Abstract
Although the primary sequence of kinases shows substantial divergence between unrelated eukaryotes, variation in the motifs that are actually phosphorylated by eukaryotic kinases is much smaller. Hence arrays developed for kinome profiling of mammalian cells are useful for kinome profiling of plant tissues as well, facilitating the study of plant signal transduction. We recently employed the Pepscan kinomics chip to reveal the small GTPases in plant sucrose signaling. Here we show that employing a different peptide library (the Pepscan kinase chip) largely similar results are obtained, confirming these earlier data, but such a different library also contributes new insights into the molecular details mediating plant cell responses to a sugar stimulus. Thus when studying plant signal transduction employing peptide arrays, using multiple platforms both increases the confidence of results and provides additional information.
Key words: sucrose, arabidopsis, kinome profiling, kinase, MAPK, SnRK, casein kinase, tyrosine phosphorylation, CDC2, AGC kinase
In our recently published paper1 we analyze the changes in the plant kinome after sucrose feeding compared to control sorbitol feeding of Arabidopsis. We employed kinomics chips (Pepscan Presto, The Netherlands) containing 960 different kinase consensus peptides selected for their importance in mammalian signal transduction. In addition we used kinase chips (Pepscan Presto, The Netherlands), containing 1,152 peptides covering the majority of peptides available through Phosphobase (version 2.0). These chips contain peptides derived from phosphorylation events described in many kingdoms and are taken from animals, plants, fungi and even bacteria.2,3 Full details as to the peptides spotted can be found at www. pepscanpresto.com. Results of the cluster analysis of three independent biological replicas of kinome profiling after treatment with water, sorbitol, sucrose and glucose are depicted in Figure 3A of our recent paper.1 A further analysis of this set of experiments was not given, due to a lower than expected biological reproducibility. In contrast to the >0.8 correlation observed for the set described in our recent paper, correlations of below 0.5 were seen for the experimental set analyzed on the kinase1 chips. However, we feel that some observation made based on these kinome profiles contain valuable information, if only to suggest follow-up experiments.
Comparison of the kinome profiles of Arabidopsis treated with sucrose or sorbitol for 1 h revealed a set of 93 differentially phosphorylated consensus peptides, with the majority of 59 peptides showing reduced phosphorylation after sucrose treatment (Table 1).
Table 1.
List of differentially phosphorylated consensus peptides
| Consensus | Kinase | sor vs. suc t-test | P up/down |
| ESSYSYEEI | 0.0003 | down | |
| PASPSPQRQ | Cdk5-p23 | 0.0014 | down |
| PKRGSKDG | AGC | 0.0019 | up |
| IREESPPHS | 0.0019 | down | |
| RTPPPSG | MAPK | 0.0020 | down |
| PASQTPNKTt | CDC2 | 0.0029 | down |
| STNEYMDMK | PI3 kinase | 0.0030 | down |
| SEENSKKTV | CKI | 0.0040 | down |
| APTPGGRR | 0.0043 | down | |
| RFTDTRKDE | CaM-III | 0.0046 | down |
| LSELSRRRI | ds-RNA | 0.0062 | up |
| PINGSPRTP | CDC2 | 0.0065 | down |
| TEGQYELQP | Tyr-K | 0.0068 | up |
| KRAQISVRGL | 0.0069 | down | |
| AKRISGKMA | 0.0069 | up | |
