary signaling pathways in Arabidopsis. The authors found that exogenously-applied ABA rapidly inhibits BRs’ signaling outputs in wild-type and an ABA biosynthetic mutant aba1 background, and ABA has a similar effect in the BR receptor mutant bri1 background. However, when the downstream component BIN2 was inhibited by LiCl, or in the ABA insensitive mutants abi2 and abi1, the inhibitory effect of ABA on BR signaling outputs is annihilated. They further defined the facet of their cross talk, which is after BR perception, and at or before the negative regulator, BIN2. This work provides a reasonable explanation for why a large proportion of BR-responsive genes are also regulated by ABA, and gives significant insight into the molecular mechanisms by which BRs could interact with ABA.
It is likely that one or a few components in the ABA signaling pathway directly interact or regulate some specific components in the BR signaling pathway. With more components of both pathways identified or clarified, the complicated molecular nature of the cross-talk between BRs and ABA will be disclosed.
Brassinosteroids and Auxin
Physiological interaction between BRs and auxin has been observed with either hypocotyl elongation or root development.1,34 Previous studies suggest that BRs act synergistically with auxin to promote cell elongation, and mutants of either pathway show similar developmental defects, including dramatic dwarf phenotype.1,35 Utilizing hypocotyl elongation as an assay, researchers have found that the auxin-responsive mutants axr1, axr2, axr3, tir1 and arf2 have reduced sensitivity to BRs to a considerable degree,36,37 implying that BR’s function on hypocotyl elongation relies partially on a functional auxin signal transduction pathway. Similarly, BR treatment significantly enhances auxin response in hypocotyl elongation,37 and BR signaling mutant bri1 is insensitive to temperature-induced hypocotyl elongation, which is likely mediated by auxin.36 Taken together, the above evidence suggests that auxin response is also dependent on a functional BR signal transduction pathway. The fact that BRs and auxin cross-talk was further consolidated by root-development essay: the expression of several genes, such as AXR3/IAA17 in root, is altered by BR treatment or in BR-deficient mutants;38 the transcription levels of PIN4 and PIN7, which encode auxin transporters, are also repressed in BR deficient mutant det2, and BRs stimulate plant tropisms through regulating localization of PIN2 in root and modulating the auxin polar transport.36,39,40
Several studies have been conducted to illustrate the potential molecular mechanisms of the cross-talk between BRs and auxin. Numerous microarray data show that BRs and auxin share a number of early responsive genes, and many of them are involved in plant growth.3,4,41 A potential molecular link between BRs and auxin pathways was identified by a recent research, in which authors proposed that the regulatory regions of BR-responsive genes are enriched with predicted ARF-binding sites.36,41 Nemhauser et al.36 used Arabidopsis mutants with perturbed BRs or auxin signaling and demonstrated the interplay of these two hormones in the control of hypocotyl elongation. More recently, it was demonstrated that the BR signaling component BIN2 can directly interact with an auxin signaling component ARF2, a member of the Auxin Response Factor family of transcriptional regulators.37 Phosphorylation of ARF2 by BIN2 results in a loss of ARF2 DNA binding and repression activities. With all of the above evidences considered, the authors propose that BIN2 increases the expression of auxin-induced genes by directly inactivating the repressor ARF2 and leads to the synergistic enhancement of transcription.
Most likely, this mode of cross-talk is only part, if not all, of the mechanism of the interplay between BRs and auxin.37 Apparently, other potential pathways are present to illustrate the complexity of their interaction.
Brassinosteroids and Gibberellins
GAs are a class of phytohormones that mainly promote plant growth, including promoting stem elongation, seed germination, and flowering, and inhibiting leaf and fruit senescence.42,43 However, it is poorly understood whether BRs cross-talk with GAs. Previous physiological studies have pointed out that BRs and GAs enhance plant growth in an additive manner,44 indicating that the two hormones may act independently at the cellular level.
However, there is accumulating evidence to support that cross-talk between BRs and GAs signaling pathways may also occur either synergistically or antagonistically. First, physiological study suggests that BRs could partially rescue the germination of GA biosynthetic and GA insensitive mutants, possibly via embryo expansion.27 Second, G-proteins (GCR1, GPA1 and AGB1) deficient mutants have reduced sensitivities to both BRs and GAs in seed germination.31 Finally, microarray data both in Arabidopsis and rice shows that the expression of a large number of genes is coordinately regulated by BRs and GAs.3,4,45 Studies in rice also show that OsGSR1 promotes BR biosynthesis by directly regulating a BR biosynthetic enzyme, and when OsGSR1 was knocked-down, an elevated level of endogenous GAs was observed, suggesting that OsGSR1 may mediate the two pathways.
