Abstract
We studied the kinetics of NADPH-dependent reduction of human CYP3A4 incorporated into Nanodiscs (CYP3A4-ND) and proteoliposomes in order to probe the effect of P450 oligomerization on its reduction. The flavin domain of cytochrome P450-BM3 (BMR) was used as a model electron donor partner. Unlike CYP3A4 oligomers, where only 50% of the enzyme was shown to be reducible by BMR, CYP3A4-ND could be reduced almost completely. High reducibility was also observed in proteoliposomes with a high lipid-to-protein ratio (L/P=910), where the oligomerization equilibrium is displaced towards monomers. In contrast, the reducibililty in proteoliposomes with L/P=76 did not exceed 55 ± 6%. The effect of the surface density of CYP3A4 in proteoliposomes on the oligomerization equilibrium was confirmed with a FRET-based assay employing a cysteine-depleted mutant labeled on Cys-468 with BODIPY iodoacetamide. These results confirm a pivotal role of CYP3A4 oligomerization in its functional heterogeneity. Furthermore, the investigation of the initial phase of the kinetics of CYP3A4 reduction showed that the addition of NADPH causes a rapid low-to-high spin transition in the CYP3A4-BMR complex, which is followed by a partial slower reversal. This observation reveals a mechanism whereby the CYP3A4 spin equilibrium is modulated by the redox state of the bound flavoprotein.
1. Introduction
The process of electron transfer from NADPH-cytochrome P450 reductase to cytochromes P450 constitutes an essential step in the catalytic cycle of microsomal cytochromes P450. Its critical importance in determining the catalytic efficiency and coupling of the microsomal monooxygenase has prompted a sustained interest in the mechanistic and kinetic aspects of the NADPH-dependent electron flow to cytochromes P450. However, although the kinetics of NADPH-dependent reduction has been studied for over 40 years, [1–8], the detailed mechanisms remain obscure.
The reduction kinetics in microsomal membranes and in most model systems are biphasic, with the rate constant of the two exponential phases differing by over an order of magnitude [2, 3, 6, 8]. Although there is no commonly accepted hypothesis on the mechanism of this biphasicity, its close connection to the position of P450 spin equilibrium is beyond doubt. In most cases, displacement of the spin equilibrium towards the high spin state caused by addition of Type I substrates increases the fraction of the fast phase [2, 4, 5, 7]. Furthermore, in some cases, the first exponential phase of reduction was shown to represent solely the reduction of the high-spin heme protein [5, 6]. These observations are difficult to explain in view of the extremely rapid rate of spin transitions [9–11], so that the spin equilibrium is expected to remain unchanged during the reduction. It appears likely that this apparent discrepancy reveals a persistent conformational heterogeneity of the heme protein [6, 12] that causes a divergence of the enzyme into two stable populations with different positions of the spin equilibrium and different rates of reduction.
Recent advances in X-ray crystallography of membranous cytochromes P450 have provided solid support for a vast conformational mobility in several of these heme proteins (see [13] for a review). This mobility is thought to play a pivotal role in substrate-dependent modulation of the functional properties of the enzyme and in the mechanisms of the substrate-induced spin shift and cooperativity in particular [14]. However, despite the fact that the functional catalytic unit of the monooxygenase system is represented by a complex of cytochrome P450 with (reduced) NADPH-cytochrome P450 reductase (CPR) and possibly cytochrome b5 (b5), little is known about conformational effects of P450 interactions with these redox partners. Moreover, the effect of the redox state of the electron carrier partners of P450 on their interactions with P450 remains completely unexplored. This fact calls into question the relevance of known effects of the interactions of P450 with oxidized CPR and b5 on spin equilibrium of P450 and its affinity for substrates (see [15] for a review) to the functionally competent monooxygenase system, where both CPR and b5 are presumed to be represented by their reduced states.
Another important question that remains unresolved is the structural and functional consequences of association of several P450 molecules to yield P450 oligomers. Numerous experimental observations reveal a considerable degree of homo- and hetero-oligomerization of cytochromes P450 in the endoplasmic reticulum (see [16] for review). In particular, oligomerization of cytochromes P450 in membranes was evidenced by rotational diffusion measurements in various implementations [17–22], freeze-fracture electron microscopy [20], immunochemical cross-linking [23], and intermolecular FRET studies employing the P450 enzymes fused with fluorescent proteins [24]. Homo- and hetero-oligomerization of cytochromes P450 in the membrane may play an important functional role (see [14, 16, 25] for review). According to our hypothesis [14], the conformational and/or steric consequences of the P450-P450 interactions in the oligomers may represent the primary cause of persistent conformational heterogeneity of the enzyme in the membrane, which is known to be tightly related to the mechanisms of heterotropic cooperativity in CYP3A4 [26–29]. Such oligomerization-promoted heterogeneity may have a prominent impact on the kinetics of electron transfer to cytochromes P450 [12, 29, 30].
An important impediment in interpretation of the kinetics of electron transfer in membranous systems at varying surface density of proteins is the difficulty in discrimination of the effects on the kinetics of intracomplex electron transfer from the changes in the concentration of the electron-transfer complexes. Membrane-incorporated systems provide no simple and adequate way to monitor and vary the concentration of the protein-protein complexes. A mixed artificial system, where the electron donor partner of a membrane-incorporated P450 is localized in the aqueous phase, may represent a model of choice for these studies. The use of a soluble flavoprotein partner, such as the flavin domain of P450BM-3 from Bacillus megaterium (BMR), eliminates the need for co-incorporation of the electron donor protein into the membrane and allows using a simple titration approach to determine the dissociation constant of the electron transfer complex. Localization of BMR in the aqueous phase also makes it possible to ensure an unchanged degree of P450 saturation with the flavoprotein at changing P450 surface density and to study the process of electron transfer at complete saturation of the heme protein with the reductase.
BMR shares 35% identity and 56% similarity to rat CPR with highly conserved segments involved in the binding of the flavins and pyridine nucleotide. The cluster of negative charges located at positions 207–215 in the FAD domain, which is believed to participate in the interactions of the rat microsomal reductase with cytochromes P450 [31], is also conserved in BMR (residues 647–655 in the FAD domain of the P450BM-3 sequence). A close resemblance of the mechanisms of interactions of CPR and BMR with microsomal cytochromes P450 is also supported by the finding that these two flavoproteins exhibit similar affinities and identical ionic strength dependencies in their interactions with rabbit microsomal cytochrome P450 2B4 (CYP2B4) [32]. An important distinction between P450-BM3 and mammalian cytochrome P450-reductase pairs is the difference in the electron-donating species of the flavins. In the microsomal systems the twoelectron reduced state of FMN, FMN hydroquinone (FMNH2), is thought to serve as the electron donor for cytochrome P450. However, electron transfer in the bacterial cytochrome P450-BM3 takes place from the one-electron reduced form of FMN, the anionic semiquinone (FMN·−) [33]. Despite this difference, BMR was found to be an efficient electron donor to several mammalian cytochromes P450 [34, 35]. However, it is unclear whether the FMN semiquinone anion of BMR is the electron donating species in these unnatural redox pairs, or whether the electrons are rather transferred from the FMN hydroquinone of BMR, similar to the natural pairs of CPR with microsomal cytochromes P450.
An artificial fusion protein consisting of BMR and human CYP3A4 domains has been recently constructed and shown to be active in the catalysis of CYP3A4-specific monooxygenation reactions. The turnover numbers exhibited by this catalytically self-sufficient chimera are close to those characteristic of CYP3A4 in reconstituted systems with rat CPR [34]. Therefore, BMR appears to be an appropriate substitute for the native flavoprotein of CYP3A4 in the model systems.
In a recent study of the kinetics of electron transfer in the complexes of BMR with oligomeric CYP3A4 in solution we demonstrated that the overall reducibility of CYP3A4 by BMR did not exceed 50% either in the absence of substrates or in the presence of bromocriptine, pyrenebutanol, or testosterone. At the same time, the reducibility of the high-spin fraction of the enzyme was always complete, so that the ferric CYP3A4 at the end of reduction process was represented by the low-spin form only [29]. Interestingly, the allosteric activator of CYP3A4 α-naphthoflavone (ANF) increased CYP3A4 reducibility, supporting a link between functional and conformational heterogeneity of CYP3A4 and heterotropic cooperativity. We also noticed a rapid displacement of the spin equilibrium of the ferric hemoprotein caused by the addition of NADPH to the complexes of CYP3A4 with BMR. This observation may reveal an important modulation of the spin state and redox properties of the heme protein by the state of reduction of the bound flavoprotein partner [29].
In the present study we probe the effect of the state of CYP3A4 oligomerization on the reducibility of the heme protein by BMR by use of nanoscale lipoprotein bilayer constructs (Nanodiscs), where the membrane-incorporated CYP3A4 is monomeric [36]. We also took advantage of the functionally homogenous and monomeric Nanodisc system to investigate the effect of the reduction of BMR flavins on the spin state of CYP3A4 associated with the flavoprotein. In our studies of the effect of P450-P450 interactions on its reducibility we also employ CYP3A4-containing proteoliposomes with different surface density of the heme protein. Whereas in liposomes with high surface density of CYP3A4 the enzyme is presumed to be severely oligomerized, decreased surface density of CYP3A4 in lipid-rich liposomes must result in a displacement of the oligomerization equilibrium towards monomers. Dissociation of CYP3A4 oligomers in the membranes of lipid-rich liposomes is demonstrated here by lifetime FRET measurements using a mutant of CYP3A4 with a single SH-reactive cysteine (CYP3A4(C468)) that was labeled with a BODIPY probe.
2. Materials and Methods
2.1. Materials
NADPH, 3,4-dihydroxybenzoic (protocatechuic) acid, protocatechuate-3,4-dioxygenase from Pseudomonas sp., bromocriptine mesylate, 3-sn-phosphatidic acid (PA) sodium salt from egg yolk, 3-sn-phosphatidylethanolamine (PE) from bovine brain, and L-α-phosphatidycholine (PC) from egg yolk were the products of Sigma Aldrich Chemicals (St. Louis, MO). α-Naphthoflavone (ANF) was from Indofine Chemical Company (Hillsborough, NJ). Octyl-β-D-glucopyranoside (octylglucoside, OG) was obtained from Fluka (Switzerland). 2-Decanoyl-1-(O-(11-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl) amino) undecyl)-sn-glycero-3-phosphocholine (BODIPY-PC), 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine (pyrene-PC), N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)methyl)iodoacetamide (BODIPY-FL iodoacetamide), and 1-pyrenebutanol (1-PB) were purchased from Invitrogen/Molecular Probes Inc. (Eugene, OR). 4,4-Difluoro-1-carboxymethyl-4-bora-3a,4a-diaza-s-indacene (CM-BODIPY) was a generous gift of Prof. Gelii Ponomariov (Institute of Biomedical Chemistry RAMN, Moscow, Russia). All other chemicals were of ACS grade and were used without further purification.
