Abstract
In plants, cytokinin (CK) perception and signaling pathway is composed by a histidine kinase receptor (HK) and a response regulator (RR), the signal being mediated by a histidine phosphotransfer (HPt), as described in Arabidopsis, maize and rice. From database searches we identified in grapevine three HKs, three HPs, four A-type RRs and six B-type RRs, suggesting a common mechanism for grapevine. The phylogenetic analysis of these Vitis genes showed a variable but high degree of homology with Arabidopsis sequences. When sulfate was withdrawn from the culture medium (−S) of in vitro Vitis shoots, we assessed a significant reduction in shoot branching. To ascertain the crosstalk of S status with CK signaling in grapevine, control and −S grown shoots and control, −S and −CK cell suspensions were used as experimental systems. Real-time PCR was elected to quantify the expression of key genes. The expression of CK receptor genes was downregulated in −S cells while not affected in −CK cells. In differentiated shoots no response to −S was observed on those genes. A-type VvRRa4 was downregulated in −S or −CK cells while Vitis B-type RRs did not respond either to CK or S starvation. The results suggest that Vitis CK signaling pathway is affected by −S, although differently according to the model system. Transcription of Vitis apical meristem-identity genes VvWUS, VvCLV and VvSTM and axillary meristem genes VvBRC1, VvBRC2, VvLAS, VvRAX and VvREV was estimated and VvSTM and VvLAS showed to be downregulated in −S. Then, the expression levels of VvSTM and VvLAS make them strong candidates to be associated with the branching pattern of Vitis shoots in −S.
Key words: apical meristem genes, axillary meristem genes, cytokinin signaling genes, sulfur, vitis
Introduction
Cytokinins are essential plant hormones that control various aspects of plant growth and development such as cell division, shoot formation, senescence and chloroplast development.1
The analysis of the mechanism of cytokinin perception and signaling in Arabidopsis referred to as a “two-component system”,1,2 resulted in a model that distinguishes four major steps: (1) cytokinin sensing and initiation of signaling by histidine kinase receptor (HKs); (2) phosphoryl group transfer to histidine phosphotransfer (HPts) and their translocation to the nucleus; (3) phosphotransfer to nuclear B-type response regulator (RRs), which activate transcription; and (4) negative feedback through cytokinin-inducible A-type RRs, which are the products of early cytokinin target genes.3 Three Arabidopsis HK genes (AHK4/CRE1, AHK2 and AHK3),2 three maize HK genes (ZmHK1, ZmHK2 and ZmHZ3a)4 and five HK in rice (OHK1, OHK2, OHK3, OHK4 and OHK5)5 were described. The identification of orthologs for cytokinin signaling components in other plant species suggests evolutionary conservation of this pathway.3 The large majority of HKs are membrane-bound, homodimeric proteins with a C-terminal cytoplasmic kinase domain6 and a extracytoplasmic CHASE domain which is the putative recognition site for cytokinin.7,8 The Arabidopsis triple mutant for the three HK genes showed no cytokinin response, but is not lethal, suggesting that either cytokinin is not essential to Arabidopsis growth or another unknown signaling pathway of cytokinin may exist.9
The HPts translocate the phosphate group perceived from HKs from the cytoplasm to the nucleus, where it is transferred to RRs.10 RRs can be broadly classified into two groups, A-type and B-type RR. The B-type RRs have a phosphorelatable receiver domain at their N-terminus and a GARP DNA-binding domain in the midpoint of the sequence; when phosphorylated the activated B-type RRs induce or repress the expression of target genes.2,11 Arabidopsis B-type RRs were found in de nucleus and the structural analysis confirmed their binding to DNA. Unlike the expression pattern of A-type RR genes, the steady-state levels of B-type RR transcripts are apparently not affected by the application of cytokinin or other plant hormones. Conversely, expression of A-type RRs genes are induced by cytokinin and display properties for cytokinin primary-response.12
The primary response of most plant systems under sulfur (S) deficiency is a clear upregulation (or de-repression) of sulfate transporters at the transcription level13–17 which is strongly downregulated (or repressed) by sulfate repletion.16,17 The evidence for the involvement of cytokinin signaling under S deficiency conditions came from the genetic study of the Arabidopsis cytokinin receptor mutant cre1.18 The mutant in CRE1 displayed a lower repression of both the SULTR 1;2 expression and S uptake in S full conditions and the authors proposed the role of cytokinin as a negative regulator and suggested two independent modes of regulation for sulfate acquisition, one induced by sulfate depletion and the other cytokinin dependent. If cytokinins were the sole mediators of sulfur starvation signal for regulating the expression of sulfur responsive genes, their concentration in −S status tissues was expected to increase. However in leaf tissues, cytokinin concentration did not increase significantly after two days of sulfate starvation suggesting that it is unlikely that, at least in leaves, cytokinins mediate directly sulfur-deficiency responses.19
Other macronutrients, e.g., phosphate (P), may respond to the application of cytokinin. It has been shown that exogenous application of cytokinin represses the induction of many P starvation-responsive genes in Arabidopsis,20 and this effect is attenuated in cre1 mutants, implicating cytokinin in the negative regulation of P starvation responses.21
Previous work in our group (unpublished) has shown that sulfate starvation impairs shoot growth and branching of in vitro grapevine, therefore suppresses the reversion of apical dominance inhibition as triggered by routinely added cytokinin to in vitro multiplication media.22,23 Two hypothesis for such symptoms can be proposed: the effect of sulfur deficiency on cytokinin signal pathway (CSP) genes or its interference with meristem identity genes. Therefore, and to elucidate those hypothesis, the crosstalk between sulfur status, cytokinin signaling and meristem genes prompted further investigation.
