Abstract
The mechanisms of long-term adaptation to low oxygen environment are quite well studied, but little is known about the sensing of oxygen shortage, the signal transduction and the shortterm effects of hypoxia in plant cells. We have found that an RNA helicase eIF4A-III, a putative component of the Exon Junction Complex, rapidly changes its pattern of localisation in the plant nucleus under hypoxic conditions. In normal cell growth conditions GFPeIF4A-III was mainly nucleoplasmic, but in hypoxia stress conditions it moved to the nucleolus and splicing speckles. This transition occurred within 15–20 min in Arabidopsis culture cells and seedling root cells, but took more than 2 h in tobacco BY-2 culture cells. Inhibition of respiration, transcription or phosphorylation in cells and ethanol treatment had similar effects to hypoxia. The most likely consequence is that a certain mRNA population will remain bound to the eIF4A-III and other mRNA processing proteins, rather than being transported from the nucleus to the cytoplasm, and thus its translation will be suspended.
Key words: eIF4A-III, nucleolus, splicing speckles, hypoxia, exon junction complex
Introduction
In order to survive and successfully reproduce, plants have to adapt to a continually changing environment and its challenges. Saving energy and resources by minimizing anabolism and protein turnover and slowing down metabolic rates is one of the strategies that can help an organism to survive through a transient stress situation. Reduction in ribosomal protein mRNA translation is an essential energy conserving mechanism which itself does not necessarily require additional synthesis of mRNA or protein, but can rather rely on reversible mRNA and protein sequestration or modification. Many studies have shown that gene regulation in response to oxygen deprivation is mediated at both transcriptional and post-transcriptional levels in plants and that stress pathways primarily control uniform responses.1,2 It is still largely unknown what signal transduction pathways are responsible for alterations in gene expression and metabolism and which proteins play roles in these pathways. We identified an Arabidopsis orthologue of mammalian eIF4A-III, the putative anchor protein of the Exon Junction Complex (EJC) to mRNA and found that this protein changes its localization in response to stress.3 Depending on the growth conditions, GFP-eIF4A-III was localized to different sub-nuclear domains: nucleoplasm, nucleolus and splicing speckles. In normal cell growth conditions GFP-eIF4A-III was found mainly as diffuse nucleoplasmic staining. In response to stress conditions such as hypoxia, the GFP-eIF4A-III was observed to concentrate in the nucleolus and splicing speckles.
It is known that in addition to its roles in the maturation, assembly and export of ribonucleoprotein particles, the nucleolus has a range of other functions. In animal cells, the nucleolus has been proposed to act as a sensor of cellular stress.4 Our analysis of the plant nucleolus has shown that it contains mRNAs, including fully spliced, aberrantly spliced and single exon gene transcripts.5 Aberrant mRNAs were much more abundant in the nucleolus, while correctly spliced products were more abundant in nucleoplasm, and moreover, the nonsense-mediated decay (NMD) factors UPF3 and UPF2 were localized to the nucleolus. Taken together, these results suggest that the Arabidopsis nucleolus is involved in mRNA biogenesis and the NMD process in particular.5 In addition, our co-localization studies with the SR proteins RNPS1 and Cyc64 clearly demonstrated the identity of eIF4A-III-containing nuclear foci as splicing speckles, sites for storage/processing of mRNA splicing factors.3 As revealed by FRAP analysis, the nucleoplasmic fraction was highly mobile, while the speckles were the least mobile fractions, and nucleolar fraction had an intermediate mobility,3 implying that the eIF4A-III protein becomes more immobilized under hypoxia stress.