2.2. Expression and purification of CYP3A4 and BMR
CYP3A4 was expressed as the His-tagged protein in Escherichia coli TOPP3 [37]. The enzyme was purified to electrophoretic homogeneity by the three-column procedure described earlier [38] except that the Macro-Prep High S resin was replaced with Macro-Prep CM Support (Bio-Rad Laboratories, Hercules, CA), which yielded better purity and yield of the protein. This procedure yields enzyme with a ratio of absorbance at 418 and 280 nm (A418/280) of 1.50–1.55, which is close to the theoretical value of 1.62 expected for the pure low-spin CYP3A4 holoprotein. The preparations were stored at −80 °C as 100 – 150 µM solutions in 100 mM HEPES buffer (pH 7.4), containing 10% glycerol (v/v), 3 mM tris(2-carboxyethyl)phosphine (TCEP), and 1 mM ethylenediaminetetraacetic acid (EDTA). Expression of BMR in E. coli BL21(DE3) cells and purification of the protein was performed as described earlier [29].
2.3. Preparation of the cysteine-depleted C58T/C64A/C98S/C239S/C377S mutant of CYP3A4 (CYP3A4(C468))
CYP3A4(C468) mutant was generated using the QuikChange site-directed mutagenesis kit and a template consisting of the N-terminally truncated (Δ3–12) and tetrahistidine-tagged CYP3A4 wild-type cDNA by consecutive single-cysteine replacements to yield C58T/C64A/ C98S/C239S/C377S as described in [38]. The protein was expressed in E. coli and purified as described above for the wild-type enzyme.
2.4. Labeling of CYP3A4(C468) by BODIPY(FL) iodoacetamide
Labeling of the protein was performed essentially as described earlier [38]. After removal of TCEP from the stock solution of CYP3A4(C468) by dialysis against 100 volumes of 100 mM Na-Hepes buffer, pH 7.4, containing 10% glycerol (v/v)) under constant argon bubbling, the protein was diluted to the concentration of 10 µM by addition of the same buffer containing 0.2 % Igepal CO-630. Labeling was performed by incubation of the protein at constant stirring under argon atmosphere in the presence of 20µM BODIPY-FL iodoacetamide and 20 µM CM-BODIPY at room temperature. Addition of the latter non-reactive water-soluble BODIPY derivative was found to increase the degree of labeling (see below). The process of modification, which was monitored by increase in the fluorescence of the label, took about 40 minutes, after which time no further increase in the fluorescence intensity was observed. The reaction was stopped by addition of a 1 M solution of DTT to the final concentration of 2 mM. The detergent and the unbound probe were removed from the sample by washing on a Ni-agarose column (1 ml of the gel per 40 nmol of the enzyme). As determined from the UV-Vis absorbance spectra, the degree of labeling of the protein achieved with the above procedure was usually around 50%. It should be noted that the degree of labeling that could be achieved in the absence of CM-BODIPY, a non-reactive water-soluble BODIPY derivative, did not exceed 25%–30%. This effect of CM-BODIPY may be explained by its competition with BODIPY-FL for the interactions with the CYP3A4 substrate-binding site and/or hydrophobic patches on the protein surface. Displacement of BODIPY-FL from the complexes with the enzyme by CM-BODIPY may promote the modification by increasing the actual concentration of the SH-reactive probe.
2.5. Preparation of Nanodiscs containing CYP3A4
Soluble nanoscale membrane bilayer particles (Nanodiscs) containing CYP3A4 were obtained by a detergent-removal technique from the mixture of CYP3A4, phospholipid and a membrane scaffold protein (MSP1D1), as described previously [39]. Briefly, the membrane scaffold protein (MSP1D1) had been incubated with TEV protease to remove its histidine tag [39] and mixed with purified CYP3A4, palmitoyl oleoyl phosphatidyl choline (POPC), and sodium cholate at a 10:1:650:2000 molar ratio. Removal of detergent by treatment with Amberlite beads initiated a self-assembly process, which resulted in formation of a CYP3A4-containing discoidal POPC bilayer particles (~10 nm in diameter) stabilized by the amphipathic MSP1D1 belt [40, 41]. The discs were purified over a Sephadex G-200 size exclusion column to remove any partially formed discs, and on a Ni-NTA column to remove any empty Nanodiscs.
2.6. Preparation of proteoliposomes
Proteoliposomes were prepared by incorporation of CYP3A4 into pre-formed liposomes prepared with the octylglucoside/dialysis technique. We used a 2:1:0.6 mixture of PC, PE and PA with addition of 1 µg of BODYPY-PC per mg of lipid mixture. BODIPY-labeled phospholipid was included in the mixture for easy detection of the liposomes during their separation by gel filtration. This fluorescent phospholipid derivative was replaced with pyrene-PC in the preparation of the liposomes for the experiments with BODIPY-labeled CYP3A4(C468). Lipids (10 mg) were mixed as chloroform solutions, and the solvent was removed by evaporation under a stream of argon gas and subsequent drying under vacuum for 2 hours. The suspension of lipids in 2.5 ml of 100 mM HEPES, 150 mM KCl, 0.5 mM EDTA, containing 10% (v/v) glycerol, pH 7.4 (Buffer A) containing 1.54% octylglucoside was prepared using a vortex mixer and incubated for 30 minutes at room temperature under argon. The mixture was then diluted with the same buffer containing no detergent to the final concentration of octylglucoside of 0.43%. The mixture was dialyzed at 4 °C under constant gentle bubbling of argon gas against three changes of 500 ml of buffer A, each containing 0.5 g of Bio-Beads hydrophobic adsorbent (Bio-Rad Laboratories, Hercules, CA). After 72 hours of dialysis (24 hours per each portion of the buffer) the mixture was concentrated on 300 kDa cut-off Diaflo membranes (Millipore, Billerica, MA) to a phospholipid concentration of 8–10 mM and stored at −80 °C under argon. To incorporate cytochrome P450 into pre-formed liposomes a solution of purified CYP3A4 (100–150 µM) was added to an 8 mM suspension of the liposomes in Buffer A containing 1 mM DTT to reach a protein-to-lipid molar ratio of 1:50 (LPS-76) or 1:500 (LPS-910). Separation of proteoliposomes from unincorporated CYP3A4 was performed by gel filtration on a 1.2 × 40 cm column of Toyopearl HW-75 resin (Tosoh Bioscience GmbH, Stuttgart, Germany). The ratio of P450 to phospholipid in the column throughput solution was assessed by the ratio of optical densities at 418 and 506 nm (or 418 and 343 nm in the case of the liposomes containing pyrene-PC). The fractions with a ratio differing by no more than 25% were pooled and concentrated to a CYP3A4 concentration of 20–30 µM. The molar ratio of phospholipids to cytochrome P450 in the final preparation was estimated based on the determination of total phosphorus in chlorophorm/methanol extract according to Bartlett [42]. The preparations of the proteoliposomes were stored at −80 °C under argon atmosphere.
2.7. Scanning stop-flow kinetic experiments
We used a rapid scanning MC2000-2 CCD-spectrometer (Ocean Optics, Inc., Dunedin, FL) combined with a SF-MiniMixer Stopped-Flow apparatus (KinTek Corporation, Austin, TX) as described earlier [12]. Anaerobiosis was achieved using an oxygen scavenging system consisting of protecatechuate-3,4-dioxygenase (0.5 units/ml) and 5 mM protocatechuic acid. The reaction was initiated by mixing equal volumes of (i) the solution of CYP3A4 and BMR and (ii) a 0.4 mM solution of NADPH containing glucose-6-phosphate dehydrogenase (2 units/ml) and 20 mM glucose-6-phosphate. When the reduction was studied in the presence of substrates, 1-PB or ANF was added to both syringes of the stop-flow apparatus to the concentration of 30 µM. All experiments were performed at 25 °C in 100 mM HEPES buffer (pH 7.4) containing 1 mM DTT and 1 mM EDTA.
2.8. Fluorescence spectroscopy
Time-resolved fluorescence spectroscopy (TRFS) measurements were performed in a 100 µl 10 × 2 mm ultra-micro fluorescence cell (Hellma GmbH & Co. KG, Müllheim, Germany, Product #105.250) with a FLS 920 fluorescence lifetime and spectrometer from Edinburgh Instruments (Edinburgh, UK). The spectrometer was equipped with a PDL 800-D pulsed diode laser driver and a LDH405 laser head from Picoquant GmbH (Berlin, Germany) as a light source. Fluorescence excitation was performed at 405 nm and fluorescence decay curves were monitored at 520 nm with a 100 ns time span. Fitting of the decay traces was made with the F900 Fluorescence Spectrometer Software (Edinburgh Instruments) using the deconvoluted bi-exponential fit mode, which takes into account the instrument response function (IRF). IRF was measured by registering the decay traces at the wavelength of excitation with a Teflon diffuser placed into the cell holder.
Steady-state fluorescence measurements were done using an MC2000-2 multi-channel CCD rapid scanning spectrometer (Ocean Optics, Inc., Dunedin, FL, USA) equipped with one fluorescence and one absorbance channels. Excitation of BODIPY fluorescence was performed at 405 nm using an LDH405 pulsed diode laser. Simultaneous control of the changes in the absorbance of the sample was performed employing the second (absorbance) channel of the spectrometer and using an OSRAM 64614 UV-enhanced tungsten halogen lamp (OSRAM, Germany) as a light source. Alternating the light sources during the measurements was achieved with computer-controlled FOS-2-inline fiber optic switch from Avantes (Eerbeek, The Netherlands).
In order to prevent the oxidation of the lipids of the membrane during our lengthy FRET experiments with the liposomes, these experiments were performed under anaerobiosis, which was attained with argon bubbling and the use of the oxygen-scavenging system with protecatechuate-3,4-dioxygenase, as described above.
2.9. Data processing
The series of absorbance spectra obtained in kinetic experiments were analyzed using principal component analysis (PCA) combined with the fitting of the principal component spectra to a set of spectral standards, as described previously [12, 43, 44]. Briefly, the PCA procedure was applied to a set of difference spectra obtained by subtraction of the first spectrum in the series (zero time point) from all subsequent spectra. To interpret the resulting series in terms of the changes in concentrations of the ferric low-spin, ferric high-spin, and the ferrous carbonyl complexes of P450 and P420 states of the heme protein, the principal vectors were approximated with a linear combination of the respective spectral standards combined with a low-order (≤4) polynomial function. The latter was introduced to compensate for the baseline fluctuations during the experiment. The high-ranking components characterized by a square correlation coefficient (ρ2) above the threshold, which was set to 0.5 in most cases, were considered significant and used to calculate the changes in the concentrations of the P450 species. This technique was described in detail earlier [43, 44]. The set of spectral standards consisted of the spectra of ferric low- and high- spin P450 species of CYP3A4 [28] and the spectra of the carbonyl complexes of ferrous P450 and P420 states of CYP3A4. The latter were refined by studying the pressure-induced P450-to-P420 conversion of the CYP3A4 ferrous carbonyl complex, similarly to the technique used for CYP2B4 and CYP1A2 [45, 46]. The complete set of the standards used in this study are available on-line as Supporting Information to our recent publication [29]. These kinetic curves were fitted to a multiexponential equation using a combination of Marquardt and Nailder-Mead algorithms. All data fitting and spectral analysis procedures were performed using our SPECTRALB software [43].
3. Results
3.1 Interaction of BMR with CYP3A4
In designing our kinetic experiments we sought to examine the kinetics of intracomplex electron transfer under conditions of complete saturation of CYP3A4 with the flavoprotein. This setup requires knowledge of the dissociation constant of the complex of BMR with CYP3A4-incorporated into Nanodiscs or proteoliposomes. Our approach to determination of these dissociation constants was based on the known fact that the formation of the complexes of cytochromes P450 with CPR results in a displacement of the spin equilibrium of the heme protein, which is reflected in the changes in UV-Vis absorbance similar to those observed for the binding of Type-I substrates [47–49].