Post-embryonic development in higher plants is characterized by the reiterative formation of lateral organs from the flanks of the apical meristems.24,25 The shoot apical meristem (SAM) is composed of three clonally distinct layers of cells,24 which contain a population of pluripotent stem cells, with three primary functions: formation of lateral organs, such as leaves; shoot axis growth; the pool of stem cells required for further growth.24,25 Several groups of transcription factors have been shown to take part in SAM differentiation. The gene Shootmeristemless (STM) is required for the initiation and maintenance of the SAM in Arabidopsis.26 Embryos homozygous for strong loss-of-function mutations in the STM form cotyledons and other embryonic structures but fail to establish a population of self-renewing stem cells.27 Another early gene expressed is WUSCHEL (WUS), a gene required to produce the stem cell maintenance signal.28 WUS expression is under negative control by the CLAVATA genes (CLV1, CLV2 and CLV3).29,30 Homeostasis of the stem cell population may be achieved through feedback regulation, whereby changes in stem cell number result in corresponding changes in CLV3 expression level, and adjustment of WUS expression.31
Secondary axillary meristems (AM) formed in the axil of each leaf are initiated in an acropetal order, from the axils of mature leaves to younger leaves axils.32 Genes such as LATERAL SUPPRESSOR (LAS)33 or REGULATOR OF AXILLARY MERISTEMS (RAX)34,35 maintain the meristematic potential of leaves and allow the organization of stem cells.35 Genes promoting local bud arrest have been described in monocots, e.g., teosinte branched1 (tb1) from maize36 and its rice ortholog, Os tb1,37,38 which are expressed in AM of arrested buds.38,39 Other gene also involved in early stages of axillary meristem is REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV/IFL1).40 In 2007 Aguilar-Martinez et al.41 described BRANCHED1 (BRC1) and BRANCHED2 (BRC2) as the genes most closely related to tb1 in Arabidopsis.
In the present work, Vitis cell suspensions and in vitro shoots were used as experimental models to assess the effects of sulfur deficiency through the analysis of growth parameters. Taking advantage of Vitis genome sequencing,42,43 we performed a database search of genes associated to the CSP and meristem identity and the expression of candidate genes was analysed by quantitative real time PCR in the different biological systems under sulfur sufficient and deficient conditions and in the presence or absence of cytokinin. The discussion of the results obtained so far allowed fostering tentative, certainly far from definitive, conclusions: −S status disrupted markedly the expression of different CSP genes while the downregulation of VvRRas in −CK status confirmed their role as CK primary response genes; growth and branching pattern of Vitis shoots in −S status is mostly explained by the expression profiles of VvSTM and VvLAS.
Results and Discussion
Effects of sulfur and cytokinin deficiency on cell and plantlet growth. The measurement of physiological parameters is a valuable mean to assess the effect of imposed conditions on the functioning of biological experimental systems. To determine if grapevine (Vitis vinifera L.) cytokinin pathway and meristematic genes were affected by sulfate deficiency, two experimental models were used: cell cultures and in vitro plantlets.
The effect of sulfate or cytokinin withdraw on the growth of Vitis cells, was determined through cell biomass along the cell growth cycle, on a weekly basis. The fresh weight (FW) of Vitis cells grown in full MS medium as control (+S), without sulfate (−S) or without cytokinin (−CK) is presented in Table 1 and shows that the biomass of Vitis cells was significantly affected by cytokinin depletion only at the seventh day. Considering cell number and cell dimension measured at that point (Suppl. Table 1), mitosis was not blocked so far but Vitis cells adjusted to −CK status at the level of cell expansion.44 Conversely, shoot FW from in vitro plantlets was significantly affected by the sulfur deficient conditions: FW of in vitro control shoots (+S) almost doubled in 2 weeks while no increase was detected in FW of −S shoots. The multiplication rate of control shoots (+S) increased along the 2 weeks culture, while the number of branches maintained constant in −S status (Table 2).
Table 1.
Fresh weight of Vitis cells grown for 7 days in control medium, without sulphur (−S) and in the absence of cytokinin (−CK)
| Fresh weight (g) | |||
| Days | Control | −S | −CK |
| 0 | 3.03fgh ± 0.37 | 3.03fgh ± 0.37 | 3.68efg ± 0.25 |
| 1 | 2.78gh ± 0.02 | 2.69h ± 0.09 | 3.22fgh ± 0.32 |
| 3 | 3.79ef ± 0.78 | 4.22e ± 0.40 | 4.4de ± 0.5 |
| 4 | 4.47de ± 0.71 | 5.93bc ± 0.78 | 5.15cd ± 0.62 |
| 6 | 6.26b ± 0.33 | 7.53a ± 0.30 | 6.12b ± 0.42 |
| 7 | 8.14a ± 1.21 | 7.72a ± 0.43 | 6.68b ± 0.25 |
Values of FW are means of three samples taken randomly ±SD. Different letters indicate significant differences at p < 0.05.
Table 2.
Fresh weight and number of shoots of grapevine in vitro shoots in −S and +S growing conditions
| Fresh weight (g) | Number of shoots | |||
| Weeks | +S | −S | +S | −S |
| 0 | 0.12b | 0.12b | 1b | 1b |
| 2 | 0.19a ± 0.02 | 0.09b ± 0.03 | 1.75a ± 0.49 | 1.03b ± 0.16 |
In vitro shoots were grown for 2 weeks in medium with (+S) and without (−S) sulphur. Each value is the mean of the FW of ten in vitro shoots ±SD. Different letters indicate significant differences at p < 0.05.