Localization of GFP-eIF4A-III in Cells Treated with Inhibitors
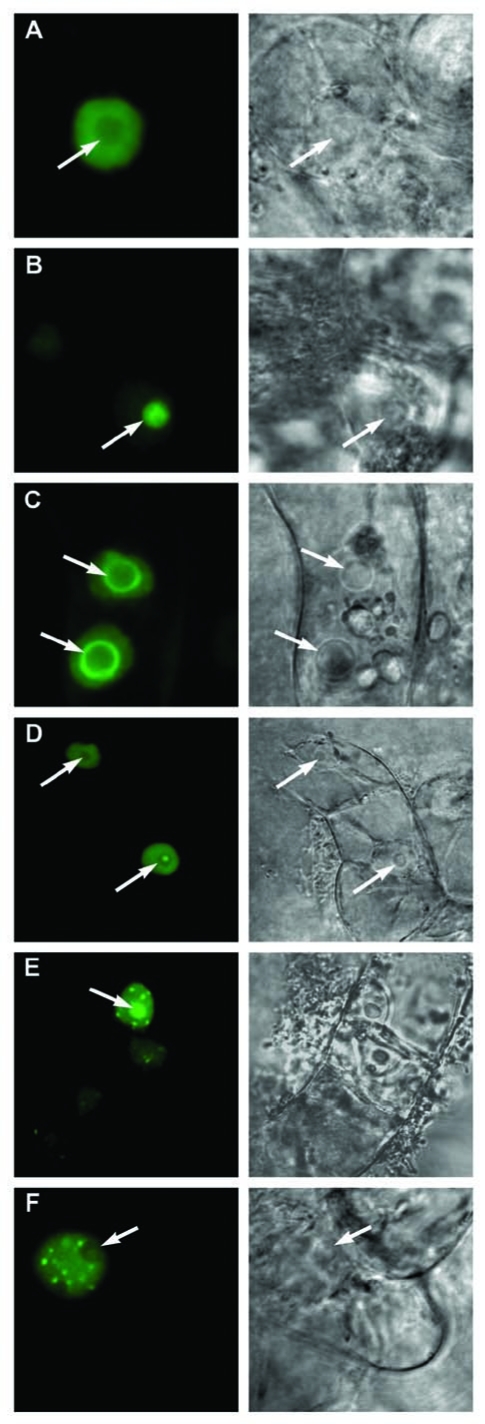
In order to dissect the processes and mechanisms controlling the localization and dynamics of eIF4A-III, we studied the effects of inhibitors specifically targeting transcription, phosphorylation or the proteasome, and compared their effects to a general stress response caused by either 20 min of limited oxygen supply (hypoxia) or treatment with 5% ethanol (Fig. 1, Table 1). Single confocal sections and corresponding bright-field images with a selection of the range of observed types of localisation (diffuse staining of nucleoplasm, nucleoli, nucleolar rim, nucleoplasmic speckles, speckle in the nucleolar cavity) are shown in Figure 1. Compared to the predominantly diffuse staining of the nucleoplasm in control cells, the most dramatic and fastest relocation of the GFP-eIF4AIII to the nucleolus and splicing speckles was observed in cells under hypoxia or treated by the respiratory inhibitor sodium azide.3 Treatment with the proteasome inhibitor MG132 caused a very specific nucleolar accumulation of GFP-eIF4AIII, probably indicating that the movement of this protein from the nucleolus to the nucleoplasm and/or its accumulation in the splicing speckles requires proteolysis. Treatment by 5% ethanol had a milder effect (Table 1). The phosphorylation inhibitors roscovitine and staurosporine had a similar effect to ethanol treatment, and the majority of cells treated by the inhibitors of transcription, actinomycin D and DRB, had signal in the nucleolus and splicing speckles. Rapid formation of a bright speckle in the nucleolar cavity (Fig. 1) was noticed in some cells, usually shown by not more than 5% of the total cell population, under a variety of treatments. In general, the effect of the inhibitors developed slowly; no changes were observed after 1 h of treatment and small and variable changes after 2 h. More consistent changes were observed between 3 and 5 h of treatment by inhibitors and therefore all statistical data were collected during this period. The effect of inhibitors of transcription and phosphorylation was reversible.
Figure 1.
Patterns of localisation of GFPeIF4A-III observed in cells treated with inhibitors. Single confocal sections and corresponding bright-field images are shown. (A) diffuse staining of nucleoplasm; (B) nucleolar; (C) nucleolar rim; (D) speckle in the nucleolar cavity and diffuse staining of nucleoplasm; (E) nucleolus and nucleoplasmic speckles; (F) nucleoplasmic speckles. Arrows indicate positions of nucleoli.
Table 1.