In order to increase sensitivity of the assays and resolve the absorbance bands of CYP3A4 from those of the flavoprotein we employed the technique of a tandem cell (“counter-flow”) continuous variation titration [50], similar to that described in our study with CYP3A4 in solution [29]. At the beginning of these experiments the first and the second compartments of a tandem cell contained equal volumes of equimolar solutions of BMR and either CYP3A4-ND or CYP3A4 incorporated into the proteoliposomes. Mixing of the interacting proteins was achieved by reciprocal gradual displacement of small aliquots of the solutions between compartments, so that the molar fraction of CYP3A4 in the first compartment (F1) increased from 0 to 0.5, concomitant with a decrease in this fraction from 1 to 0.5 in the second compartment. As discussed in [50], in the case of the formation of 1:1 complexes between the interacting proteins, the titration curves obtained in this way are equivalent to the ascending branch of the symmetric bell-shaped curve characteristic of the traditional Job’s titrations. In addition to probing the stoichiometry of complex formation, this setup considerably increases the accuracy of titrations by eliminating the spectral signal of the reductase, so that the observed spectral changes reflect the changes in the absorbance of the heme protein only.
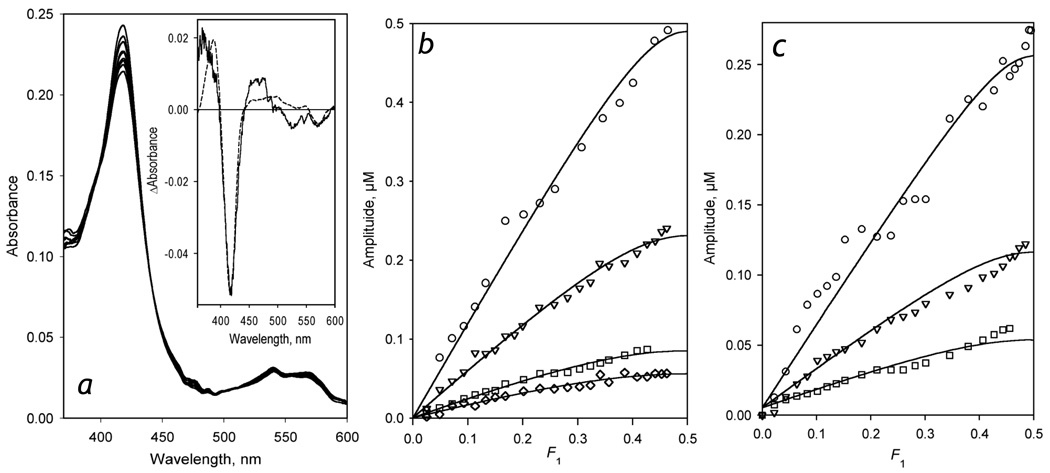
A series of spectra obtained in a counter-flow continuous variation experiment with 5 µM CYP3A4-containing proteoliposomes (LPS-76) is shown in Fig. 1a. As seen from this figure, a stepwise mixing of the equimolar solutions of LPS-76 and BMR results in a decrease of the amplitude of the low-spin CYP3A4 Soret band at 418 nm concomitant with increase in the absorbance band at 380 – 400 nm. This BMR-induced transition in CYP3A4 is attributable to a low- to- high spin shift, as expected according to published observations [47–49]. However, detailed analysis of the spectrum of the first principal component of the changes (Fig 1a inset) reveals some deviations from the shape characteristic of a canonical low-to-high spin shift. In particular, the position of the main maximum of the spectrum is shifted to wavelength shorter than expected (388 nm) and the amplitude of this maximum is lower than that anticipated from the amplitude of the trough at 420 nm. It should be noted, however, that the spectra reported by French and co-authors for the interactions of CYP2B4 with CPR in the absence of substrate ([47], Fig. 5A, inset) may be also indicative of a similar deviation.
Figure 1.
Formation of the CYP3A4 complexes with BMR studied by counter-flow (tandem cell) continuous variation titration. The interactions were monitored by the changes in the concentration of P450 low spin and high spin states. Panel a shows a series of spectra obtained in the experiment with CYP3A4 containing liposomes (LPS-76) at a concentration of 5 µM. The inset represents the spectrum of the first principal component obtained by PCA. The dashed line shows the approximation of this spectrum by the set of CYP3A4 absorbance standards. Panel b shows a series of titration curves obtained with LPS-76 and BMR taken at the concentration of 5 (circles), 2.6 (triangles), 1.1 (squares) and 0.8 µM (diamomds). Panel c represents a series obtained with CYP3A4-ND and BMR at a concentration of 5.3 (circles), 2.6 (triangles), and 1.3 µM (squares). Solid lines represent the results of the fitting of the data sets to the equation for the equilibrium of bimolecular association suited to the case of the counter-flow titration (eq. (3) in [50]).
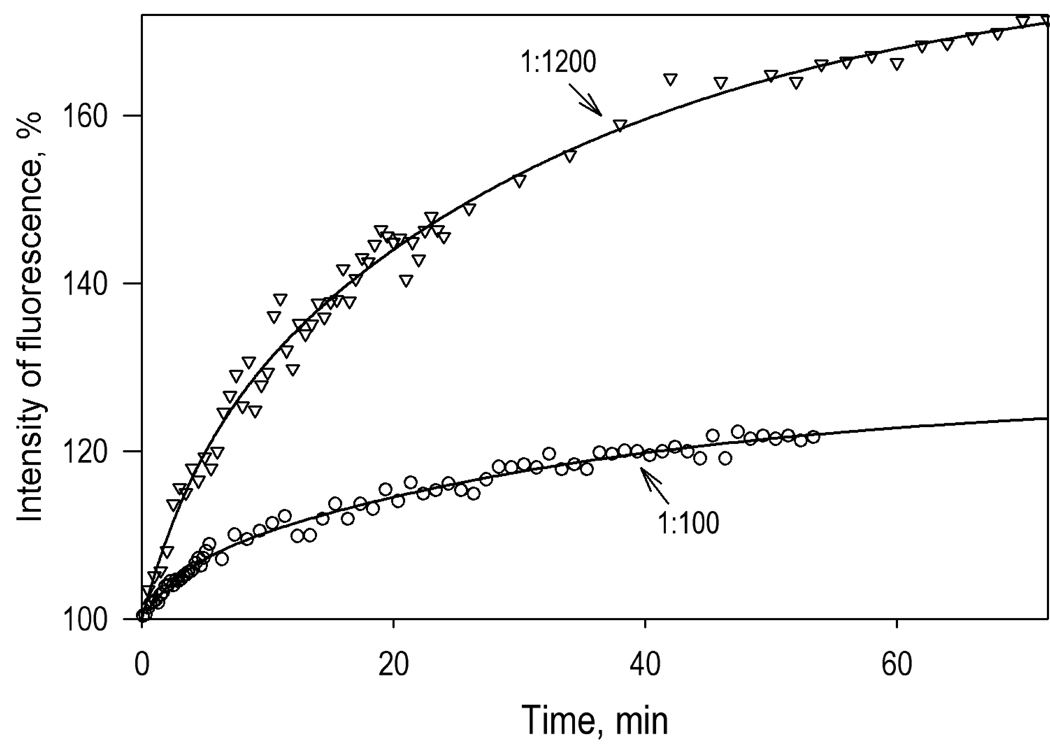
Figure 5.
Kinetics of the changes in the intensity of fluorescence of CYP3A4(C468)-BODIPY upon its interactions with liposomes. Conditions: 1 µM CYP3A4(C468)-BODIPY in 0.1 M Na-HEPES buffer, 1 mM DTT, and 1 mM EDTA. Process was initiated by the addition of the liposomes to yield the L/P ratio of 100 (circles) and 1200 (triangles). Solid lines show the results of fitting of the kinetic curves with a bi-exponential equation, which gives the maximal amplitudes of the observed changes of 26% and 78% respectively.
A series of titration curves obtained with various concentrations of LPS-76 are shown in Fig. 1b. Here the abscissa represents the molar fraction of BMR in the first compartment of the tandem cell, while the ordinate shows the amplitude of the observed decrease in the mean concentration of the low-spin heme protein in the two compartments. The shape of the titration curves is indicative of the formation of a 1:1 complex. Global fitting of this data set to the equation for the equilibrium of bimolecular association suited to the case of the counter-flow titration (eq. (3) in [50]) gives the KD values of CYP3A4-BMR complex of 0.14 ± 0.08 µM and the maximal amplitude of the BMR-induced spin shift of 26.5 ± 6 %. Similar results were also obtained with CYP3A4-ND (Fig. 1c), where the dissociation constant and the maximal spin shift amplitude were equal to 0.10 ± 0.05 µM and 11.5 ± 0.5%
Therefore, similar to CYP3A4 in solution [29], the heme protein incorporated in to the lipid bilayer in either proteoliposomes or Nanodiscs interacts with BMR with the formation of 1:1 complexes. The values of dissociation constants of these complexes suggest that at the concentrations of CYP3A4 and BMR used in our kinetic experiments (1.5–3 and 3–6 µM respectively), the whole pool of the heme protein is represented by the complexes with its flavoprotein partner.
3.2 Spectral changes in BMR during its NADPH-dependent reduction
Since flavins exhibit absorbance spectra overlapping those of cytochrome P450 and that are highly dependent on the state of reduction, spectral studies of the process of reduction of cytochrome P450 in the presence of excess BMR require a detailed knowledge of the spectral transitions of the flavoprotein upon its reduction. In order to obtain a set of spectral standards characterizing these changes we examined the kinetics of NADPH-dependent reduction of the flavoprotein by rapid-scanning stop-flow spectrometry. It should be noted that we did not intend to repeat the analysis of the process of BMR reduction, which was done in fine detail by Sevrioukova and co-authors [51]. The only purpose of our studies of BMR reduction was to obtain a set of prototypical differential spectra (spectral standards) which may be used to discriminate the transitions in cytochrome P450 from those in the reductase.