The enhanced branching verified in in vitro multiplication is based on the inhibition of apical dominance by exogenous cytokinin.22,23 Then, the decrease in branching rate by −S nutrition could result from an effect on cytokinin signal transduction and lead us to investigate the effect of sulfur depletion on changes in the transcription of cytokinin signaling-related genes.
Identification of vitis genes associated to cytokinin signaling under sulfur stress conditions.
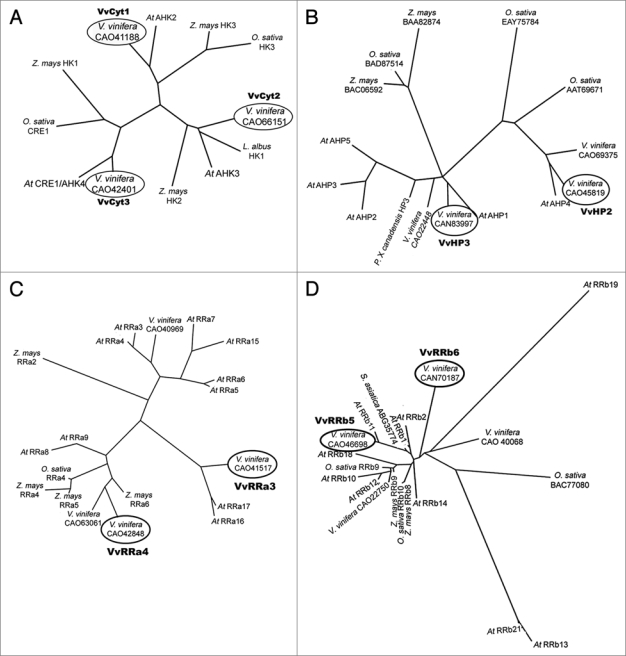
The initial approach was to evaluate the phylogenetic relationship between Vitis cytokinin signaling genes and annotated genes in other species and to register them at NCBI databases. Using the cytokinin receptors annotated in Arabidopsis AHK2, CRE1/AHK4, and AHK3, as initial queries we identified three Histidine Kinases (HK) in Vitis genome, VvCyt1, VvCyt2 and VvCyt3 with a high degree of similarity to Arabidopsis,2 maize4 and rice5 HKs and registered VvCyt1 (FJ822975) and VvCyt3 (FJ822976). More specifically, VvCyt1 shares 65% identity with AHK2, VvCyt2 is similar to CRE1/AHK4 in 69% of the amino acids and VvCyt3 shares 68% homology with AHK3 (Fig. 1A). Four Vitis HPt genes, VvHP1, VvHP2, VvHP3 and VvHP4 were identified from which VvHP2 (FJ822977), VvHP3 (FJ822978) and VvHP4 (FJ822979) were registered. The amino acid sequence of VvHP3 shares 59% identity with AHP1 while the homology of VvHP2 is similar to AHP4 in 72% of the amino acids and even higher to some rice sequences (Fig. 1B). Using Arabidopsis RRa17, described as responding to mineral stress,45 four Vitis RRas amino acid sequences were identified corresponding to the coding genes VvRRa1 (FJ822980), VvRRa2 (FJ822981), VvRRa3 (FJ822982) and VvRRa4 (FJ822983). The presence and homology of Vitis A-type RRs to other species, e.g., rice5 (Fig. 1C), suggests that they may play a role as A-type RRs. With Arabidopsis RRb18 protein45 as query, six RRbs genes were identified VvRRb1, VvRRb2, VvRRb3, VvRRb4, VvRRb5 and VvRRb6, from which VvRRb1 (FJ822984), VvRRb3 (FJ822985), VvRRb4 (FJ822986), VvRRb5 (FJ822987) and VvRRb6 (FJ822988) were registered. The 56% identity of VvRRb5 to ARRb11 and 45% homology of VvRRb6 to ARR18, Z. mays and rice RRb9,5 (Fig. 1D) suggest that these genes may play a role as B-type ARRs. These results suggest that the “two-component system” predicted for cytokinin signaling pathway in Arabidopsis2 and rice5 also applies to Vitis. According to Müller and Sheen3 cytokinin signaling pathway is conserved in all plant species, suggesting an evolutionary conservation of this pathway.
Figure 1.