Studies also showed that BRs and GAs antagonistically regulate the accumulation of mRNAs of a GA-responsive gene GASA1 and GA-suppressed gene GA5.46 Additionally, BRs and GAs regulate the expression of GA-responsive gene γ-TIP in an antagonistic manner, and the mRNA level of γ-TIP accumulates ectopically in BR biosynthetic and responsive mutants.47 Given that γ-TIP encodes a tonoplast-intrinsic aquaporin, it is reasonable to suspect that BRs and GAs regulate turgor pressure or solute flow antagonistically.
Unlike the cross talk between BRs and ABA or BRs and auxin, our understanding on the mode of the cross-talk between BRs and GAs stays at the physiological level. Whether their interaction is through the modification or interaction of their primary signaling cascades, or parallel pathways, remains unclear.
BR and Other Hormones
Ethylene is a gaseous hormone that regulates various plant developmental processes, such as seed germination, abscission, senescence, plant defense and fruit ripening.48,49 It was found that the expression of BRP, a BR-repressed reporter gene, can be activated by ethylene.50 BRs regulate the biosynthesis of ethylene via stabilizing the ACC synthase that catalyzes a ratelimiting step in ethylene biosynthesis.51 Other research implies that ethylene and BR may promote the biosynthesis of each other.52 Furthermore, both ethylene and BRs can promote the growth of cotton fiber. However, ethylene could overcome the inhibitory effect of BRZ on fiber cells, while BRs counteract the effect of AVG, which is the ethylene biosynthesis inhibitor, on fiber elongation to a much lesser degree, suggesting BRs act on the upstream of ethylene in cotton fiber elongation.52
There are a few studies indicating that BRs may also interact with other hormones. A previous study implies that BRs may influence stress responses of plants by stimulating synthesis of JA. The expression of the OPR3, encoding a 12-oxo-phytodienoic acid reductase, is induced by BRs and JA, depending on environmental and developmental conditions, which drops a hint on the possible relationship between BRs action and JA synthesis.53
Perspectives
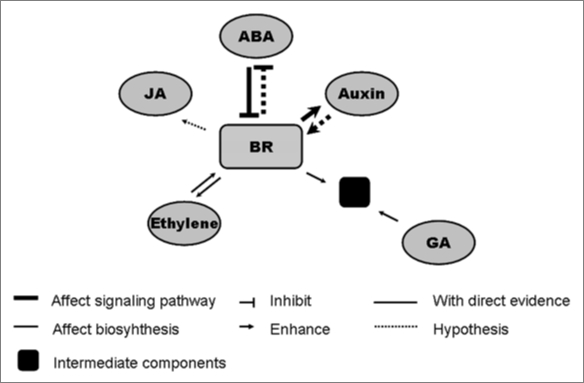
In summary, a proposed network of BRs interacting with multiple hormones is summarized in Figure 1. Many great progresses have been made in hormonal interaction, but most of these studies remain at the physiological level. For this reason, a shift of priority in researches from physiological to molecular evidences, is bound to be intensified. Within the foreseeable future, studies on the relationship between BRs and other hormones at molecular and biochemical level will draw us a systematic and clarified picture depicting the sophisticated interactive network of BRs and multiple hormones.
Figure 1.
A proposed network of brassinosteroids interacting with multiple plant hormones. BR signaling pathway is primarily inhibited by ABA signaling through certain unknown components, and BR may also regulate the ABA signaling outputs. BRs regulate auxin signaling via BIN2, while, reciprocally, BR signaling may also be regulated by auxin. BR affects the biosynthesis of ethylene and, in all probability, that of JA. Also, BR co-regulates physiological processes (black boxes), such as solute flow, with GA through primary signaling pathway or the level of phytohormones.
Acknowledgements
This work was supported by a start-up fund of Fudan University to X.W., grants of National Natural Science Foundation of China (Grant: 30671118; 30871330; 90817004) to X.W., a grant of Shanghai Pujiang Project (Grant: 07pj14014) to X.W.
