Abstract
The signaling molecule auxin plays a central role in several aspects of plant growth and developmental processes. Underlying optimization of these processes are complex mechanisms orchestrating the expression of genes involved in controlling auxin level, movement and signalling. The small auxin-up RNA (SAUR) family comprises a large set of genes whose expressions are early auxin-responsive. However, the function of these genes is largely unknown. Loss-of-function mutants in a number of Arabidopsis SAUR genes did not show any marked phenotypic differences to wild-type plants likely due to compensatory functions of conserved members of the SAUR gene family. We have recently shown that a rice SAUR39 gene negatively regulates auxin synthesis and transport in rice. Here we propose a model that constitutive induction of SAUR39 gene expression reduces growth and seed yield in rice plants due to the presence of a lower auxin level, reduced polar auxin transport, less chlorophyll and increased sugar and anthocyanin contents. In wild-type plants, the SAUR39 gene is expressed at a low level and is transiently induced by changes in external auxin or other environmental stimuli, but within hours of this change its expression is reduced to the low constitutive level. This homeostatic mechanism is essential for optimal plant growth and seed yield and its disruption due to the constitutive overexpression of SAUR39 leads to a set of negative pleiotropic phenotypes.
Key words: auxin synthesis, polar auxin transport, anthocyanin, chlorophyll, sugar
Auxin plays a key regulatory role for optimal plant growth and development by modulating the expression of genes involved in several physiological processes.1–3 This includes the regulation of genes from three different early auxin-responsive gene families, through the rapid and transient induction of these genes in response to exogenous auxin supply. These gene families are auxin/indoleacetic acid (Aux/IAA), Gretchenhagen-3 (GH3) and small auxin-up RNA (SAUR).3 Several genes from the Aux/IAA and GH3 families have been functionally characterized. However, the function of any SAUR gene remained unknown.4 Several unsuccessful previous attempts were made to find functional phenotypes in loss-of-function mutants in different SAUR genes. This could be due to several reasons such as functional redundancy and a compensatory role through other mechanisms.4–6 Most of these functional analysis studies were done in the model plant Arabidopsis which has 78 SAUR genes (www.arabidopsis.org/).5,6 A large number of these genes are located in tandem with high sequence similarity and identity at the cDNA and amino acid levels amongst them, with the potential for them having overlapping functions. In rice there are 58 SAUR genes and most of the genes are staggered on different chromosomes, except for the clustering of some genes on chromosome number 9.4 However, the sequence identity and similarity among rice SAUR genes is less when compared to the Arabidopsis SAUR genes. Therefore, there is a higher probability of finding some SAUR genes in rice that have unique functions.
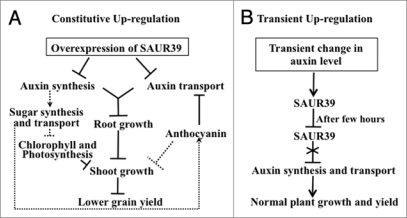
In our microarray analysis conducted under different nitrogen regimes in rice, one SAUR39 (Os09g0545300 or LOC_Os09g37330.1) gene was induced by changing nitrogen levels.7 In addition, this gene was also upregulated by transient application of cytokinin, anoxia, and salt stress (Gene expression omnibus profiles, www.ncbi.nlm.nih.gov/) and was also responsive to exogenous auxin supply.7 The transgenic lines overexpressing this SAUR39 were developed using a constitutively expressed ubiquitin promoter and interestingly these overexpresser plants were smaller and had less vegetative growth and lower yield as compared to wild-type plants.7 A model for the role of SUAR39 gene in this effect is shown in Figure 1 and the reasoning behind consists of the following. The free Indole-3-acetic acid (IAA) level in overexpresser plants was lower than wild-type which could be due to negative feedback inhibition by the constitutively induced SAUR39 gene. In contrast, in wild-type plants, the SAUR39 gene likely is repressed due to the presence of destabilizing elements in 3′ UTR region, whereas under the constitutive induction of the SAUR39 gene the plant might sense a sufficient auxin level and give signals to produce less auxin. In turn, a lower auxin level would lead to an increased sugar level which would repress the expression of photosynthetic genes and chlorophyll production (Fig. 1A). An inverse relation between a lower auxin level and higher sugar content in Arabidopsis has been reported by Ohto et al.8 Furthermore, higher sugar levels in plants can repress several genes associated with photosynthesis and induce genes associated with anthocyanin accumulation.9–11 Less chlorophyll and downregulation of photosynthetic genes in overexpresser plants