| VVGGSLRGA | AGC | 0.0071 | down |
| KRPSNRAKA | 0.0072 | up | |
| ERQKTQTKL | SnRK, MLCK | 0.0073 | down |
| EEGISQESS | 0.0080 | up | |
| PVPEYINQS | EGFR (Tyr-K) | 0.0082 | down |
| FGHNTIDAV | 0.0082 | down | |
| ARVFSVLRE | CaM-II | 0.0085 | down |
| SNDDSDDDD | CKII | 0.0085 | down |
| GGVDYKNIH | Tyr-K | 0.0094 | up |
| SRSRSRSRS | 0.0103 | up | |
| SPSLSRHSS | GSK3 | 0.0107 | down |
| RAKRSGSV | 0.0120 | down | |
| RRASLG | AGC 1/2 | 0.0128 | down |
| GRASSHSSQ | S6K | 0.0129 | down |
| SGYISSLEY | CKII | 0.0139 | down |
| FFRRSKIAV | AGC | 0.0140 | up |
| STNDSPL | beta-ARK | 0.0145 | down |
| LRRASPG | 0.0149 | up | |
| SAVASNMRD | GRK | 0.0154 | up |
| KRPSGRAKA | 0.0160 | up | |
| KRSNSVDTS | AGC | 0.0165 | down |
| RQLRSPRRT | CDC2 | 0.0171 | up |
| GRALSTRAQ | CDPK, PhK | 0.0172 | down |
| VSRTSAVPT | AGC | 0.0173 | down |
| TRKISQTAQ | AGC | 0.0174 | down |
| STTVSKTET | 0.0180 | down | |
| ESPASDEAE | 0.0184 | up | |
| LSYRGYSL | PhK | 0.0185 | down |
| DDINSYEAW | 0.0186 | up | |
| PNVSYIASR | 0.0191 | down | |
| KQPIYIVME | FES (Tyr-K) | 0.0195 | up |
| LVVASAGPT | 0.0198 | down | |
| TGFLTEYVA | MAPKK | 0.0198 | up |
| TEDQYSLVE | Src | 0.0212 | up |
| SSSSSPKAE | MAPK | 0.0213 | up |
| EKAKSPVPK | 0.0221 | down | |
| RRRASVA | AGC1/2 | 0.0221 | down |
| APVASPAAP | MAPK | 0.0225 | down |
| LRRLSTKYR | AGC1/2 | 0.0234 | down |
| EKHHSIDAQ | 0.0256 | down | |
| VRKRTLRRL | SnRK, AGC | 0.0266 | down |
| DLPGTEDFV | GRK2 | 0.0277 | down |
| LSEHSSPEE | CKII | 0.0278 | down |
| KREASLDNQ | AGC | 0.0279 | down |
| TKKQSFKQT | AGC | 0.0280 | up |
| VRLRSSVPG | autoP | 0.0285 | down |
| KRPSLRAKA | 0.0293 | up | |
| PGPQSPGSP | 0.0308 | down | |
| YSGHSMSDP | 0.0309 | up | |
| ADGVYAASG | FES (Tyr-K) | 0.0311 | up |
| ENQASEEED | CKII | 0.0317 | down |
| TLASSFKRR | AGC | 0.0324 | up |
| TVKSSKGGP | AGC | 0.0326 | down |
| GVLRRASVA | 0.0327 | up | |
| SPRKSPRKS | sperm-specific | 0.0328 | down |
| PRRDSTEGF | SnRK, AGC | 0.0332 | down |
| RRRRAASVA | 0.0346 | down | |
| SRKDSLDDS | GRK | 0.0371 | down |
| ENPEYLGLD | Tyr-K | 0.0380 | down |
| KAKTTKKRP | 0.0382 | up | |
| RRPSV | 0.0392 | down | |
| QKAQTERKS | AGC | 0.0401 | down |
| AKAKTTKKR | 0.0404 | up | |
| GSDVSFNEE | CKII | 0.0409 | down |
| DEPSTPYHS | GSK3 | 0.0409 | down |
| SSRPSSNRS | CDPK, AGC | 0.0411 | up |
| GGRASDYKS | AGC | 0.0413 | up |
| YMAPYDNYV | Tyr-K | 0.0420 | up |
| LELSDDDD | CKII | 0.0422 | down |
| THVASVSDV | SnRK AMPK | 0.0423 | down |
| SMANSFVGT | PDKI | 0.0427 | down |
| DLLTSPDVG | CDC2 | 0.0441 | down |
| RGKSSSYSK | AGC | 0.0441 | up |
| SSSNTIRRP | AGC | 0.0453 | up |
| RRDSV | 0.0457 | down | |
| TKAASEKKS | 0.0469 | up | |
| DRLVSARSV | CDPK, SnRK, AGC | 0.0480 | down |
| RLSISTESQ | AMPK | 0.0489 | up |
Arabidopsis seedlings were incubated in a solution of 100 mM sucrose or sorbitol for 1 hour after which extracts were made as described before (Plos One) and incubated on Kinase1 PepChips (1152 consensus peptides spotted twice per slide; Pepscan). The averaged phosphorylation intensities obtained from three independent experiments were analyzed using a Student’s t-test. Indicated is whether phosphorylation of consensus peptides is higher (up) or lower (down) after sucrose treatment, compared to sorbitol. Kinase annotation according to Pepscan Presto (www.pepscanpresto.com), or our own analysis (see main text).