Pylogenetic tree of (A) HKs, accession numbers from NCBI: Arabidopsis: At CRE1/AHK4, BAB33310; At AHK2, NP_568532; At AHK3, NP_564276; Lupines: L. albus HK1, ABJ74169; Maize: Z. mays HK2, BAD01584; Z. mays HK3, BAD01586; Z. mays HK1, NP_001104859; Rice: O. sativa HK3, AAP53311; O. sativa CRE1, ABF89563; Vitis: V. vinifera, CA O42401; V. vinifera, CA O41188; V. vinifera, CAO66151, (B) HPts, accession numbers from NCBI: Arabidopsis: At AHP1, NP_188788; At AHP2, NP_850649; At AHP3, NP_198750; At AHP4, NP_566544; At AHP5, NP_563684; Maize: Z. mays, BAC 06592; Z. mays, BAA82874; Populus: P. x canadensis HP3, CAH 56501; Rice: O. sativa, ATT69671; O. sativa, BAD87514; O. sativa, EAY75784; Vitis: V. vinifera, CAO71051; V. vinifera, CAN83997; V. vinifera, CAN69375; V. vinifera, CAO45819; (C) A-type RRs, accession numbers from NCBI: Arabidopsis: At RRa3, NP_176202; At RRa4, NP_172517; At RRa5, NP_190393; At RRa6, NP_201097; At RRa7, NP_17339; At RRa8, NP_181663; At RRa9, NP_191263; At RRa15, NP_177627; At RRa16, NP_181599; At RRa17, NP_567037; Maize: Z. mays RRa2, BAA 85113; Z. mays RRa4, BAB20579; Z. mays RRa6, BAB20581; Z. mays RRa5, BAB20580; Rice: O. sativa RRa4, NP_001045420; Vitis: V. vinifera, CAO41517; V. vinifera, CA O42848; V. vinifera, CA O40969; V. vinifera, CAO63061 and (D) B-type RRs accession numbers from NCBI: Arabidopsis: At RRb1, NP_850600; At RRb2, NP_193346; At RRb10, NP_194920; At RRb11, NP_176938; At RRb12, NP_180090; At RRb13, NP_180275; At RRb14, NP_178285; At RRb18, Q9FGT7; At RRb19, NP_175345; At RRb21, NP_196338; Maize: Z. mays RRb8, BAB41137; Z. mays RRb9, BAB55874; Rice: O. sativa RRb10, AAN52739; O. sativa RRb9, BAD72541; O. Sativa, BAC77080; S. asiatica, ABG35774; Vitis: V. vinifera, CAO22750; V. vinifera, CA O40068; V. vinifera, CAN70187; V. vinifera, CAO46698. The circled assignments correspond to sequences analyzed by real time PCR for Vitis gene expression. The phylogenetic tree based on the amino acid sequence was constructed using the PHYLIP programs, PRODIST and NEIGHBOR.55
Expression of identified CK signaling genes: Effect of −S and −CK conditions.
To investigate the effect of sulfur or cytokinin deficiency on the expression level of CK signaling expressed genes, the transcription of VvCyt1, VvCyt2, VvCyt3, VvHP2, VvHP3, VvRRa3, VvRRa4, VvRRb5 and VvRRb6 was quantified in −S and −CK culture cells and in −S in vitro shoots. The transcripts levels were measured by real-time PCR and expressed in fold change as compared to control conditions. Real-time PCR is increasingly used in plants to study the expression levels of particular genes, allowing the detection of a given target gene in a rapid, specific and sensitive manner.46
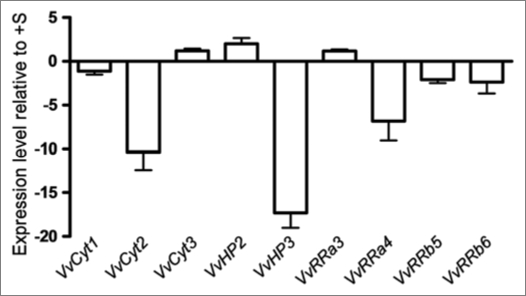
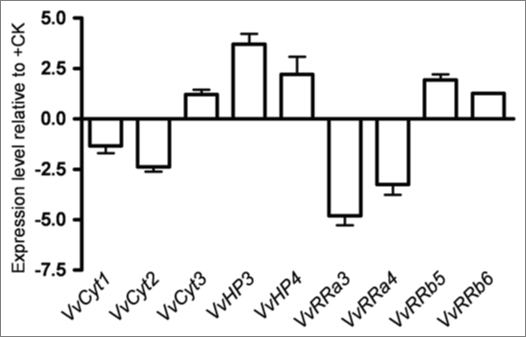
Cultured cells. In Figure 2 are shown the expression levels of the expressed genes in Vitis cells at day 7 of growth in −S conditions as quantified by real time PCR. VvCyt2, VvHP3 and VvRRa4 were significantly downregulated in −S status cells while in −CK cells only VvRRa3 is significantly below the threshold (Fig. 3) suggesting that the transcription of cytokinin receptors and HP genes is independent of the hormone availability. In Arabidopsis the triple mutation in the cytokinin receptors causes CK resistance, a strong growth inhibition and a loss of CSP signal.9 The downregulation of cytokinin receptor in −S status cells could depict the same type of effect as described by Higuchi et al.9 Regarding A-type RRs, VvRRa4 was downregulated in −S grown cells (Fig. 2) and the expression of VvRRa3 and VvRRa4 also decreased in −CK cells (Fig. 3). Hwang and Sheen2 propose A-type ARRs as negative regulators of CSP because these genes are able to disconnect CK signal by a feedback loop. In Vitis cells cytokinin depletion seems to interrupt the CSP at RRa level, confirming A-type RRs as cytokinin primary-response12 and negative regulator2 genes. From Arabidopsis results, the B-type RR genes are apparently not induced by cytokinin but can be involved in the transcription of cytokinin primary target genes.2,12,47 Vitis B-type RRs did not respond either to cytokinin or to S starvation. Taken the Vitis cell results as a whole, only VvRRas respond to CK withdraw, confirming their role as CK primary response genes2,12 while −S status disrupted markedly genes of different steps of the CSP.
Figure 2.
Expression levels of VvCyt1, VvCyt2, VvCyt3, VvHP2, VvHP3, VvRRa3, VvRRa4, VvRRb5 and VvRRb6 as quantified by real time PCR in Vitis cells at day 7 of growth in −S conditions.
Figure 3.