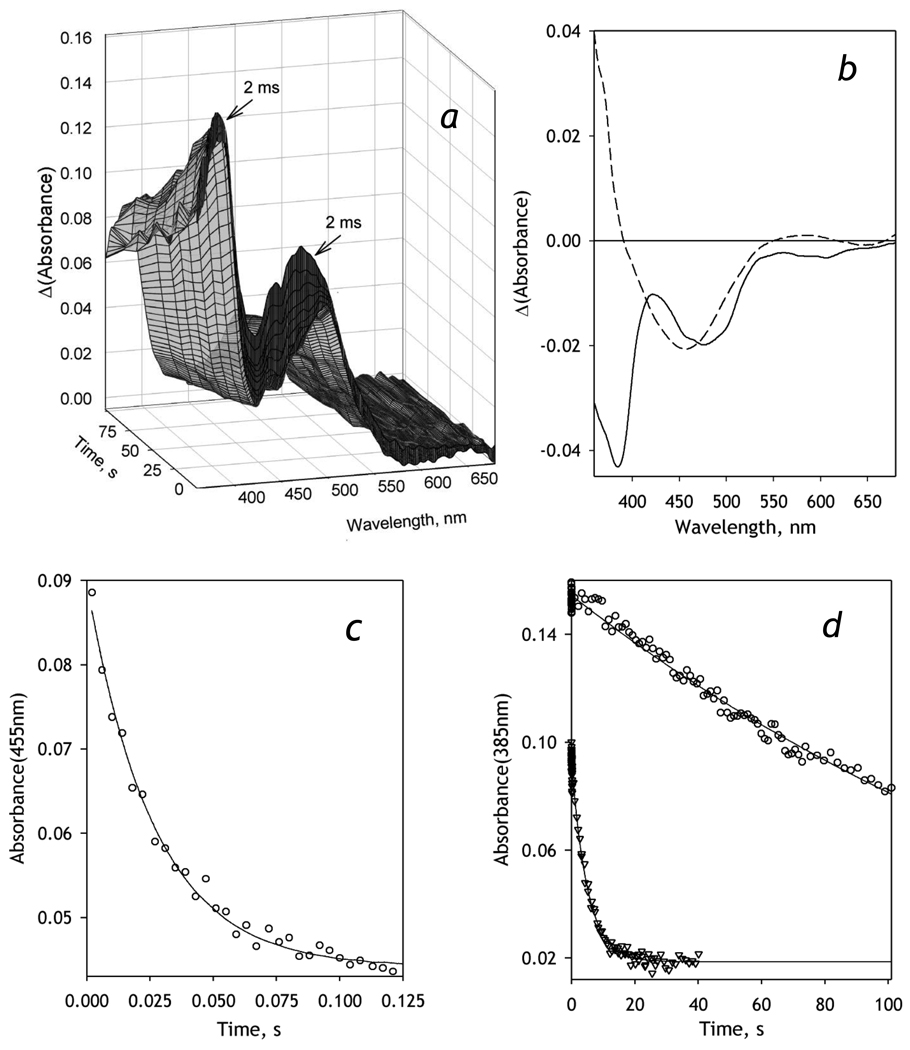
As the process of BMR reduction is very rapid [51], these experiments were performed at 5° C for better resolution of spectral changes. As seen from Fig 2a, the first spectrum of the series is already characterized by a substantially decreased absorbance band of the flavoprotein at 455 nm, as compared with the absorbance spectra of the oxidized BMR (see Fig. 1A in [51]). This decrease in the ratio of the flavin bands at 380 and 456 nm is indicative of formation of the reduced anionic red semiquinone form of FMN [51], which takes place during the dead time of the instrument (~2 ms). During the initial phase of monitoring, a decrease in the absorbance band at 456 nm continues, while the absorbance at 380 nm remains virtually unchanged. During the following, slower phase of the reduction, a decrease in the absorbance at 380 nm indicates the formation of the flavin hydroquinone. No considerable absorbance band of the neutral blue flavin semiquinone around 550–600 nm was observed in our experiments, demonstrating that the anionic flavin semiquinone is the predominant semiquinone state of FMN in BMR, in agreement with previous observations [51, 52]. It should be noted that, due to very low extinction coefficients of the blue semiquinone absorbance band (λmax ≈ 550 nm) and the charge transfer band of the BMR complex with NADPH (λmax ≈ 750 nm), we made no attempt to resolve negligibly small alterations of BMR absorbance caused by the changes in the concentration of these species. These changes, which were characterized in detail by Sevrioukova and co-authors [51], are negligible compared with the changes in CYP3A4 absorbance and may be ignored in the spectral analysis of the CYP3A4 reduction process.
Figure 2.
Kinetics of NADPH-dependent reduction of BMR recorded with a rapid scanning stop-flow technique. Conditions: 20 µM BMR, 0.2 mM NADPH in 0.1 M Na-Hepes buffer, pH 7.4, 1 mM DTT, 1 mM EDTA containing NADPH-generating and oxygen-scavenging systems (see Materials and Methods), 5 °C. Spectra recorded in a stop-flow cell with 5 mm optical path length. a: Absorbance spectra of BMR taken during the reduction. The first spectrum corresponds to ~2 ms after addition of NADPH. b: The spectra of the first (solid line) and the second (dashed line) principal components found by PCA applied to a manifold of 5 series of spectra obtained in independent experiments. Panels c and d show the changes in absorbance at 455 and 385 nm respectively. Data points in circles correspond to the data set obtained at 5 °C shown in panel a. The data points shown in triangles in panel d represent a similar experiment at 25 °C. Solid lines show the results of fitting of the kinetic curves by a first order kinetic equation.
Application of PCA reveals two significant principal vectors, which cover over 99.5% of the spectral changes (Fig. 2b). These principal vectors were obtained by global analysis of a series of sets of spectra obtained in four independent measurements and were used to resolve BMR- and CYP3A4-related spectral changes in the analysis of the results of our CYP3A4-reduction experiments (see below). The first principal component reflects a decrease in the amplitudes of both 380 and 450–470 nm bands, which is indicative of the formation of the hydroquinone or neutral semiquinone forms of the flavin. In contrast, the long-wavelength and short-wavelength branches of the second principal vector have opposite signs, so that a simultaneous increase in the loading factors of both principal vectors represents a decrease in the 456-nm band only. This simultaneous increase observed during the initial phase of the reduction represents the formation of red, anionic semiquinone forms of the flavins. This rapid initial phase (Fig. 2c) obeys a simple first-order reaction equation with a rate constant of 23 ± 8 s−1, which is consistent with the value of 27 s−1 reported by Sevrioukova and co-authors as a rate constant of the formation of [NADPH+, FADH•, FMN•−] state [51]. The slow phase of the reduction, which is characterized by a rate constant of 0.019 ± 0.08 s−1 is attributable to the appearance of the three- and/or four-electron reduced BMR that contains flavin hydroquinone(s). At 25 °C this rate constant increases 10 times and reaches 0.16 ±0.03 s−1 (Fig. 2d), while the rate constant of the fast phase increases to approximately 30 – 60 s−1 and becomes too high to be determined with our technique (data not shown).
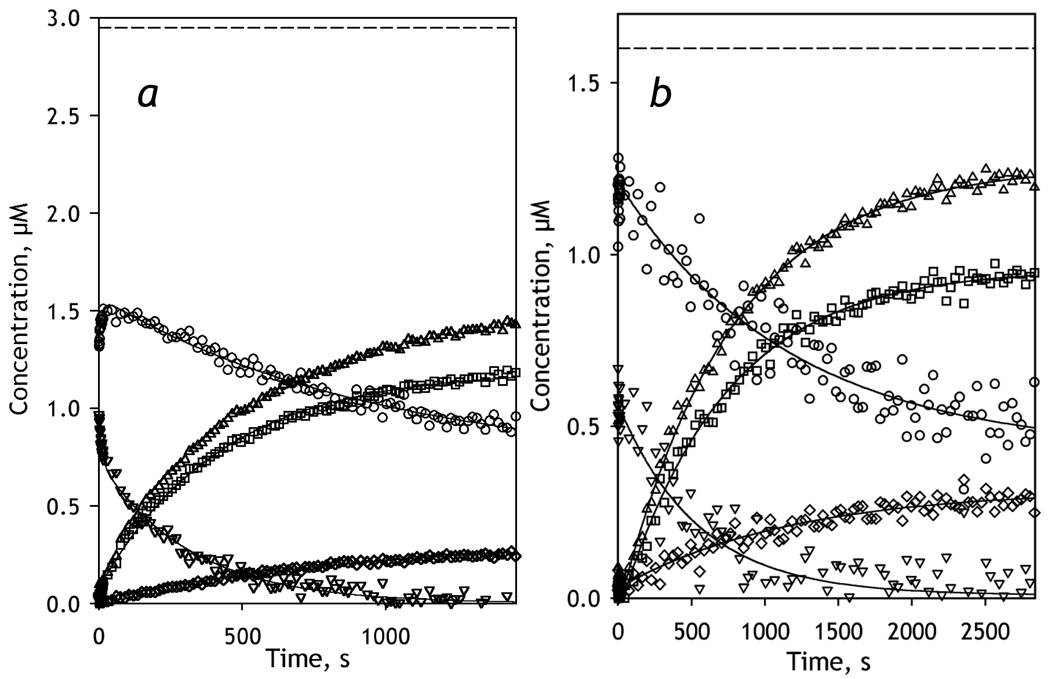
3.3 Kinetics of CYP3A4 reduction in Nanodiscs
A series of spectra taken after addition of NADPH to the mixture of CYP3A4-ND with BMR reflects both the reduction of BMR flavins and a consequent electron transfer to the heme protein (see Fig. S1a in the (Supporting Material). In order to analyze the changes in the concentrations of the individual P450 states (ferric high- and low-spin states and the ferrous carbonyl complexes of both P450 and P420 forms of CYP3A4) we used a PCA-based procedure designed to separate the spectral changes in the heme protein from those in the flavins. Application of PCA to the series of raw spectra taken in the process of reduction resulted in two principal components which cover over 99.5% of the spectral changes. The principal components were approximated by a linear combination of the standard differential spectra of CYP3A4 reduction complemented with the two differential spectra reflecting the process of BMR reduction (see above). The results of this approximation (ρ2≥0.99) were used to eliminate the changes in the flavin-specific bands from the experimental spectra.