Kinome Profiling
When the results obtained from the set of experiments analyzed on the Kinomics chip are compared to those of the Kinase1 chip similarities can be noted. Tyrosine kinase consensus peptides are consistently more phosphorylated after sucrose and phosphorylation levels of CDC2 consensus substrates is increasing. However, also differences are apparent; phosphorylation of Casein Kinase (CK) consensus peptides is consistently going down over this set of experiments, whereas in the set of experiments analyzed before in the Kinomics chips CK activity is going up. This might be due to a difference in experimental set-up. In these experiments plants were submerged during sugar incubation, whereas in the other set of experiments the sugar solution was added to the filter on which plants were grown in vitro. Many AGC-protein kinase consensus substrates are present on the Kinase1 chip. This family of kinases consists of many family members. It is reported that AGC2 phosphorylates several targets of which consensus sequences were described by Anthony et al.4 According to the analysis described in this paper peptides RRRASVA and LRRLSTKYR should be considered AGC2 kinase consensus peptides. Both show reduced phosphorylation after sucrose. In addition, peptide RRASLG is the classical PKA substrate called kemptide that can also be used to measure AGC1/2 activity.5 Its phosphorylation also goes down after sucrose. Judged from the many AGC consensus peptides that show increased phosphorylation levels, other members of the AGC kinase family are probably activated by sucrose.
Kinome Analysis Indicates MAPK Signaling
The enhanced phosphorylation of peptide TGFLTEYVA after sucrose treatment points to a role of a MAPK cascade in the signaling. Since MAPK pathways are identified in signaling of many compounds, ranging from salt, water, pathogens and hormones, involvement of MAPKs is sugar signaling is not surprising.6 The TGFLTEYVA peptide contains the TEY signature, which includes the Thr (T) and Tyr (Y) residues phosphorylated in MAPKs.7 Phosphorylation of these residues increases the activity of MAPKs. The TEY triplet is present in 12 of the 20 Arabidopsis MPKs, the others have TDY.8 At the same time the peptide TGFLTEYVA is also a MAPKK consensus peptide, since it represents the Thr and Tyr phosphorylation sites in MAPK that are phosphorylated by the dual-specificity kinase MAPKK.9 Since TGFLTEYVA shows higher phosphorylation upon sucrose feeding we presume that MAPKK activity, and with that MAPK activity, is increased by sucrose.
Also some MAPK consensus peptides can be found among the significantly differentially phosphorylated peptides, APVASPAAP, SSSSSPKAE, RTPPPSG, are annotated as such. In contrast to our prediction, two out of three show lower phosphorylation after sucrose, this could be due to either differential activities of a subset of MAPKs, or phorphorylation of these substrates by other kinases.10
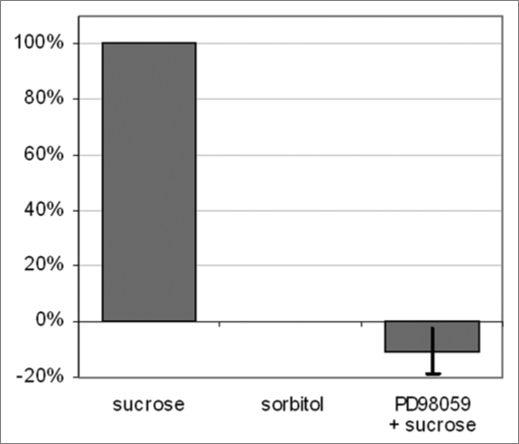
Based on these data it is difficult to access the importance of MAPK cascades in sucrose signaling, we performed another inhibitor assay using PD98059, an inhibitor of MAPKK. This inhibitor could repress sucrose induction of the fructosyltransferase promoter completely (Fig. 1), providing evidence that MAPK cascades are important in sucrose signaling. As MAPK cascades are the classical effectors of small GTPases, this observation would further support our hypothesis that such small GTPases are central orchestrators of the sugar response in plants.
Figure 1.
Role of MA P kinases in sucrose signaling. Activity of the MA PKK inhibitor PD98059 in sucrose-induced GUS activity as quantified using MUG as a substrate.
Kinome Analysis Reveals SnRK Involvement in Sucrose and Glucose Signal-Transduction
SnRK is the plant homologue of AMPK; Peptide THVASVSDV is both AMPK and SnRK consensus peptide and its phosphorylation is decreased after sucrose feeding. We searched for additional SnRK consensus peptides on the PepChip by using the peptides identified by Kleinow et al.11 and found the peptides VRKRTLRRL, PRRDSTEGF, ERQKTQTKL and DRLVSARSV to resemble SnRK consensus peptides. Phosphorylation of these peptides is lower upon sucrose addition compared to sorbitol (Table 1). The reduced phosphorylation of these peptides upon sugar feeding is in agreement with the negative role that SnRK and its yeast and mammalian homologous play in carbon signaling.