Expression levels of VvCyt1, VvCyt2, VvCyt3, VvHP2, VvHP3, VvRRa3, VvRRa4, VvRRb5 and VvRRb6 genes as quantified by real time PCR in Vitis cells at day 7 of growth in −CK conditions.
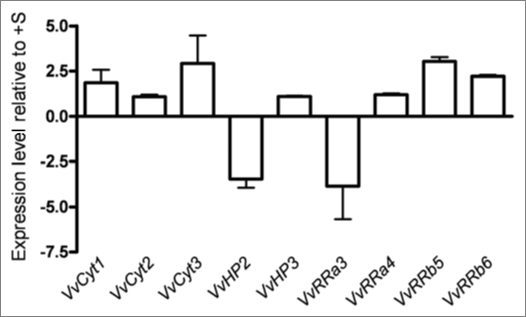
In vitro shoots. In in vitro propagated shoots under −S condition the variation of CSP genes was within the threshold limits (Fig. 4) steering the different response of CSP genes to sulfur depletion in dedifferentiated and differentiated cells. These results follow the same trend as whole plants or plant organs adjustment to nutrient supply.48
Figure 4.
Expression levels of VvCyt1, VvCyt2, VvCyt3, VvHP2, VvHP3, VvRRa3, VvRRa4, VvRRb5 and VvRRb6 genes as quantified by real time PCR in Vitis shoots after 2 weeks of −S growth conditions.
In order to explain the lack of branches in shoots under −S condition our next step was to analyze the level of expression of apical and axillary meristem genes in response to sulfur nutrition.
Expression of the identified apical and axillary meristem genes in shoots: effect of sulfur depletion.
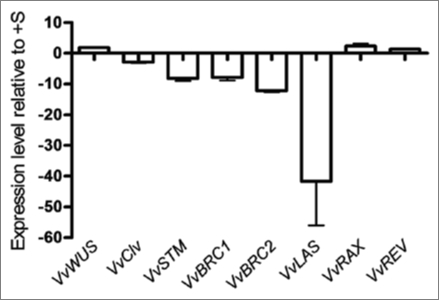
Plant shoot is derived from the primary shoot apical meristem (SAM) whose activity is regulated by environmental inputs, such as nutrient availability, that can be relayed by plant hormones.32 Although the expression of CSP genes did not vary significantly in −S in vitro Vitis shoots, a strong effect on growth and branching was evident (Table 2), what prompted to study the effect of −S status on apical and axillary meristem genes in order to investigate the effect of −S on shoot growth and branching on a different level of control. We identified three apical meristem genes VvWUS (FJ822989), VvCLV and VvSTM and found that sulfur starvation alters the expression of VvSTM by a downregulation in second week (Fig. 5). One of the earliest indicators of a switch in fate from indeterminate meristem to determinate leaf primordium is the downregulation of KNOX1 genes orthologous to STM in the incipient primordial.26 In Arabidopsis it is well demonstrated that STM is required for the establishment49 or maintenance25 of stem cell fate and that its loss-of-function results in loss of the SAM, in agreement with its role in meristem identity acquisition/maintenance.27 Since Vitis VvSTM is downregulated in −S conditions, it is reasonable to assume that the apical meristem is not properly formed in sulfur starvation, and this may be one of the reasons for Vitis shoot growth impairment.
Figure 5.
Expression levels of Vitis apical (VvWUS, VvCLV and VvSTM) and axillary (VvBRC1, VvBRC2, VvLAS, VvRAX and VvREV) meristem genes as quantified by real time PCR in Vitis shoots after 2 weeks of −S growth conditions.
Shoot branching is the process by which axillary buds, located on the axil of a leaf, develop and form new branches. Shoot multiplication in vitro is based on the inhibition of apical dominance and forced branching by adding exogenous cytokinin. In vitro shoots might then comprise a suitable model for analyzing axillary meristem gene expression. Among the genes responsible for axillary meristem fate are Arabidopsis transcription factors BRANCHED1 (BRC1) and BRANCHED2 (BRC2)41 the transcription factor LATERAL SUPPRESSOR (LAS)33 and the REGULATOR OF AXILLARY MERISTEMS (RAX)34,35 and REVOLUTA (REV)40 genes. The amplification of the transcripts with specific primers designed for each gene allowed to identify in Vitis the homologous VvBRC1, VvBRC2, VvLAS (FJ822990), VvRAX and VvREV genes. VvRAX and VvREV expression was unaffected by sulfur starvation, while the other genes were downregulated (Fig. 5). BRC1 responds to developmental and environmental stimuli controlling branching by arresting bud development when expressed in developing buds.41 Genes promoting bud arrest have been described in monocots, e.g., teosinte branced1 (tb1) from maize36 and their homologs from rice Os tb1.37,38 In Arabidopsis when BRC1 and BRC2 are downregulated the number of rosette branches increase, indicating that BRC1 and BRC2 retard all stages of bud development. LAS is necessary for axillary meristem initiation and exerts important roles in diverse processes such as signal transduction, meristem maintenance and development.50 LAS transcripts accumulate in the axils of all primordial derived from the SAM and mutations in this gene showed a severe reduction in the number of axillary shoots.33 Comparison of the phenotypes of tomato and Arabidopsis lateral suppressor mutants revealed that the described control mechanism is conserved during evolution.39 We forward the hypothesis that the decrease in Vitis branching is a consequence of VvLAS downregulation (Fig. 5). The apparent contradiction between the expression levels of VvBRC1, VvBRC2 and VvLAS may be solved by the VvLAS function upstream of VvBRC1 and VvBRC2, preventing any phenotype expression due to downstream genes. Taken together, the expression profiles of VvSTM and VvLAS could explain the growth and branching pattern of Vitis shoots in response to S deficiency.