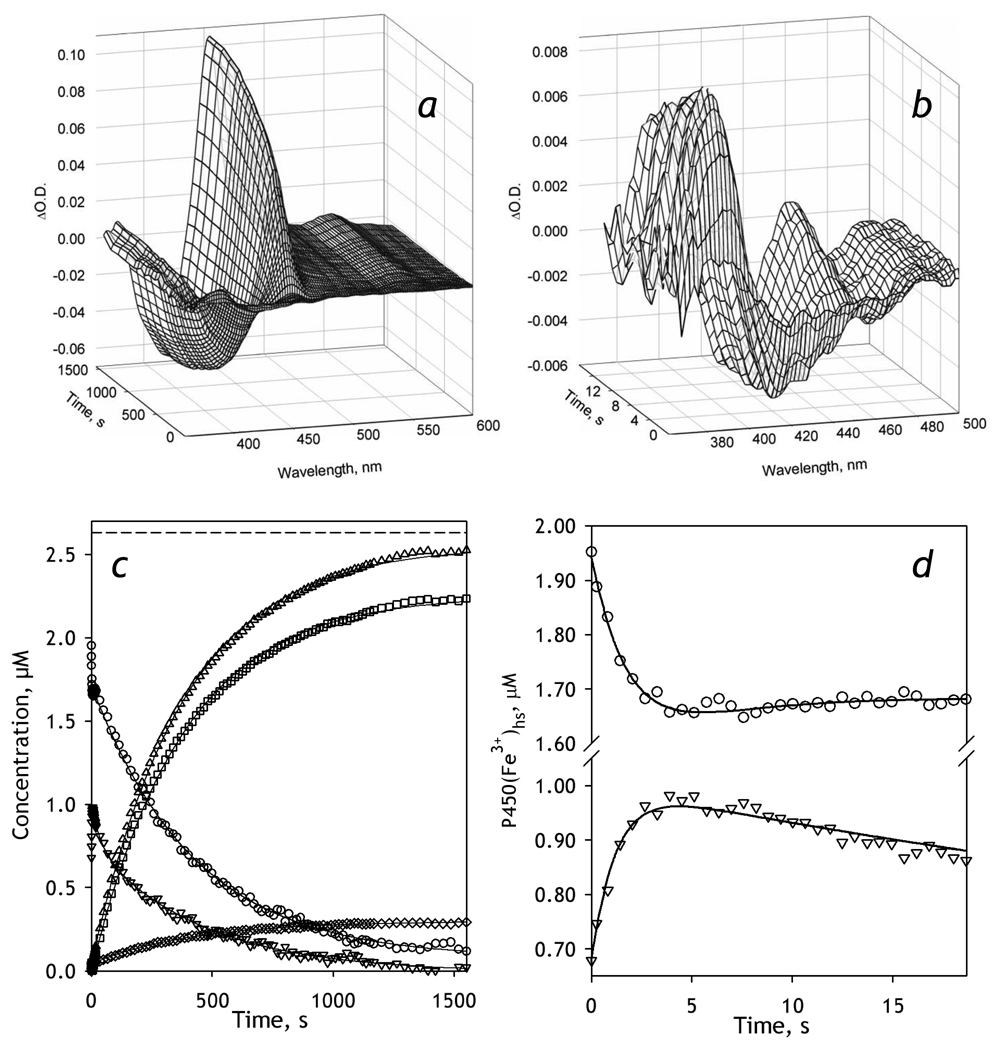
A series of differential spectra reflecting the changes in CYP3A4-ND, which was obtained in this way, is exemplified in Fig. 3a and 3b. A preliminary visual analysis of this series suggests that the appearance of the band of absorbance of ferrous carbonyl CYP3A4 at 448 nm is associated with a profound decrease in the band centered around 390 nm, which suggests a major decrease in the concentration of the high-spin P450. This is quite unexpected, as the content of the high-spin heme protein in the presence of BMR at no substrate added was found to be as low as 5% (Fig. 1). Therefore, the first spectrum of the series, which was taken during the first 2–3 ms after the mixing (the dead time of the instrument) and used as a base for the difference spectra shown in Fig. 3, appears to be highly enriched in the high-spin state compared with the oxidized enzyme without NADPH. Furthermore, the spectra taken during the initial phase of reduction (~5 seconds after the addition of NADPH) reflect rapid kinetics of further conversion of the low-spin P450 to the high-spin state (Fig. 3b). Correspondingly, the analysis of time changes in the concentration of the individual states of CYP3A4 (Fig. 3c) indicates that the content of the high-spin state of P450 at the end of the dead-time of mixing was as high as 41 ± 2%. The initial phase of the changes in the concentration of the high-spin state (Fig. 3d) exhibits a further increase, which obeys an exponential equation with a rate constant of 1.0 ± 0.2 s−1 (based on the average of the results of 3 individual experiments). This initial increase paralleled a decrease in the low-spin content (Fig. 3d), signifying a low-to-high spin transition taking place in the CYP3A4-BMR complex in response to the reduction of BMR.
Figure 3.
Kinetics of BMR-dependent reduction of CYP3A4-ND in the absence of substrate. Conditions: 2.6 µM 3A4, 5.2 µM BMR, 0.2 mM NADPH, CO-saturated 0.1 M Na-Hepes buffer, pH 7.4, 1 mM DTT, 1 mM EDTA containing oxygen-scavenging and NADPH-generating systems (see Materials and Methods), 25 °C. Spectra recorded in a stop-flow cell with 5 mm optical path length. a, b: Changes in absorbance in the Soret region during the reduction. The spectrum measured at time of origin is subtracted. Panel b scales up the spectra representing the first 14 seconds after mixing. c: The time course of the changes in the concentrations of the carbonyl complexes of CYP3A4(Fe2+) P450 (squares) and P420 states (diamonds), their total (triangles), as well as the low-spin (circles) and the high-spin (inverted triangles) states of CYP3A4(Fe3+). The dashed line indicates the total concentration of CYP3A4. Panel d shows the initial part of the kinetic curves for the low-spin (circles) and the high-spin (inverted triangles) states of CYP3A4(Fe3+).
As seen in Fig. 3d this initial spin shift changes its direction at ~5 sec after mixing, so that the concentration of the high-spin enzyme starts to decrease, concomitant with an increase in the low-spin content. This partial reversal of the initial spin shift is followed by a continuous decrease in both ferric states due to the reduction of the heme protein to ferrous carbonyl complexes (Fig. 3c). The process of reduction reflected in the appearance of the reduced states of the heme protein obeys a single-exponential equation with the rate constant of (2.5 ± 0.2)·10−3 s−1. In contrast, the decrease in the concentration of the high-spin state is bi-exponential, with a rate constant of the fast and slow phases of 0.037 ± 0.010 s−1 and (2.4 ± 0.2)·10−3 s−1, respectively. The dynamics of the low-spin fraction after the initial rapid spin-shift also reflects two exponential terms, although in this case the signs of these terms are opposite. While the increase in the low-spin between 7 and 20 s after mixing is a counterpart of the corresponding high-spin decrease, the slow phase (k = (2.2 ± 0.3)·10−3 s−1 ) reflects the reduction of the heme protein.
Therefore, our analysis resolved three consecutive processes reflected in the changes in the concentration of P450 states after addition of NADPH. The fastest process (k = 1.0 ± 0.2 s−1) reflects a low-to-high spin transition, which is thought to be a result of the formation of a red flavin semiquinone state of BMR (presumably [NADPH+, FADH•, FMN•−]), which takes place during the first milliseconds after addition of NADPH (k ≥ 30 s−1, at 25 °C, see above). The following slow partial reversal of the above spin shift (k = 0.037 ± 0.010 s−1) is apparently caused by the formation of the completely reduced state of BMR (and, as a result, disappearance of the anionic semiquinone state), which has a rate constant of 0.16 ± 0.03 s−1 at 25 °C (see above). The third, highest amplitude process reflects the reduction of CYP3A4 with formation of the carbonyl complexes of the ferrous heme protein.
As seen from Fig. 3 and Table 1, the reducibility of CYP3A4-ND by BMR is close to 100%, in contrast to incomplete reducibility observed in the enzyme oligomers in solution. [29]. As seen from Fig. 3c, in addition to the formation of the ferrous P450 state, reduction of CYP3A4-ND results in the formation of some amount (≤15%) of P420. At the end of reduction the concentration of the carbonyl complexes of these two ferrous states of the enzyme constitute 94 ± 4% of the total concentration of the heme protein (Table 1). Addition of the CYP3A4 substrates 1-PB or ANF had virtually no effect on the kinetics or extent of reduction of CYP3A4-ND by BMR (Table 1). In contrast to enzyme oligomers in solution, where the addition of ANF improved the reducibility considerably [29], this effector rather caused a marginal decrease in the maximal level of reduction in CYP3A4-ND (Table 1).
Table 1.
Parameters of BMR-dependent reduction of CYP3A4 in Nanodiscs and in proteoliposomes with various content of the heme protein.*
| System | Substrate | Fh, %a | kf·103, s−1b | ks·103, s−1 | Ff, % | Total reducibility, % |
|
|---|---|---|---|---|---|---|---|
| CYP3A4-ND | none | 41 ± 2 | 37 ± 10 | 2.4 ± 0.2 | 20 ± 10 | 93 ± 4 | |
| 1-PB | 38 ± 1 | 37 ± 5 | 2.2 ± 0.5 | 28 ± 4 | 91 ± 2 | ||
| ANF | 49 ± 4 | 23 ± 16 | 3.2 ± 0.9 | 35 ± 10 | 79 ± 6 | ||
| LPS-910 | none | 36 ± 6 | 1.8 ± 0.7 | 83 ± 7 | |||
| ANF | 32 ± 7 | 0.87 ± 0.5 | 85 ±12 | ||||
| LPS-76 | none | 29 ± 5 | 111 ± 34 | 2.4 ± 0.4 | 33 ± 8 | 55 ± 6 | |
| ANF | 55 ± 4 | 50 ± 13 | 2.7 ± 0.4 | 25 ± 4 | 47 ± 2 | ||
The values given in the table represent the averages of 3–7 individual measurements and the “±” values show the confidence interval calculated for p = 0.05. The kinetic curves of the decrease in the high spin state in Nanodiscs and in LPS-76 were fitted to a bi-exponential equation. In all other cases a mono-exponential equation was used. The values of kf and ks represent the rate constants of the fast and the slow phases of the decrease in the concentration of the high-spin ferric state, while the value of Ff represent the relative amplitude of the fast phase. The rate constants of the changes in the low-spin ferric and both (P450 and P420) ferrous states were characterized by values indistinguishable from those of ks within the accuracy of the confidence interval.
The fraction of the high spin state at the beginning of the reduction, which is calculated from the amplitude of the decrease in the high-spin ferric state in the reduction.
The reducibility is calculated from the increase in the concentrations of the carbonyl complexes of ferrous P450 and P420 states.
3.4 Kinetics of CYP3A4 reduction in proteoliposomes as a function of the surface density of the enzyme
In order to probe the effect of CYP3A4 oligomerization in the membrane on its reducibility by BMR we studied the kinetics of reduction in proteoliposomes with two different lipid-to-protein ratios. In the first preparation, which we designate as LPS-76, the molar ratio of CYP3A4 to phospholipids was as low as 1:76, whereas in the second preparation of proteoliposomes, designated as LPS-910, this ratio was equal to 1:910. This decreased surface density of CYP3A4 in LPS-910 is presumed to cause a displacement of the oligomerization equilibrium towards monomers, as compared with LPS-76 where the enzyme is severely oligomerized. Kinetic curves of the changes in the concentration of ferric and ferrous states of CYP3A4 after addition of NADPH to the liposomes are shown in Fig. 4. As seen from this figure and Table 1, these two preparations differ considerably in the reducibility of CYP3A4 with an amplitude of BMR-dependent reduction of 55 ± 10 % of the total CYP3A4 content in LPS-76 and a reducibility in LPS-910 as high as 83 ± 10 %. This increase in reducibility upon decreased surface density of the heme protein is attributed to a high degree of CYP3A4 monomerization in the diluted membranes as in CYP3A4-ND.
Figure 4.
Kinetics of BMR-dependent reduction of CYP3A4 incorporated in proteoliposomes at a 1:76 (LPS-76, a) or 1:910 (LPS-910, b) molar ratio of CYP3A4 to phospholipids. Reaction mixtures contained 2.95 (a) and 1.6 µM µM (b) CYP3A4 in proteoliposomes and a two-fold molar excess of BMR. Other conditions were as indicated for Fig. 3. The panels show the time course of the changes in the concentrations of the carbonyl complexes of CYP3A4(Fe2+) P450 (squares) and P420 states (diamonds), their total (triangles), as well as the low-spin (circles) and the high-spin (inverted triangles) states of CYP3A4(Fe3+). The dashed line indicates the total concentration of CYP3A4.
The rapid changes in the spin state of ferric CYP3A4 caused by reduction of BMR, which we discussed above for the case of CYP3A4-ND, are also clearly seen in LPS-76 (Fig 4a). Similar to CYP3A4-ND, in LPS-76 we observed bi-exponential kinetics of the decrease in the high-spin content, which follows a very rapid initial low-to-high spin burst. In contrast, changes in the concentration of the low-spin state exhibit an intermittent increase, which is followed by a monoexponential reduction phase. In the case of diluted liposomes (LPS-910), this initial spin shift could not be resolved due to high level of noise in this turbid system (Fig. 4b). However, even in this case the amplitude of the observed decrease in the concentration of high-spin state of the enzyme was considerably higher than the amplitude that could be expected based on the high-spin content in LPS-910 mixture with oxidized BMR, which did not exceed 5% (data not shown).