Abbreviations
- BR
brassinosteroid
- ABA
abscisic acid
- GA
gibberellin
- JA
jasmonate
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9903
References
- 1.Teale WD, Ditengou FA, Dovzhenko AD, Li X, Molendijk AM, Ruperti B, et al. Auxin as a model for the integration of hormonal signal processing and transduction. Mol Plant. 2008;1:229–237. doi: 10.1093/mp/ssn006. [DOI] [PubMed] [Google Scholar]
- 2.Weiss D, Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007;144:1240–1246. doi: 10.1104/pp.107.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 4.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 5.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haubrick LL, Assmann SM. Brassinosteroids and plant function: some clues, more puzzles. Plant Cell Environ. 2006;29:446–457. doi: 10.1111/j.1365-3040.2005.01481.x. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 8.He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 9.Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 10.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROIDINSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 17.Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 19.He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 23.Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 25.Assmann SM. Plant G proteins, phytohormones and plasticity: three questions and a speculation. Sci STKE. 2004;2004:20. doi: 10.1126/stke.2642004re20. [DOI] [PubMed] [Google Scholar]
- 26.Perfus-Barbeoch L, Jones A, Assmann S. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001;125:763–769. doi: 10.1104/pp.125.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leubner-Metzger G. Brassinosteroids and gibberellins promote tobacco seed germination by distinct pathways. Planta. 2001;213:758–763. doi: 10.1007/s004250100542. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Cai Z, Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey S, Chen J-G, Jones AM, Assmann SM. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 2006;141:243–256. doi: 10.1104/pp.106.079038. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, et al. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–915. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y, Wang S, Asami T, Chen JG. Loss-offunction mutations in the Arabidopsis heterotrimeric G-protein alpha subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant Cell Physiol. 2008;49:1013–1024. doi: 10.1093/pcp/pcn078. [DOI] [PubMed] [Google Scholar]
- 33.Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta. 2007;225:353–364. doi: 10.1007/s00425-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 34.Hardtke CS. Transcriptional auxin-brassinosteroid crosstalk: who’s talking? Bioessays. 2007;29:1115–1123. doi: 10.1002/bies.20653. [DOI] [PubMed] [Google Scholar]
- 35.Halliday KJ. Plant hormones: the interplay of brassinosteroids and auxin. Curr Biol. 2004;14:1008–1010. doi: 10.1016/j.cub.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:1460–1471. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H, Park PJ, Hwang HJ, Lee SY, Oh MH, Kim SG. Brassinosteroid signals control expression of the AXR3/IAA17 gene in the cross-talk point with auxin in root development. Biosci Biotechnol Biochem. 2006;70:768–773. doi: 10.1271/bbb.70.768. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura A, Goda H, Shimada Y, Yoshida S. Brassinosteroid selectively regulates PIN gene expression in Arabidopsis. Biosci Biotechnol Biochem. 2004;68:952–954. doi: 10.1271/bbb.68.952. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Xu J, Xu ZH, Xue HW. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell. 2005;17:2738–2753. doi: 10.1105/tpc.105.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15:1591. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swain S, Singh D. Tall tales from sly dwarves: novel functions of gibberellins in plant development. Trends Plant Sci. 2005;10:123–129. doi: 10.1016/j.tplants.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Gregory L, Mandava N. The activity and interaction of brassinolide and gibberellic acid in mung bean epicotyls. Physiol Plant. 1982;54:239–243. [Google Scholar]
- 45.Yang GX, Jan A, Shen SH, Yazaki J, Ishikawa M, Shimatani Z, et al. Microarray analysis of brassinosteroids- and gibberellin-regulated gene expression in rice seedlings. Mol Genet Genomics. 2004;271:468–478. doi: 10.1007/s00438-004-0998-4. [DOI] [PubMed] [Google Scholar]
- 46.Bouquin T, Meier C, Foster R, Nielsen M, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127:450–458. [PMC free article] [PubMed] [Google Scholar]
- 47.Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- 48.Wang K, Li H, Ecker J. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14:131–151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grbic V, Bleecker A. Ethylene regulates the timing of leaf senescence. Plant J. 1995;8:595–602. [Google Scholar]
- 50.Gendron J, Haque A, Gendron N, Chang T, Asami T, Wang Z. Chemical Genetic Dissection of Brassinosteroid-Ethylene Interaction. Mol Plant. 2008;1:368–379. doi: 10.1093/mp/ssn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen M, Chae H, Kieber J. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009;57:606–614. doi: 10.1111/j.1365-313X.2008.03711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y, Zhu S, Mao X, Feng J, Qin Y, Zhang L, et al. Transcriptome profiling, molecular biological and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müssig C, Biesgen C, Lisso J, Uwer U, Weiler E, Altmann T. A novel stress-inducible 12-oxophytodienoate reductase from Arabidopsis thaliana provides a potential link between brassinosteroid-action and jasmonic-acid synthesis. J Plant Physiol. 2000;157:143–152. [Google Scholar]