Material and Methods
Plant material and growth conditions.
Cell suspensions and in vitro shoots of Vitis vinifera var. Touriga Nacional were used in all experiments. Cell suspensions were obtained by adapting to liquid culture Vitis vinifera callus grown as described in Jackson et al.51 Ca 4 g callus tissue was dispersed in 50 ml of liquid MS52 (Duchefa Biochemie, Haarlem, NL) supplemented with 2.5 µM 2,4-D, 1 µM kinetin, 5 g l−1 PVP-40T, 20 g l−1, sucrose, pH 5.7, in 250 ml flasks. The cultures growing in a rotary shaker at 100 rpm, in the dark, at 25°C were sub-cultured weekly by diluting 25 ml culture into 25 ml of fresh medium.
The growth medium and conditions for cells in the absence of cytokinin (−CK) were the same as above except that kinetin was withdrawn from the medium. For sulfate deprivation (−S), after two cycles in full MS medium (1.5 mM sulfate) a modified MS medium where sulfates were substituted for chlorates was used.
In vitro shoots of Vitis vinifera L., var. Touriga Nacional were used as explants for in vitro multiplication as described in Neves et al.53 Explants were sub-cultured every four weeks into MS basal medium supplemented with 0.5 µM NAA, 5.0 µM BA, 30 g L−1 sucrose, pH 5.8, and 2 g L−1 Gelrite. Cultures of five explants in 250 mL Magenta vessels (Sigma-Aldrich St. Louis, MO) were maintained in a growth chamber under light from cool-white fluorescent lamps with 16/8 h photoperiod at 50 ± 5 µmol m−2 s−1at 25 ± 1°C (light) and 22 ± 1°C (dark) in full MS medium (control, +S) or in a modified MS as described above for sulfate deprivation (−S).
Determination of growth parameters.
FW was registered at day 0, 1, 3, 4, 6 and 7 of culture from samples of at least three flasks per treatment. At the second week of culture shoot FW and the number of new branches formed in vitro were recorded.
To count the number of cells present in suspension cultures 1 ml of cell suspension was mixed with 0.5 ml 10% cellulose and 0.5 ml 5% pectinase and incubated for 30 minutes at 25°C, under 100 rpm.54 Isolated cells obtained by disaggregation of cell clusters were counted in hemacitometer chambers under the microscope. The larger linear dimension of isolated cells was measured under a microscope equipped with a micrometric ocular.
Sequence identification.
Arabidopsis protein sequences extracted from the NCBI databases (www.ncbi.nlm.nih.gov/) were used as queries, researched using Vitis proteins as queries and subsequently reconfirmed in Genoscope database (www.genoscope.cns.fr/spip/). After obtaining the protein sequence for each hit, the full-length cDNAs of all predicted genes was extracted from NCBI database. The phylogenetic trees were constructed using the PHYLIP, PRODIST and NEIGHBOR programs.55
Gene expression.
RNA extraction and cDNA preparation. Total RNA was extracted from Vitis cells using Rneasy plant Mini Kit (Qiagen, Hilden, Germany). All RNA samples were treated with RNase free DNase I (Qiagen, Hilden, Germany) according to the manufacturer protocol and quantified using absorption of U.V. light at 260 nm. Reverse transcription was carried out using superscript III Rnase H-reverse transcriptase priming with oligo-d(T) (Invitrogen) according to the manufacturer’s recommendations.
Total RNA was extracted from Vitis shoots using the method described by Reid et al.56 and treated as above.
To further examine gene expression by real-time PCR, specific primer pairs were designed to amplify fragments of 100 to 200 bp (Suppl. Table 2) using the Clone Manager program (Scientific & Educational Software, NC). To confirm the amplification of the targeted genes, a PCR reaction was conducted (Master Cycler Gradient termocycler, Eppendorf, Germany) under the following conditions: 95°C, 3 min (initial polymerase activation); 94°C, 3 min; 40 cycles at 95°C, 15 s (denaturation); 61°C, 30 s (annealing); 72°C, 20 s (extension). PCR products were resolved on 2% (w/v) agarose gels. The PCR products were purified using the Wizard® SV Gel and PCR Clean up system (Promega, Madison) and sequenced by STABVIDA (Oeiras, Portugal) or Macrogen (Seoul, Korea). The sequence alignments were carried out in BioEdit program and their identity confirmed by searches in NCBI database, using the translated protein product.
Gene expression analysis by real-time PCR. Real-time PCR was performed in 20 µL of reaction mixture composed of cDNA, 0.5 µM gene-specific primers and master mix iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) using an iQ5 Real Time PCR (BioRad). Reaction conditions for thermal cycling were: 95°C for 3 min; 40 cycles of 95°C for 10 sec; 61°C for 25 sec; 72°C for 30 sec followed by melt curve analysis. The detection of PCR product was monitored by measuring the fluorescence after each extension step caused by the binding of SYBR green dye to dsDNA. Each run was completed with a melting curve analysis to confirm the specificity of amplification and confirm the absence of primer dimmers. Relative amounts were calculated and normalized with respect to actin (Act) mRNA levels. Data were analyzed with the iQ5 optical system software (Bio-Rad, Hercules, CA), which calculates the threshold cycle (CT) and exported into a MS Excel workbook (Microsoft Inc.,) for further analysis. Each reaction was done in triplicate and corresponding CT values were determined. The method described by Bovy et al.57 was applied to compare the level of gene expression in −S status to +S status and −CK to +CK conditions.