Surprisingly, in contrast to CYP3A4 in solution, addition of ANF to LPS-76 or LPS-910 had no effect on either reducibility of the enzyme or the kinetic parameters of reduction. One of the possible explanations for this contrast may be the effect of polar groups of phospholipids on the interactions of CYP3A4 with BMR, a soluble enzyme which accesses CYP3A4 from the aqueous phase. These interactions, which may differ from those of membranous CPR, may interfere with the effect of substrate in our model system.
3.5 Design of a FRET-based method for probing CYP3A4 oligomerization
In order to probe the state of CYP3A4 oligomerization in liposomes and validate our conclusion that the differences in the kinetics of CYP3A4 reduction in LPS-76 and LPS-910 are due to different degrees of oligomerization of the enzyme, we established a new FRET-based assay for monitoring the equilibrium of CYP3A4 oligomerization in the membrane. In these studies we used BODIPY-FL iodoacetamide as the FRET donor. The spectrum of emission of this probe is characterized by a broad asymmetric band (λmax= 513 nm), which has a profound overlap with the α- and β-bands of the CYP3A4 heme, so that the value of R0 of FRET in BODIPY-heme pair is as large as 62 Å (κ2= 2/3). The label is therefore anticipated to be an efficent donor of FRET to the heme chromophores located both in the same protein molecule as well as in the neighboring CYP3A4 subunits in the oligomer. Dissociation of oligomers is therefore expected to result in a considerable increase in the intensity and the lifetime of the fluorescence.
To ensure uniform site-specific labeling of the enzyme we used our recently developed cysteine-depleted mutants of CYP3A4 [38]. In our preliminary studies (data not shown) we labeled three mutants that contain a single reactive cysteine CYP3A4(C64), CYP3A4(C239) and CYP3A4(468) with BODIPY and found that the highest amplitude of inter-molecular FRET from the label to the heme in CYP3A4 oligomer is achieved with CYP3A4(C468). The interactions of this mutant with BCT, 1-PB, and ANF were very close to those of the wild type CYP3A4 (data not shown).
Measurement of the amplitude of the increase in the domor fluorescence caused by destruction or abstraction of the acceptor chromophore (the heme in our case) provides the most accurate way to probe the efficiency of FRET. Destruction of the porphyrin ring by H2O2 is known as an efficient and gentle approach to deplete cytochromes P450 completely of the heme group [38, 53, 54]. Therefore, in order to estimate the efficiency of FRET from BODIPY to the heme chromophore we employed the heme depletion (bleaching) of CYP3A4(C468)-BODIPY in the presence of H2O2. These experiments, which are illustrated in Supporting Material, indicated that the efficiency of FRET in the oligomers in solution is equal to 82% (see Fig. S3). Monomerization of the enzyme oligomers by addition of 0.5% non-ionic detergent Igepal CO-630 results in a 2.5-fold increase in the fluorescence intensity (Fig. S4). This increase was accompanied by an increase in the mean fluorescence lifetime, so that the fraction of the fast phase for bi-exponential approximation of the decay traces decreases from 81% to 22% upon monomerization of the enzyme. Consequently, the efficiency of FRET from BODIPY to the heme in the enzyme monomer is decreased to 49%, as determined in our bleaching experiments (Fig S4). Therefore, the BODIPY fluorophore in CYP3A4(C468)-BODIPY is involved in FRET to the heme groups of the neighboring P450 molecules in the oligomer, and the increase in FRET efficiency may be used to monitor the formation of the enzyme oligomers.
3.5. Probing the equilibrium of CYP3A4 oligomerization in proteoliposomes with FRET
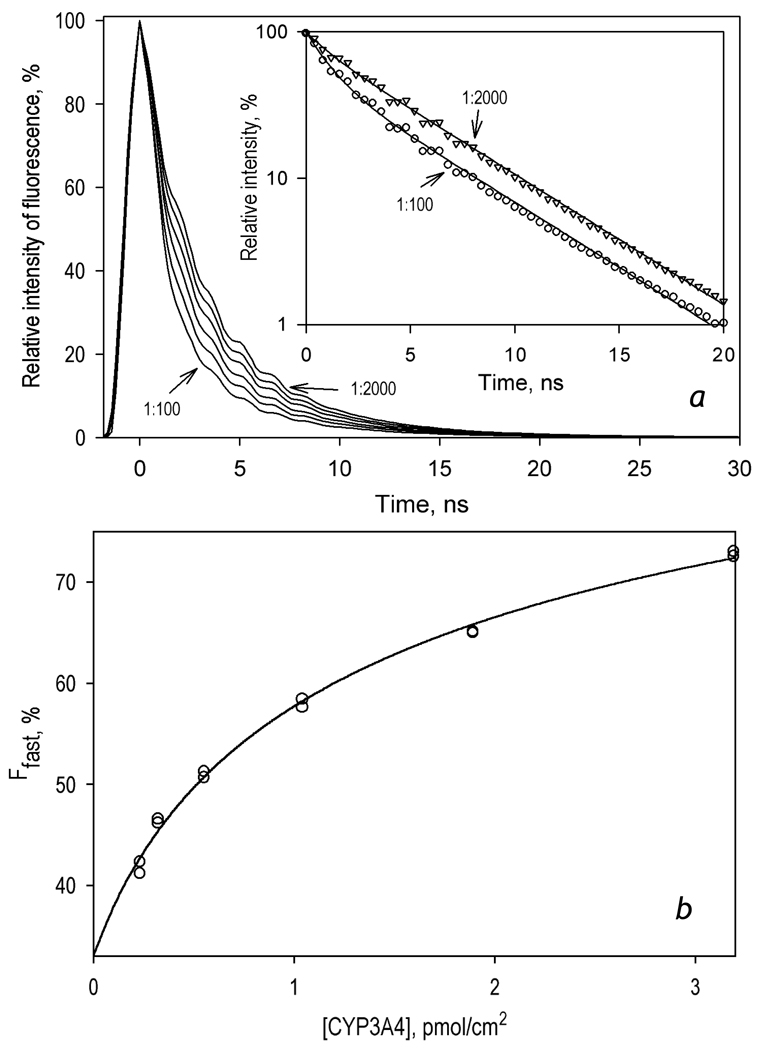
To probe the monomerization CYP3A4 oligomers upon its incorporation into the proteoliposomes we studied the changes in the intensity of BODIPY fluorescence during co-incubation of CYP3A4(C468)-BODIPY with different amounts of the liposomes. As shown in Fig. 5, CYP3A4(C468)-BODIPY interactions with liposomes result in a time-dependent increase in the intensity of fluorescence, which reflects the process of the protein incorporation into the membrane and a consequent dissociation of its oligomers. Increase in the lipid-to-protein ratio results in a considerable augmentation of the effect. The amplitude of the observed changes in the fluorescence intensity increases from 26 to 78% upon increase of the L/P ratio from 100 to 1200 (Fig. 5). This difference is consistent with higher degree of CYP3A4 oligomerization in the liposomes with high surface densities of the heme protein.
In order to further characterize the degree of oligomerization and determine the apparent dissociation constant for the enzyme oligomers in proteoliposomes we employed a series of time-resolved fluorescence measurements in the liposomes with different L/P ratio. The changes in the efficiency of FRET were assessed from the fraction of the fast phase (Ffast) in bi-exponential approximations of the decay traces (τ1 = 1.1 ns, τ2 = 4.9 ns). As seen in Fig. 6a, decrease in the L/P ratio from 2000 to 100 boosts Ffast from 40 to 70%. The dependence of Ffast on the surface density of the heme protein obtained from these experiments (Fig. 6b) was used to estimate the apparent dissociation constant of CYP3A4 oligomers. Fitting of the experimental results to the equation for the equilibrium of bimolecular association ([55], p 73, eq. II-53) yields the value of the apparent Kd of 1.35 ± 0.06 pmol/cm2, which is equivalent to the L/P ratio of 296 . It should be noted, however, that the use of the above equation in this case is arbitrary. Strictly speaking, the formalism deduced for the case of multi-molecular reactions in 3-dimensional space is not directly applicable for the case, where the diffusion of the reagents takes place in two dimensions of the membrane [56]. Nevertheless, the value of apparent Kd determined above may be used for a rough estimation of the oligomerization state of CYP3A4 in the membrane.
Figure 6.
Effect of the surface density of CYP3A4(C468)-BODIPY in proteoliposomes on the lifetime of fluorescence of the BODIPY probe. The preparations of the proteoliposomes were obtained by incubation of 1 µM CYP3A4-(C468)-BODIPY with various amounts of pre-formed liposomes for 1 hour in 0.1 M Na-HEPES buffer, 1 mM DTT, and 1 mM EDTA at 25 °C. Panel a: A series of normalized fluorescence decay traces (excitation an 405 nm, emission at 520 nm with the band with of 18 nm) obtained with L/P ratios of 100, 200, 400, 800, 1400 and 2000. All the curves of the set may be approximated by a sum of two exponents with characteristic times of 1.1 and 4.9 ns (ρ2 > 0.95). The inset shows the traces obtained with L/P ratio of 100 (circles) and 2000 (triangles) in semi-logarithmic coordinates with fitting curves shown in solid lines.
b: Dependence of Ffast on the concentration of CYP3A4(C468)-BODIPY in the membrane of proteoliposomes. Data points represent the results of two independent experiments. The solid line illustrates the fitting of the data set with the equation for the equilibrium of bimolecular association (KD = 1.35 ± 0.06 pmol/cm2).
To probe the possible effect of CYP3A4 interactions with BMR on the state of oligomerization of the heme protein we monitored the fluorescence of CYP3A4(C468)-BODIPY in liposomes with different L/P ratios as a function of added BMR. The considerable overlap of both excitation and emission spectra of BODIPY with those of the flavins precluded the use of the life-time FRET technique in these experiments. Therefore, we probed the effect of BMR on the intensity of fluorescence of CYP3A4(C468)-BODIPY (excitation at 495 nm). In this setup the band of BODIPY can be resolved spectrally from the flavin fluorescence. These experiments showed that a stepwise increase in the concentration of BMR from 0 to 20 µM results in a nearly linear low-amplitude decline in the intensity of BODIPY fluorescence (data not shown). The amplitude of this decrease does not exceed ~10–15% at the BMR concentrations used in our stop-flow experiments. As this effect was identical in the liposomes with L/P ratios of 150:1 and 1200:1, there is no obvious change in the CYP3A4 oligomerization state in the presence of BMR.
4. Discussion
The principal conclusion derived from our results concerns the relationship between the state of aggregation of CYP3A4 and its reducibility by the flavoprotein. As we demonstrated in our recent study on BMR-dependent reduction of CYP3A4 in solution, the reducibility of the enzyme in its oligomers in the absence of substrates does not exceed 50 ± 9 % [29]. We interpreted this finding as an indication of conformational heterogeneity of the enzyme caused by unequal location and/or orientation of the subunits in the oligomer. The observation that this incomplete reducibility may be rescued by the addition of ANF offers a clue to the mechanism of action of this heterotropic activator [29]. In this study we sought to probe the effect of CYP3A4 monomerization on the functional heterogeneity revealed in incomplete reducibility by BMR. In good agreement with our initial model, monomerization of the enzyme by its incorporation into lipoprotein Nanodiscs eliminates the heterogeneity and makes the enzyme completely reducible.