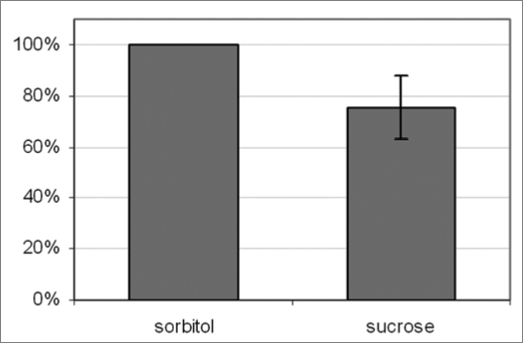
Both human AMPKα as well as Arabidopsis SnRK1.1 are able to complement a yeast SNF1 mutant.12 Therefore it is proposed that all three kinases work in the same manner and react to similar activation signals, such as phosphorylation of a Thr residue in a conserved region of the protein. For that reason antibodies against the phosphorylated fragment SDGEFLRpTSCGSPNY of human AMPKα (Cell Signaling Technologies) were used to detect phosphorylated SnRK from Arabidopsis (Fig. 2). SnRK1.1, SnRK1.2 and SnRK1.3 harbor sequences which are highly homologous to the fragment used for antibody production, therefore the phosphorylated state of these kinases is most likely the detected by this antibody. Quantification of western blot analysis revealed that slightly less phosphorylated SnRK1 is present after sucrose and glucose compard to sorbitol (Fig. 2). In analogy to AMPK, less phosphorylation would mean lower SnRK activity, which is in agreement with the data we obtained from kinome profiling (Table 1). Although contrasting views are present, inactivation of Arabidopsis SnRK1 upon sucrose was reported before.13
Figure 2.
Role of AM PK homologues in sucrose signaling. Western blot analysis of putative SnRK phosphorylation, see main text for explanation on the antibody used.
Conclusions
In view of limited tools available with respect to signal transduction research in plants, the advent of kinome profiling using peptide arrays seems useful, despite the absence of dedicated plant platforms. Nevertheless, it seems that changes of finding interesting phenomena increase by using multiple peptide libraries. With respect to sugar signaling it can be said that disregarding the exact platform used, data are still best explained by postulating a central role for small GTPases in the regulation of the molecular events evoked by a sucrose stimulus. As such GTPases are especially important in the regulation of cytoskeletal events,13 it should prove interesting to investigate actin reorganization in response to sugar stimulus as well.
Acknowledgements
The authors are grateful to grant 3.103 from the Top Institute Pharma that supports their research.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10022
References
- 1.Ritsema T, Brodmann D, Diks SH, Bos CL, Nagaraj V, Pieterse CM, et al. Are small GTPases signal hubs in sugar-mediated induction of fructan biosynthesis? PloS ONE. 2009;4:6605. doi: 10.1371/journal.pone.0006605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diks SH, Parikh K, van der Sijde M, Joore J, Ritsema T, Peppelenbosch MP. Evidence for a minomal eukaryotic phosphoproteome? PLoS ONE. 2007;2:777. doi: 10.1371/journal.pone.0000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritsema T, Joore J, van WW, Pieterse CM. Kinome profiling of Arabidopsis using arrays of kinase consensus substrates. Plant Methods. 2007;3:3. doi: 10.1186/1746-4811-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony RG, Henriques R, Helfer A, Mészáros T, Rios G, Testerink C, et al. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 2004;23:572–581. doi: 10.1038/sj.emboj.7600068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zwerger K, Hirt H. Recent advances in plant MAP kinase signalling. J Biol Chem. 2001;382:1123–1131. doi: 10.1515/BC.2001.142. [DOI] [PubMed] [Google Scholar]
- 6.Nuhse TS, Peck SC, Hirt H, Boller T. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J Biol Chem. 2000;275:7521–7526. doi: 10.1074/jbc.275.11.7521. [DOI] [PubMed] [Google Scholar]
- 7.MAPK Group, authors. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 8.Meskiene I, Hirt H. MAP kinase pathways: molecular plug-and-play chips for the cell. Plant Mol Biol. 2000;42:791–806. doi: 10.1023/a:1006405929082. [DOI] [PubMed] [Google Scholar]
- 9.Jonak C, Okresz L, Bogre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002;5:415–424. doi: 10.1016/s1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 10.Glinski M, Weckwerth W. Differential multisite phosphorylation of the trehalose-6-phosphate synthase gene family in Arabidopsis thaliana: a mass spectrometry-based process for multiparallel peptide library phosphorylation analysis. Mol Cell Proteom. 2005;4:1614–1625. doi: 10.1074/mcp.M500134-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchert K, Koncz C. Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 2000;23:115–122. doi: 10.1046/j.1365-313x.2000.00809.x. [DOI] [PubMed] [Google Scholar]
- 12.Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 13.Peppelenbosch M, Boone E, Jones GE, van Deventer SJ, Haegeman G, Fiers W, et al. Multiple signal transduction pathways regulate TNF-induced actin reorganization in macrophages: inhibition of Cdc42-mediated filopodium formation by TNF. J Immunol. 1999;162:837–845. [PubMed] [Google Scholar]