First, the CT values were normalized to the CT value of Act2, a housekeeping gene found to be expressed at constant level in the conditions tested. Next, the expression level of each gene in −S condition was expressed relative to its expression in +S conditions according to the equation:
Finally, the relative expression of the genes in −S conditions was expressed according to the expression 2 exp ΔΔCT(+S/−S), where exp = exponential. The same calculations were made to assess the expression level of −CK relative to +CK. Taking into account the calculated standard deviations, the threshold to consider a significant fold variation in gene expression was set at +3 or −3.
Statistics.
For each parameter, S treatment and control were performed at least twice. Data are presented as mean values ± standard deviation (SD). The results were statistically evaluated by variance analysis (ANOVA) comparing the treatments applied (+S/−S status and +CK/−CK) using MS Excel workbook (Microsoft Inc.) software and Statistica 8.
Supplementary Material
Acknowledgements
This work was funded by ‘Fundação para a Ciência e Tecnologia’ (FCT), Portugal, projects POCTI/AGG/46607/2002 and ERANET 006 2006. J.F. received a training grant (2007) from IEFP and S.T. received the PhD grant SFRH/BD/18904/2004 from FCT.
Abbreviations
- 2,4-D
2,4-dichlorophenoxy-acetic acid
- AM
axillary meristem
- BA
6-benzyladenine
- HK
histidine kinase receptor
- HPt
histidine phosphotranfer protein
- CK
cytokinin
- CSP
cytokinin signal pathway
- FW
fresh weight
- MS
murashige and skoog medium
- NAA
α-naphthaleneacetic acid
- PVP-40T
polyvinylpyrrolidone
- RR
response regulator protein
- S
sulfur
- SAM
shoot apical meristem
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9942
References
- 1.Heyl A, Schmülling T. Cytokinin signal and transduction. Curr Opin Plant Biol. 2003;6:480–488. doi: 10.1016/s1369-5266(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 2.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 3.Müller B, Sheen J. Advances in cytokinin signaling. Science. 2007;318:68–69. doi: 10.1126/science.1145461. [DOI] [PubMed] [Google Scholar]
- 4.Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. Molecular characterization of cytokininresponsive Histidine Kinases in maize. Differential ligand preferences and response to cis-Zeatin. Plant Physiol. 2004;134:1–8. doi: 10.1104/pp.103.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito Y, Kurata N. Identification and characterization of cytokinin-signaling gene families in rice. Gene. 2006;382:57–65. doi: 10.1016/j.gene.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 6.West AH, Stock AM. Histidine Kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 7.Ueguchi C, Koizumi H, Suzuki T, Mizuno T. Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:231–235. doi: 10.1093/pcp/pce015. [DOI] [PubMed] [Google Scholar]
- 8.Du L, Jiao F, Chu J, Jin G, Chen M, Wu P. The twocomponent signal system in rice (Oryza sativa L.): A genome-wide study of cytokinin signal perception and transduction. Genomics. 2007;89:697–707. doi: 10.1016/j.ygeno.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi MH, Pischke M, Mähönen A, Miyawaki K, Hashimato Y, Seki M, et al. In plant functions of Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 20. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka Y, Suzuki T, Yamashino T, Mizuno T. Comparative studies of the AHP histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in Arabodopsis thaliana. Biosci Biotech Biochem. 2004;68:462–465. doi: 10.1271/bbb.68.462. [DOI] [PubMed] [Google Scholar]
- 11.Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, et al. ARR1, a transcription factor for genes immediately responsive to cytokinin. Science. 2001;294:1519–1522. doi: 10.1126/science.1065201. [DOI] [PubMed] [Google Scholar]
- 12.D’Agostino IB, Deruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124:1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT. Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA. 1995;92:9373–9377. doi: 10.1073/pnas.92.20.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, van den Berg PJ, Belcher AR, et al. Regulation of expression of a cDNA from barley roots enconding a high affinity sulfate transporter. Plant J. 1997;12:875–884. doi: 10.1046/j.1365-313x.1997.12040875.x. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi H, Yamazak M, Sasakura N, Watanabe A, Leustek T, de Almeida-Engler J, et al. Regulation of sulfur in higher plants: a sulfate transporter induced in sulfate starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:11102–11107. doi: 10.1073/pnas.94.20.11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K. Two distinct high-affinity sulfate transporter with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 2002;29:465–473. doi: 10.1046/j.0960-7412.2001.01231.x. [DOI] [PubMed] [Google Scholar]
- 17.Tavares S, Sousa C, Carvalho LC, Amancio S. De-repressed transporters are strongly repressed after sulfate addition to sulfur depleted Vitis cells. Int J Plant Sci. 2008;169:987–997. [Google Scholar]
- 18.Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H. A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 2004;38:779–789. doi: 10.1111/j.1365-313X.2004.02079.x. [DOI] [PubMed] [Google Scholar]
- 19.Ohkama N, Takei K, Sakakibara H, Hayashi H, Yoneyama T, Fujiwara T. Regulation of sulfur-responsive gene expression by exogenously applied cytokinin in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:1493–1501. doi: 10.1093/pcp/pcf183. [DOI] [PubMed] [Google Scholar]
- 20.Martín A, del Pozo J, Iglesias J, Rubio V, Solano R, de la Peña A, et al. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- 21.Franco-Zorrilla JM, Marín AC, Solano R, Rubio V, Leyva A, Paz-Ares J. Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 2002;32:353–360. doi: 10.1046/j.1365-313x.2002.01431.x. [DOI] [PubMed] [Google Scholar]