To probe further the effect of CYP3A4 oligomerization in the membrane on its reducibility we compared two preparations of liposomes with different lipid:protein ratios. Assuming the average area of bilayer membrane per one phospholipid molecule to be equal to 72 Å2 (mean value for egg phosphatidylcholine membrane at 30 °C from Table 3 in [57]) and the footprint of one CYP3A4 molecule to be around 16 nm2, we may calculate that the liposomes LPS-910 contained one molecule of CYP3A4 per ~340 nm2 of the lipid bilayer, which corresponds to the concentration of 0.49 pmol/cm2. Similar calculations for LPS-76 result in the value of 43 nm2 per CYP3A4 molecule (or the concentration of 3.9 pmol/cm2). For comparison, according to studies of Watanabe and co-authors [58], the total membrane area per one P450 molecule in the endoplasmic reticulum of mouse hepatocytes is in the range of 170–260 nm2 in the absence of P450 induction and decreases to 60–130 nm2 upon induction with phenobarbital.
According to our results, the reducibility of CYP3A4 in the liposomes with low surface density of CYP3A4 (LPS-910) was as high 87 ± 9 %, while the liposomes with the high protein density (LPS-76), where the enzyme is thought to be highly oligomerized, showed total reducibility of only 55 ± 5 %. Therefore, in good agreement with data obtained with CYP3A4 oligomers and CYP3A4-ND in solution, our experiments with CYP3A4-containing liposomes suggest that functional heterogeneity of the enzyme reflected in its incomplete reducibility by BMR is caused by the formation of CYP3A4 oligomers in the membrane.
To probe the reliability of this interpretation we studied the effect of the surface density of CYP3A4 in the membrane on the state of its oligomerization. Using a newly developed method based on FRET from BODIPY fluorophore to the heme groups of the neighboring P450 molecules in the oligomer of CYP3A4(C468)-BODIPY we confirmed the concentration-dependent formation of the enzyme oligomers in the liposomal membrane and estimated the value of the apparent Kd of the oligomer to be of 1.35 pmol/cm2. According to these results, the fraction of monomeric CYP3A4 in LPS76 (3.9 pmol CYP3A4 per cm2) and in LPS900 (0.5 pmol/cm2) may be estimated to be around 45% and 82% respectively. A notable congruence between these values and the estimates of the enzyme reducibility in these liposomes (55% and 87% respectively) suggests that the efficient transfer of electrons from BMR to membrane-bound CYP3A4 takes place only in the complexes of BMR with the monomers of the heme protein. According to the results of our continuous variation titration experiments with BMR and LPS-76, the stoichiometry of the BMR-CYP3A4 interactions in these liposomes is equal to 1:1. The value of Kd for the BMR-CYP3A4 complex (0.14 ± 0.08 µM ) suggests that in the conditions of our experiments (1.5–3 µM CYP3A4 and a two-fold molar excess of BMR) the saturation of the heme protein with BMR was virtually complete, regardless of its oligomeric state. This finding suggests that some part of the complexes of the redox-partners is not functionally competent. This inference is consistent with the observation of two different types of complexes of rabbit microsomal cytochrome P450 2B4 with CPR, only one of which is governed by electrostatic interactions, whereas the second contact region is hydrophobic [59]. Our observation is also consistent with the recent indication of multiple orientations of the flavoprotein in the complexes of the heme domain of P450BM-3 with the flavodoxin from Desulfovibrio vulgaris (which served as a model of the FMN domain of BMR) [60].
A plausible hypothesis on the molecular basis of the differences in the reducibility (or the rate of intracomplex electron transfer) between the monomers and oligomers of CYP3A4 may be based on our observation of the effect of CYP3A4 interactions with partially reduced BMR on the spin state of the heme protein. The results suggested that the formation of CYP3A4 oligomers, both in solution and in the membrane, may prevent this flavoprotein-induced spin shift (at least in some subunits of the oligomeric enzyme) by restricting the conformational mobility, which is required for this effect to take place.
It should be noted that the finding that a decrease in the surface density of CYP3A4 in the liposomes and a subsequent dissociation of CYP3A4 oligomers renders the reducibility of the enzyme similar to that observed in the Nanodiscs provides an additional support to our interpretation of incomplete reducibility as a result of CYP3A4 oligomerization. A close match of the kinetics of reduction of CYP3A4-ND with that observed in diluted liposomes also suggests that the difference in the lipid composition between the liposomes used in these studies, which were composed of a 2:1:0.6 PC:PE:PA mixture, and the POPC-containing Nanodiscs, is not essential for the properties of CYP3A4-ND, which lateral mobility is severely restricted due to immobilization within a thin belt of phospholipids.
The availability of homogeneous monomeric CYP3A4 in Nanodiscs allowed us to analyze some remarkable intermittent rapid spin-shift processes in the CYP3A4 complex with BMR that precede the heme protein reduction. Similar transitions were noticed earlier with CYP3A4 in solution [29] but were not investigated in any detail. Modulation of the P450 spin state by the state of reduction of the interacting flavoprotein is consistent with recent indications of a large-scale conformational transition in NADPH-cytochrome P450 reductase involved in the intra-domain electron transfer [61, 62]. The remarkable conformational flexibility of some microsomal cytochromes P450 [63], close distance (12–18 Å) between the heme moiety and the isoalloxazine ring of FMN [61, 64], and important role of the dynamics of intramolecular salt bridges in spin transitions of the heme protein [27, 65, 66] are all consistent with the hypothesis that the reduction of FMN may induce a conformational transition in the heme protein that changes the position of its spin equilibrium. In particular, such a transition may be triggered by the appearance of a negative charge on the isoalloxazine ring due to the formation of the red anionic semiquinone of FMN in BMR.
A transient spin shift in P450 in response to the reduction of the flavoprotein observed in this study may reveal a novel mechanism of mutual coordination of substrate binding and the interactions of the enzyme with the reduced electron donor. In particular, displacement of the spin equilibrium of the substrate-free enzyme after its binding to a partially reduced flavoprotein would foster the binding of type-I substrates, thus preventing a futile cycling of the system. Furthermore, a conversion from the hexa-coordinated low-spin to the penta-coordinated high-spin state, which is triggered by reduction of the reductase flavins may have an important impact on the catalytic efficiency and coupling of the P450. This transition is known to increase the rate of the electron transfer [2, 4, 5, 7] due to a considerable decrease in the reorganization energy associated with the reduction process [67, 68] along with a positive displacement of the redox potential of cytochromes P450 [69, 70], and 3A4 [71] in particular. At the same time, the modulation of the spin state of cytochromes P450 in response to the changes in the redox state of the reductase may explain an apparent lack of correlation between the rate of NADPH-dependent reduction or the rate of substrate turnover and the spin-state for certain microsomal cytochromes P450 [8], which was considered as a disproof of the theory of the spin-state control of the rate of electron transfer. Our results show that the spin state of the substrate complexes of P450 associated with the reduced reductase may differ considerably from the spin state of a “resting” enzyme (reductase-free or associated with the oxidized flavoprotein), which was employed in probing these correlations.
Therefore, the newly discovered modulating effect of the reduction of the flavoprotein on the spin state may play an important role in fine tuning of the electron flow from CPR to P450 during the catalytic cycle. The slow reversal of this transition observed in our experiments might be specific for the case of BMR, which is characterized by a very slow rate of electron transfer to CYP3A4. We may expect that in the case of complexes of CYP3A4 with its native microsomal reductase this reversal would be too slow to compete with the intracomplex electron transfer from the reduced flavoprotein to P450.
Further studies with microsomal CPR would provide proof of the physiological relevance of the newly discovered modulation of the spin state of CYP3A4 by the state of reduction of the flavoprotein. A critical question is whether the electron-donating species of BMR and CPR to CYP3A4 are different (FMN·− as opposed to FMNH2, respectively) or whether the role of the anionic semiquinone is a unique feature of P450-BM3, so that the reduction of CYP3A4 by BMR occurs through the formation of FMN hydroquinone similar to CPR [33, 51]. The fact that the rate of CYP3A4 reduction by BMR (see Table 1) is lower than the rate of the formation of semiquinone-containing species of the flavoprotein in the process of its NADPH-dependent reduction (see section 3.2) is consistent with FMNH2 as electron donor CYP3A4. Therefore, a similar spin-modulating mechanism of CYP3A4 by CPR as observed with BMR is anticipated.
In summary, our current results on complete reducibility of CYP3A4 upon monomerization of the hemeprotein and on the spin transitions in the enzyme during its reduction may provide a missing clue to the mysterious difference in the kinetics of reduction of the low- and high-spin fractions of cytochromes P450. This difference, which runs counter to the paradigm of rapid spin equilibrium in the enzyme, remains controversial since its first observation over 25 years ago [4–7, 72, 73]. Our results may also shed light on the reasons for striking differences in the kinetics of reduction of various cytochromes P450 and the effects of divergent P450 substrates on such kinetics [8]. The use of BMR as a model electron donor partner of CYP3A4 facilitated considerably the resolution of the changes in the spectra of CYP3A4 from those of the flavoprotein due to the fact that the intracomplex electron transfer from CPR to CYP3A4 is slow. Further studies on the relationship between FMN reduction and the spin state of cytochrome P450 in the complexes of the enzyme with CPR in Nanodiscs and in liposomal and/or microsomal membranes are necessary to probe the validity of the newly discovered modulatory mechanisms for the physiologically relevant microsomal electron transfer system. Studies of the kinetics of electron transfer in CPR-containing liposomes at different surface density of the flavoprotein-saturated CYP3A4 are now in progress.
Supplementary Material
Acknowlegments
This research was supported by NIH grants GM54995 (JRH) GM33775 (SGS), and GM31756 (SGS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This research was supported by NIH grants GM54995 (JRH), GM33775 (SGS), and GM31756 (SGS).
Supplementary data
Supplementary data associated with this article include a series of spectra reflecting the spectral changes in the process of BMR-dependent reduction of CYP3A4-ND and the results of its analysis with the method of principal components. The effect of monomerization of CYP3A4(C468)-BODIPY on the intensity of BODIPY fluorescence and the efficiency of FRET to the heme as determined in the bleaching assay are also included in the supplementary data. These materials may be found in the online version.