Distribution of localisation patterns of GFP-eIF4A-III observed in cells treated for 3C5 h with inhibitors, 5% ethanol or 20 min anoxia, in comparison to control cells
| Treatment | Expressing cells, % | Total cells, % (number) | |||
| N | No | No + S | S | ||
| *Control | 85 | 14 | 1 | 1 | 100 (354) |
| *MG132 100 µM | 51 | 48 | 0 | 0 | 100 (338) |
| *Hypoxia 20 min | 0 | 1 | 78 | 21 | 100 (102) |
| Ethanol 5% | 37 | 35 | 17 | 11 | 100 (46) |
| Roscovitine 50 µM | 36 | 18 | 28 | 19 | 100 (297) |
| Staurosporine 10 µM | 36 | 43 | 18 | 4 | 100 (168) |
| DRB 100 µg/ml | 65 | 32 | 3 | 0 | 100 (387) |
| ActinomycinD 5 µg/ml | 43 | 42 | 15 | 1 | 100 (177) |
Each column presents % (number) of cells with a specific pattern of localisation: N, diffuse nucleoplasm; No, nucleoli; No + S, nucleoli and nucleoplasmic speckles; S, speckles. *data presented in Koroleva et al.3
Localization of GFP-eIF4AIII Protein in Tobacco BY-2 Cells
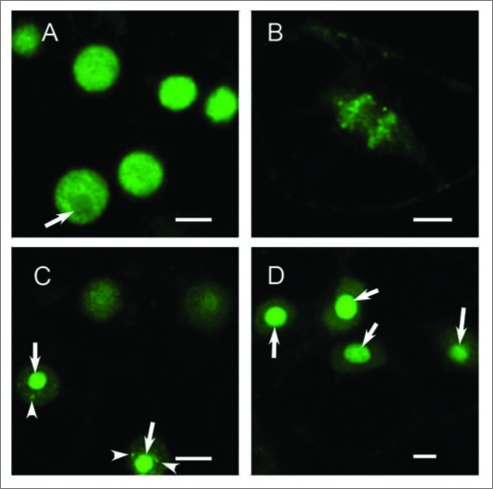
When BY-2 tobacco suspension culture cells were stably transformed by GFPeIF4AIII, using the method described by Koroleva et al.6 the patterns of localization were very similar to those observed in Arabidopsis cells. However, the general nucleoplasmic pattern (Fig. 2A) was the predominant form and did not change after 30 min hypoxia treatment. Much longer periods of anoxia, more than 2 h, were required for the appearance of a stronger signal in the nucleolus (<50% of cells) and speckles (<10% of cells). In mitotic cells in control conditions, the GFP-eIF4AIII was associated mostly with the chromatin and to a lesser extent with the spindle (Fig. 2B). After 5 h of treatment by 5% ethanol, only half of the cells in the population showed relocation of the GFP signal to the nucleolus and speckles (Fig. 2C). Treatment with the proteasome inhibitor MG132 led to uniform pattern of nucleolar accumulation of GFP-eIF4AIII (Fig. 2D), the same effect as observed in Col-0 cells, which implies involvement of proteolysis in translocation of eIF4A-III to the splicing speckles.
Figure 2.
Localization GFP-eIF4AIII in BY-2 tobacco cells. (A) control cells, six nuclei with diffuse signal in the nucleoplasm are shown; (B) mitotic control cell in anaphase: more signal is associated with the separating chromosomes than with the mitotic spindle; (C) cells treated by 5% ethanol for 5 h show localization of signal in nucleoli and speckles. (D) cells treated by MG132 for 5 h show uniform localization in nucleoli only. Bar, 10 µm. Arrows indicate position of nucleoli, arrowheads indicate splicing speckles.
The fact that much longer hypoxia and ethanol treatment were required to cause relocation of the eIF4A-III into nuclear speckles correlates well with the general ability of BY-2 culture to withstand stress and hypoxic conditions compared to Col-0 culture cells. The BY-2 cells in stationary phase of cell culture can survive for several days without fresh media supply, while Arabidopsis cell culture cells cannot. Therefore, it is possible that the sensitivity of a cell to hypoxia stress is directly related to the formation of the splicing speckles.
Other RNA-Processing Factors and a Subset of mRNAs may also Accumulate in Splicing Speckles Under Stress
Reversible relocation of the diffuse nuclear fraction of eIF4AIII into the nucleolus and speckles is caused by a variety of treatments, such as hypoxia, ethanol, inhibition of transcription, phosphorylation or proteasome function. GFP-eIF4AIII co-localized with cyclophilin 64-RFP and RNPS1-RFP in the splicing speckles, suggesting the cooperative dynamic transition of several mRNA processing proteins between subnuclear compartments. Therefore, relocation of the eIF4AIII protein (probably as part of EJC complexes together with associated mRNAs) is a complex process that is likely to involve several major signal transduction pathways. The rapid reaction to the supply of oxygen (less than 20 min) compared to the much longer times required for the effect of inhibitors (usually more than 3 h) indicates that the hypoxic stress response is likely to be mediated through a specific primary mechanism not involving global changes in phosphorylation, transcription or proteolysis in a plant cell.
