Abstract
The conjugation of small ubiquitin-related modifier (SUMO) to substrates (sumoylation) is one of posttranslational modification systems in eukaryotes. Sumoylation plays an important role in the regulation of environmental stress response, biotic stress response, and flowering control in plants. Covalent SUMO conjugation requires an E1/E2/E3 enzyme, and SUMO E3 ligase SIZ1 is essential for these regulations. This addendum summarizes our recent study in which it has been established that in Arabidopsis, SUMO E3 ligase SIZ1 negatively controls abscisic acid (ABA) signaling through the sumoylation of ABI5. The conjugation of SUMO to ABI5 represses its activity and also prevents ABI5 from undergoing degradation.
Key words: seed germination, seedling growth, SUMO, sumoylation, SIZ1, signal transduction
Many biochemical pathways are reversible; this allows the creation of on and off states, which are essential for biological regulation. A reversible system has the advantage of quick initiation and termination of a biological response that needs to be precisely regulated. SUMO modification is an example of a reversible system that is controlled by a series of on-and-off enzymes.1,2 Sumoylation is catalyzed by E1-activation, E2-conjugation and E3-ligation enzymes, whereas SUMO proteases contribute to deconjugation of SUMO from substrates.3,4
On the basis of genetic analysis and biochemical evidence, it has been established that sumoylation and desumoylation are functionally linked to ABA signaling,5 phosphate-starvation response,6 thermotolerance,7 cold acclimation,8,9 drought and salt tolerance,10,11 defense against phytopathogens, 12 growth13 and flowering.14–16 SIZ1 (SAP and Miz), the principal SUMO E3 ligase in Arabidopsis,6,13 plays an important functional role in most of these processes.6–8,11–14 Recently, HPY2/MMS21 (high proidy2/methyl methanesulfonate sensitive21) has been identified as another SUMO E3 ligase that modulates cell cycle progression and meristem maintenance.17,18 Cells having the hpy2 mutation had high ploidy and caused severe growth defects.17 On the basis of ploidy and the expression of CDKB, it has been reported that SIZ1 and HPY2 may play different roles in mediating plant growth.17
Arabidopsis SIZ1 is the plant ortholog of yeast SIZ and mammalian protein inhibitor of activated STAT (PIAS) proteins.2 SIZ/PIAS proteins perform several functions, including repression or activation of transcription factors to control signaling.19 SIZ/PIAS proteins are also reported to function as transcriptional co-regulators, controlling subnuclear relocalization of substrates and interactions of transcriptional coregulatory complexes.1,19,20
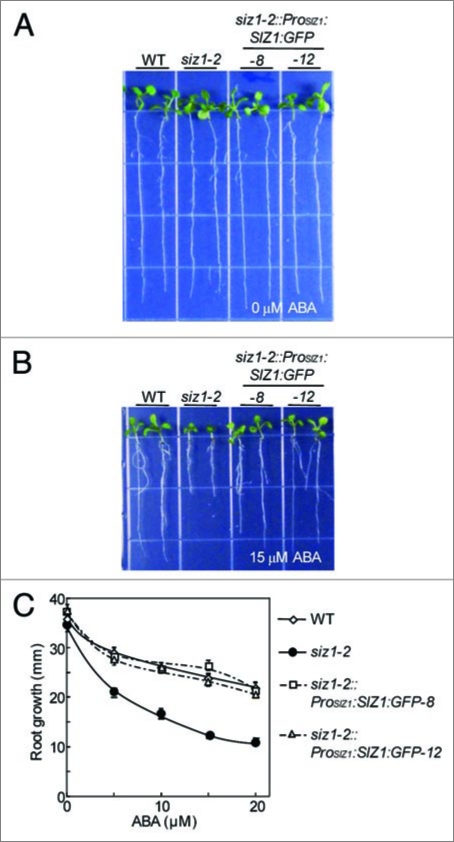
Plant sumoylation appears to be important for ABA signaling. Our research shows that in Arabidopsis, ABA signaling is negatively regulated by SIZ1 SUMO E3 ligase-mediated sumoylation of ABI5.21 The loss-of-function siz1 mutation facilitated ABA-mediated inhibition of seed germination and root growth, which were restored by introduction of ProSIZ1:SIZ1:GFP into siz1-2 mutants (Fig. 1). Furthermore, overexpression of AtSUMO1 or AtSUMO2 attenuates and co-suppression of AtSCE1 accelerates ABA-mediated inhibition of root growth;5 therefore, the SUMO reaction contributes to the regulation of ABA responses. ABI5 is an important basic leucine zipper (bZIP)-type transcription factor that controls ABA response and ABA-responsive genes.22–24 SIZ1-mediated sumoylation of ABI5 negatively regulates ABA signaling and responses.21 Furthermore, sumoylation of ABI5 blocked proteasome-mediated degradation that was facilitated by AFP (ABI five binding protein). These findings indicate that SIZ1-mediated sumoylation of ABI5 is an important regulatory process in ABA signaling and responses and in the prevention of degradation of ABI5.