- 22.Sachs T, Thimann KV. Release of lateral buds from apical dominance. Nature. 1964;201:939–940. [Google Scholar]
- 23.Cline M, Wessel T, Iwamura HP. Cytokinin/auxin control of apical dominance in Ipomoea nil. Plant Cell Physiol. 1997;38:659–667. [Google Scholar]
- 24.Lenhard M, Laux T. Shoot meristem formation and maintenance. Curr Opin Plant Biol. 1999;2:44–50. doi: 10.1016/s1369-5266(99)80009-0. [DOI] [PubMed] [Google Scholar]
- 25.Bowman JL, Eshed Y. Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 2000;5:110–115. doi: 10.1016/s1360-1385(00)01569-7. [DOI] [PubMed] [Google Scholar]
- 26.Uchida N, Townsley B, Chung K-H, Sinha N. Regulation of SHOOT MERISTEMLESS genes via an upstream-conserved noncoding sequence coordinates leaf development. Proc Natl Acad Sci USA. 2007;104:15953–15958. doi: 10.1073/pnas.0707577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 28.Mar Castellano M, Sablowski R. Intercellular signaling in the transition from stem cells to organogenesis in meristems. Curr Opin Plant Biol. 2005;8:26–31. doi: 10.1016/j.pbi.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 30.Lenhard M, Jurgens G, Laux T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development. 2002;129:3195–3206. doi: 10.1242/dev.129.13.3195. [DOI] [PubMed] [Google Scholar]
- 31.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidipsis on feedback loop regulation by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 32.Ongaro V, Leyser O. Hormonal control of shoot branching. J Exp Bot. 2008;59:67–74. doi: 10.1093/jxb/erm134. [DOI] [PubMed] [Google Scholar]
- 33.Greb T, Clarenz O, Schäfer E, Müller D, Herrero R, Schmitz G, et al. Molecular analysis of the LATERAL SUPRESSOR genes in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17:1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller T, Abbott J, Moritz T, Doerner P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell. 2006;18:598–611. doi: 10.1105/tpc.105.038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller D, Schmitz G, Theres K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell. 2006;18:586–597. doi: 10.1105/tpc.105.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 37.Hu W, Zhang S, Zhao Z, Sun C, Zhao Y, Luo D. The analysis of the structure and expression of Os TB1 gene in rice. J Plant Physiol Mol Biol. 2003;29:507–514. [Google Scholar]
- 38.Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, et al. The Os TB1 gene negatively regulates lateral branching in rice. Plant J. 2003;33:513–520. doi: 10.1046/j.1365-313x.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 39.Hubbard L, McSteen P, Doebley J, Hake S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics. 2002;162:1927–1935. doi: 10.1093/genetics/162.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otsuga D, DeGuzman B, Prigge M, Drews GN, Clark S. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- 41.Aguilar-Martínez J, Poza-Carrión C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell. 2007;19:458–472. doi: 10.1105/tpc.106.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 43.Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beemster GTS, Baskin TI. STUNTED PLANT 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 2000;124:1718–1727. doi: 10.1104/pp.124.4.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyl A, Werner T, Schmülling T. Cytokinin metabolism and signal transduction. In: Hedden P, Thomas SG, editors. Plant Hormone Signaling. Oxford, UK: Blackwell Publ Ltd; 2006. pp. 93–123. [Google Scholar]
- 46.Gachon C, Mingam A, Charrier B. Real-time PCR: what relevance to plant studies? J Exp Bot. 2004;55:1445–1454. doi: 10.1093/jxb/erh181. [DOI] [PubMed] [Google Scholar]
- 47.Kiba T, Taniguchi M, Imamura A, Ueguchi C, Mizuno T, Sugiyama T. Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:767–771. doi: 10.1093/oxfordjournals.pcp.a029604. [DOI] [PubMed] [Google Scholar]
- 48.Amtmann A, Armengaud P. Effects of N, P, K and S on metabolism: new knowledge gained from multi-level analysis. Curr Opin Plant Biol. 2009;12:275–283. doi: 10.1016/j.pbi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 49.Barton MK, Poethig RS. Formation of the shoot apical meritem in Arabidopsis thaliana: an analysis in the wild type and in the shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- 50.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 51.Jackson P, Galinha C, Pereira C, Fortunato A, Soares N, Amancio S, et al. Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalysed by a 40 kDa peroxidase. Plant Physiol. 2001;127:1065–1076. [PMC free article] [PubMed] [Google Scholar]
- 52.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue. Physiol Plant. 1962;15:493–497. [Google Scholar]
- 53.Neves C, Sá MC, Amancio S. Histochemical detection of H2O2 by tissue printing as a precocious marker of rhizogenesis in grapevine. Plant Physiol Biochem. 1998;36:817–824. [Google Scholar]
- 54.Godoy-Hernández G, Vázquez F. Growth measurements: estimation of cell division and cell expansion. In: Loyola-Vargas V, Vázquez-Flota F, editors. Plant cell culture protocols. U.S.: Humana Press INC; 2005. pp. 51–58. [Google Scholar]
- 55.Felsenstein J. PHYLIP Phylogeny Inference Package, version 3.6. Seattle: Department of Genome Sciences, University of Washington,; 2005. [Google Scholar]
- 56.Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6:27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bovy A, de Vos R, Kemper M, Schijlen E, Pertejo MA, Muir S, et al. High-Flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell. 2002;14:2509–2526. doi: 10.1105/tpc.004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.