resulted in lower shoot growth as compared to wild-type plants (Fig. 1A). Higher sugar content is thought to increase anthocyanin content in plants (Fig. 1A).8,11 The higher anthocyanin contents are known to repress auxin transport in Arabidopsis12 and the auxin transport was also lower in SAUR39 overexpresser plants.7 The lower auxin level and transport would result in less lateral root formation in plants.1,2,13 Reduced shoot and root growth ultimately resulted in lower seed yield in SAUR39 overexpresser plants compared to wild-type (Fig. 1A). In wild-type plants the expression of SAUR39 could be transiently induced by exogenous auxin supply but returns to the normal level after 24 h.7 The negative feedback regulation of auxin synthesis and transport by the transiently induced SAUR39 gene would be relieved once the SAUR39 transcript returns to its baseline level (Fig. 1B). As mentioned earlier, the expression of the SAUR39 gene can be induced by other environmental stimuli such as anoxia, salinity and application of cytokinin. The prolonged or recurring exposure of plants to these conditions could induce the SAUR39 gene for a longer period and would possibly lead to a similar downregulation of auxin synthesis and transport and lower growth and yield as was observed in SAUR39 overexpresser plants. Therefore, a detailed understanding of the molecular mechanisms of this and other SAUR genes would shed light on the involvement of SAUR genes in auxin signalling and their involvement in feedback regulation of the auxin level in plants. This future proposed work might include: (1) understanding the role of SAUR genes via interaction with calmodulin genes, since the expression of several Ca+-binding/calmodulin genes was higher in SAUR39 overexpresser plants7 and some SAUR proteins have been shown to bind with Ca+-binding/Calmodulin proteins14 (2) study the interaction of SAUR proteins with other auxin-induced and auxin response factor proteins (3) identification of other SAUR genes through transcript profiling in response to different growth conditions and functional characterization of gain-of-function and loss-of-function plants of these genes to identify their unique and non-overlapping functions in various tissues or whole plants.
Figure 1.
A model to explain SAUR39 gene function in negative regulation of auxin level in rice plants. (A) Constitutive upregulation of SAUR39 results in reduced rice plant growth and yield via reduced auxin synthesis and transport. (B) Transient change in auxin level results in temporary induction of SAUR39. The pointed arrows indicate positive regulation and blunt end arrows indicate negative regulation. Dotted lines indicate putative regulation.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10043
References
- 1.Teale WD, Paponov IA, Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- 2.Woodward AW, Bartel B. Auxin: regulation, action and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- 4.Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa) Genomics. 2006;88:360–371. doi: 10.1016/j.ygeno.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Gil P, Green PJ. Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J. 1996;15:1678–1686. [PMC free article] [PubMed] [Google Scholar]
- 6.Park J-E, Kim Y-S, Yoon H-K, Park C-M. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007;172:150–157. [Google Scholar]
- 7.Kant S, Bi YM, Zhu T, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009;151:691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohto MA, Hayashi S, Sawa S, Hashimoto-Ohta A, Nakamura K. Involvement of HLS1 in sugar and auxin signaling in Arabidopsis leaves. Plant Cell Physiol. 2006;47:1603–1611. doi: 10.1093/pcp/pcl027. [DOI] [PubMed] [Google Scholar]
- 9.Krapp A, Hofmann B, Schäfer C, Stitt M. Regulation of the expression of rbcS and other photosynthetic genes by carbohydrates: a mechanism for the sink regulation of photosynthesis. Plant J. 1993;3:817–828. [Google Scholar]
- 10.Koch KE. Carbohydrate-Modulated Gene Expression in Plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- 11.Martin T, Oswald O, Graham IA. Arabidopsis seedling growth, storage lipid mobilization and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 2002;128:472–481. doi: 10.1104/pp.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazar G, Goodman HM. MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:472–476. doi: 10.1073/pnas.0509463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chhun T, Uno Y, Taketa S, Azuma T, Ichii M, Okamoto T, et al. Saturated humidity accelerates lateral root development in rice (Oryza sativa L.) seedlings by increasing phloem-based auxin transport. J Exp Bot. 2007;58:1695–704. doi: 10.1093/jxb/erm026. [DOI] [PubMed] [Google Scholar]
- 14.Yang T, Poovaiah BW. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem. 2000;275:3137–3143. doi: 10.1074/jbc.275.5.3137. [DOI] [PubMed] [Google Scholar]