Figure 1.
the siz1-2 mutation increases aBa inhibition of primary root growth in a seedling, and this mutation is complemented by transformation of the wild-type SIZ1 gene. three-day-old seedlings were transferred onto media containing various concentrations of aBa. Pictures show representative wild-type, siz1-2 and siz1-2 transformed with ProSIZ1:SIZ1:GFP, seven days after transfer onto the media containing 0 (a) or 15 μm (B) aBa. (c) root growth values are expressed as mean ± Se (n = 12). siz1-2::ProSIZ1:SIZ1:GFP-8 and siz1-2::ProSIZ1:SIZ1:GFP-12 are representative lines illustrating that SIZ1:GFP driven by the promoter of SIZ1 was transformed into siz1-2,14 and genetically complemented the mutation.
Sumoylation may play an important role in the precise regulation of ABI5 activity. Unlike the proteasome-mediated degradation of ABI5 that is facilitated by AFP,25 sumoylation/desumoylation system is a reversible mechanism. ABI5 may be stored in the inactive form by sumoylation; subsequently, it is released by desumoylation, if necessary. Phosphorylation of ABI5 may play a role in the activation of the transcription factor because SnRK2.2 and SnRK2.3 phosphorylate ABI5 in response to ABA.26 These posttranslational modifications are required for controlling ABI5 activity precisely.
Acknowledgements
K.M. is supported by grants from, in part, Special Coordination Funds for Promoting Science and Technology from the Japanese Ministry of Education, Culture, Sports, Science and Technology, and from Grant-in-Aid for Young Scientists (B, No. 21770032). P.M.H. is supported by grants from National Science Foundation Plant Genome Program (DBI-98-13360) and the U.S. Department of Agriculture- National Research Initiative Competitive Grants Program (08-35100-04529).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10044
References
- 1.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 2.Miura K, Jin JB, Hasegawa PM. Sumoylation, a posttranslational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Meulmeester E, Melchior F. SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 4.Yeh ETH. SUMOylation and De-SUMOylation: Wrestling with life’s processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lois LM, Lima CD, Chua NH. Small ubiquitinlike modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell. 2003;15:1347–1359. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura K, Rus A, Sharkhuu A, Yokoi S, Karghikeyan AS, Raghothama KG, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miura K, Hasegawa PM. Regulation of cold signaling by sumoylation of ICE1. Plant Signal Behav. 2008;3:52–53. doi: 10.4161/psb.3.1.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, Sadanandom A. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. 2008;20:2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 13.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, et al. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/ Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, et al. SUMO E3 ligase HIGH PLOIDY 2/AtMMS21 controls endocycle onsets and meristem maintenance in Arabidopsis. Plant Cell. 2009;21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 19.Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–1392. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Shuai K. Regulation of the sumoylation system in gene expression. Curr Opin Cell Biol. 2008;20:288–293. doi: 10.1016/j.ceb.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 2003;17:410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujii H, Verslues PE, Zhu J-K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth and gene expression in Arabidopsis. Plant Cell. 2009;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]