The fast emergence of the splicing speckles in response to stress in plant cells is reminiscent of the phenomenon of formation of nuclear stress granules containing heat shock factor 1 (HSF1) in response to a heat shock and other stress stimuli in HeLa cells.7 The similarity of the splicing speckles (sites for storage/processing of splicing factors) in the nuclei of plant cells to the nuclear stress granules in mammalian cells suggests that nuclear stress granules and splicing speckles may be related nuclear structures. Although many components of animal nuclear stress granules have been identified, including heat shock factor HSF1 and pre-mRNA processing factors, including a subset of SR proteins, their function is still not known.7,8 Interestingly, not only the sub-nuclear distribution of SR factors, but also alternative splicing of certain transcripts, is differentially affected by heat shock, which is consistent with the idea that reversible sequestration of mRNA processing proteins does indeed represent a specific level of regulation of gene expression and potentially of stress-induced alternative splicing of certain transcripts.8 One possibility is that the sequestration of eIF4A-III and other mRNA processing proteins into splicing speckles is an early response to stress in plants with a similar function to movement of mRNAs and splicing factors to nuclear stress granules in animal cells. In yeast, the mRNA export factor Rat8p (also a DEAD box protein like eIF4AIII) exhibits rapid and reversible changes in localization, accumulating in the nucleus in response to stress caused by ethanol.9 The treatment of plant cells by the proteasome inhibitor MG132 led to accumulation of GFPeIF4A-III in the nucleolus, suggesting that transition of eIF4A-III between sub-nuclear domains is controlled by proteolysis-labile factors. It has also been reported that proteasome inhibitors induce the formation of HSF1-containing stress granules in the nuclei of mammalian cells,10 while our data suggest that proteasome inhibitor MG132 has caused relocation of GFP-eIF4AIII into the nucleolus only. This effect might be specific for the eIF4AIII protein or for the plant splicing speckles in general, and needs further investigation as it is intrinsically connected to the mechanisms regulating transition of proteins between sub-nuclear compartments.
Summary
We have found that the pattern of nuclear localization of eIF4AIII in Arabidopsis cells changes rapidly in response to a specific type of stress or inhibitor action. The faster relocation of eIF4A-III into nucleoli and splicing speckles in Arabidopsis Col-0 cells than in tobacco BY-2 cells may be related to the higher sensitivity to hypoxia of the Arabidopsis cells. We propose that the changes in subnuclear localization reflect the role of this protein in regulation of the mRNA processing, export and/or turnover. The eIF4AIII-dependent and -independent recruitment of RNA-binding proteins to mRNA may have a role in remodeling mRNPs involving interactions with other proteins already bound to the pre-mRNA. The sequestration of RNA-binding proteins and bound mRNA into less mobile fractions may have implications for subsequent processing steps and degradation/ nonsense-mediated mRNA decay, and may mediate stress response.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/9906
References
- 1.Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 2008;56:743–755. doi: 10.1111/j.1365-313X.2008.03642.x. [DOI] [PubMed] [Google Scholar]
- 2.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 3.Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, et al. Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell. 2009;21:1592–1606. doi: 10.1105/tpc.108.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, et al. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell. 2009;21:2045–2057. doi: 10.1105/tpc.109.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koroleva OA, Tomlinson M, Parinyapong P, Sakvarelidze L, Leader D, Shaw P, et al. CycD1, a putative G1 cyclin from Antirrhinum majus, accelerates the cell cycle in cultured tobacco BY-2 cells by enhancing both G1/S entry and progression through S and G2 phases. Plant Cell. 2004;16:2364–2379. doi: 10.1105/tpc.104.023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jolly C, Usson Y, Morimoto RI. Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc Natl Acad Sci USA. 1999;96:6769–6774. doi: 10.1073/pnas.96.12.6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takemura R, Inoue Y, Izawa S. Stress response in yeast mRNA export factor: reversible changes in Rat8p localization are caused by ethanol stress but not heat shock. J Cell Sci. 2004;117:4189–4197. doi: 10.1242/jcs.01296. [DOI] [PubMed] [Google Scholar]
- 10.Holmberg CI, Illman SA, Kallio M, Mikhailov A, Sistonen L. Formation of nuclear HSF1 granules varies depending on stress stimuli. Cell Stress Chaperones. 2000;5:219–228. doi: 10.1379/1466-1268(2000)005<0219:fonhgv>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]