References
- 1.Gigon PL, Gram TE, Gillette JR. Studies on rate of reduction of hepatic microsomal cytochrome P-450 by reduced nicotinamide adenine dinucleotide phosphate - effect of drug substrates. Mol. Pharmacol. 1969;5:109–122. [PubMed] [Google Scholar]
- 2.Peterson JA, Ebel RE, O'keeffe DH, Matsubara T, Estabrook RW. Temperature dependence of cytochrome P-450 reduction. A model for NADPH-cytochrome P-450 reductase:cytochrome P-450 interaction. J. Biol. Chem. 1976;251:4010–4016. [PubMed] [Google Scholar]
- 3.Blanck J, Smettan G, Ristau O, Ingelman-sundberg M, Ruckpaul K. Mechanism of rate control of the NADPH-dependent reduction of cytochrome P-450 by lipids in reconstituted phospholipid vesicles. Eur. J. Biochem. 1984;144:509–513. doi: 10.1111/j.1432-1033.1984.tb08495.x. [DOI] [PubMed] [Google Scholar]
- 4.Tamburini PP, Gibson GG, Backes WL, Sligar SG, Schenkman JB. Reduction kinetics of purified rat liver cytochrome P-450. Evidence for a sequential reaction mechanism dependent on the hemoprotein spin state. Biochemistry. 1984;23:4526–4533. doi: 10.1021/bi00315a004. [DOI] [PubMed] [Google Scholar]
- 5.Backes WL, Tamburini PP, Jansson I, Gibson GG, Sligar SG, Schenkman JB. Kinetics of cytochrome P-450 reduction: evidence for faster reduction of the high-spin ferric state. Biochemistry. 1985;24:5130–5136. doi: 10.1021/bi00340a026. [DOI] [PubMed] [Google Scholar]
- 6.Karyakin AV, Davydov DR. Kinetics of electron-transfer reactions in monooxygenase system. Vestn. Akad. Med. Nauk SSSR. 1988;1988:53–62. [PubMed] [Google Scholar]
- 7.Backes WL, Eyer CS. Cytochrome P-450 LM2 reduction. Substrate effects on the rate of reductase-LM2 association. J. Biol. Chem. 1989;264:6252–6259. [PubMed] [Google Scholar]
- 8.Guengerich FP, Johnson WW. Kinetics of ferric cytochrome P450 reduction by NADPH-Cytochrome P450 reductase - Rapid reduction in the absence of substrate and variations among cytochrome P450 systems. Biochemistry. 1997;36:14741–14750. doi: 10.1021/bi9719399. [DOI] [PubMed] [Google Scholar]
- 9.Cole PE, Sligar SG. Temperature-jump measurement of the spin state relaxation rate of cytochrome P450cam. FEBS Lett. 1981;133:252–254. doi: 10.1016/0014-5793(81)80517-0. [DOI] [PubMed] [Google Scholar]
- 10.Ziegler M, Blanck J, Ruckpaul K. Spin equilibrium relaxation kinetics of cytochrome P450 LM2. FEBS Lett. 1982;150:219–222. [Google Scholar]
- 11.Fisher MT, Sligar SG. Temperature jump relaxation kinetics of the P-450cam spin equilibrium. Biochemistry. 1987;26:4797–4803. doi: 10.1021/bi00389a029. [DOI] [PubMed] [Google Scholar]
- 12.Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: Heterogeneity of the enzyme caused by its oligomerization. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Q, Lu AYH. The challenges of dealing with promiscuous drug-metabolizing enzymes, receptors and transporters. Curr. Drug Metab. 2008;9:374–383. doi: 10.2174/138920008784746337. [DOI] [PubMed] [Google Scholar]
- 14.Davydov DR, Halpert JR. Allosteric P450 mechanisms: multiple binding sites, multiple conformers or both? Expert Opin. Drug Met. 2008;4:1523–1535. doi: 10.1517/17425250802500028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hlavica P, Lewis DFV. Allosteric phenomena in cytochrome P450-catalyzed monooxygenations. Eur. J. Biochem. 2001;268:4817–4832. doi: 10.1046/j.1432-1327.2001.02412.x. [DOI] [PubMed] [Google Scholar]
- 16.Backes WL, Kelley RW. Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharm. Ther. 2003;98:221–233. doi: 10.1016/s0163-7258(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 17.Greinert R, Finch SA, Stier A. Conformation and rotational diffusion of cytochrome P-450 changed by substrate binding. Bioscience Rep. 1982;2:991–994. doi: 10.1007/BF01122166. [DOI] [PubMed] [Google Scholar]
- 18.Kawato S, Gut J, Cherry RJ, Winterhalter KH, Richter C. Rotation of cytochrome P-450. I. Investigations of protein-protein interactions of cytochrome P-450 in phospholipid vesicles and liver microsomes. J. Biol. Chem. 1982;257:7023–7029. [PubMed] [Google Scholar]
- 19.Kiselev PA, Garda G, Finch SA, Stir A, Khatyleva MS, Akhrem AA. Regulation of the catalytic activity of the monooxygenase enzyme system depending of the substrate structure and phospholipid composition of the model membrane] Biochemistry (Moscow) 1990;55:1535–1544. [PubMed] [Google Scholar]
- 20.Schwarz D, Pirrwitz J, Meyer HW, Coon MJ, Ruckpaul K. Membrane topology of microsomal cytochrome P-450: Saturation transfer EPR and freeze-fracture electron microscopy studies. Biochem. Biophys. Res. Commun. 1990;171:175–181. doi: 10.1016/0006-291x(90)91373-z. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz D, Kruger V, Chernogolov A, Usanov SA, Stier A. Rotation of cytochrome P450SCC (CYP11A1) in proteoliposomes studied by delayed fluorescence depolarization. Biochem. Biophys. Res. Commun. 1993;195:889–896. doi: 10.1006/bbrc.1993.2128. [DOI] [PubMed] [Google Scholar]
- 22.Yamada M, Ohta Y, Bachmanova GI, Archakov AI, Hatta I, Kawato S. Effect of microsome-liposome fusion on the rotational mobility of cytochrome P450IIB4 in rabbit liver microsomes. J. Inorg. Biochem. 2001;83:261–268. doi: 10.1016/s0162-0134(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 23.Alston K, Robinson RC, Park SS, Gelboin HV, Friedman FK. Interactions among cytochromes P-450 in the endoplasmic reticulum. Detection. J. Biol. Chem. 1991;266:735–739. [PubMed] [Google Scholar]
- 24.Szczesna-Skorupa E, Mallah B, Kemper B. Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J. Biol. Chem. 2003;278:31269–31276. doi: 10.1074/jbc.M301489200. [DOI] [PubMed] [Google Scholar]
- 25.Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol. Appl. Pharm. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Koley AP, Robinson RC, Markowitz A, Friedman FK. Drug-drug interactions: effect of quinidine on nifedipine binding to human cytochrome P450 3A4. Biochem. Pharm. 1997;53:455–460. doi: 10.1016/s0006-2952(96)00836-2. [DOI] [PubMed] [Google Scholar]
- 27.Davydov DR, Halpert JR, Renaud JP, Hui Bon Hoa G. Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem. Biophys. Res. Commun. 2003;312:121–130. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- 28.Fernando H, Halpert JR, Davydov DR. Resolution of multiple substrate binding sites in cytochrome P450 3A4: The stoichiometry of the enzyme-substrate complexes probed by FRET and Job's titration. Biochemistry. 2006;45:4199–4209. doi: 10.1021/bi052491b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernando H, Halpert JR, Davydov DR. Kinetics of electron transfer in the complex of cytochrome P450 3A4 with the flavin domain of cytochrome P450BM-3 as evidence of functional heterogeneity of the heme protein. Arch. Biochem. Biophys. 2008;471:20–31. doi: 10.1016/j.abb.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davydov DR, Karyakin AV, Binas B, Kurganov BI, Archakov AI. Kinetic studies on reduction of cytochromes P-450 and b5 by dithionite. Eur. J. Biochem. 1985;150:155–159. doi: 10.1111/j.1432-1033.1985.tb09001.x. [DOI] [PubMed] [Google Scholar]
- 31.Shen A, Kasper C. Role of acidic residues in the interaction of NADPH-cytochrome P450 oxidoreductase with cytochrome P450 and cytochrome c. J. Biol. Chem. 1995;270:27475–27480. doi: 10.1074/jbc.270.46.27475. [DOI] [PubMed] [Google Scholar]
- 32.Davydov DR, Kariakin AA, Petushkova NA, Peterson JA. Association of cytochromes P450 with their reductases: opposite sign of the electrostatic interactions in P450BM-3 as compared with the microsomal 2B4 system. Biochemistry. 2000;39:6489–6497. doi: 10.1021/bi992936u. [DOI] [PubMed] [Google Scholar]
- 33.Sevrioukova IF, Peterson JA. NADPH-P-450 reductase - structural and functional comparisons of the eukaryotic and prokaryotic isoforms. Biochimie. 1995;77:562–572. doi: 10.1016/0300-9084(96)88172-7. [DOI] [PubMed] [Google Scholar]
- 34.Dodhia VR, Fantuzzi A, Gilardi G. Engineering human cytochrome P450 enzymes into catalytically self-sufficient chimeras using molecular Lego. J Biol. Inorg. Chem. 2006;11:903–916. doi: 10.1007/s00775-006-0144-3. [DOI] [PubMed] [Google Scholar]
- 35.Fairhead M, Giannini S, Gillam EMJ, Gilardi G. Functional characterisation of an engineered multidomain human P450 2E1 by molecular Lego. J. Biol. Inorg. Chem. 2005;10:842–853. doi: 10.1007/s00775-005-0033-1. [DOI] [PubMed] [Google Scholar]
- 36.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 37.Harlow GR, Halpert JR. Analysis of human cytochrome P450 3A4 cooperativity: construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc. Natl. Acad. Sci. USA. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsalkova TN, Davydova NE, Halpert JR, Davydov DR. Mechanism of interactions of alpha-naphthoflavone with cytochrome P450 3A4 explored with an engineered enzyme bearing a fluorescent probe. Biochemistry. 2007;46 doi: 10.1021/bi061944p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch. Biochem. Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 41.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J.Am.Chem.Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 42.Bartlett GR. Phosphorus assay in column chromatography. J. Biol. Chem. 1959;234:466–468. [PubMed] [Google Scholar]
- 43.Davydov DR, Deprez E, Hui Bon Hoa G, Knyushko TV, Kuznetsova GP, Koen YM, Archakov AI. High-pressure-induced transitions in microsomal cytochrome P450 2B4 in solution - evidence for conformational inhomogeneity in the oligomers. Arch. Biochem. Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 44.Renaud JP, Davydov DR, Heirwegh KPM, Mansuy D, Hui Bon Hoa G. Thermodynamic studies of substrate binding and spin transitions in human cytochrome P-450 3A4 expressed in yeast microsomes. Biochem. J. 1996;319:675–681. doi: 10.1042/bj3190675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davydov DR, Knyushko TV, Hui Bon Hoa G. High pressure induced inactivation of ferrous cytochrome P-450 LM2 (2B4) CO complex: evidence for the presence of two conformers in the oligomer. Biochem .Biophys.Res.Commun. 1992;188:216–221. doi: 10.1016/0006-291x(92)92372-5. [DOI] [PubMed] [Google Scholar]
- 46.Davydov DR, Petushkova NA, Archakov AI, Hui Bon Hoa G. Stabilization of P450 2B4 by Its association with P450 1A2 revealed by high-pressure spectroscopy. Biochem. Biophys. Res. Commun. 2000;276:1005–1012. doi: 10.1006/bbrc.2000.3596. [DOI] [PubMed] [Google Scholar]
- 47.French JS, Guengerich FP, Coon MJ. Interactions of cytochrome P-450, NADPH-cytochrome P-450 reductase, phospholipid, and substrate in the reconstituted liver microsomal enzyme system. J. Biol. Chem. 1980;255:4112–4119. [PubMed] [Google Scholar]
- 48.Bosterling B, Trudell JR. Association of cytochrome b5 and cytochrome P-450 reductase with cytochrome P-450 in the membrane of reconstituted vesicles. J. Biol. Chem. 1982;257:4783–4787. [PubMed] [Google Scholar]
- 49.Tamburini PP, Jansson I, Favreau LV, Backes WL, Schenkman JB. Differences in the spectral interactions between {NADPH}-cytochrome-P-450 reductase and a series of cytochrome-P-450 enzymes. Biochem. Biophys. Res. Commun. 1985;137:437–442. doi: 10.1016/0006-291x(86)91229-5. [DOI] [PubMed] [Google Scholar]
- 50.Davydov DR, Femando H, Halpert JR. Variable path length and counter-flow continuous variation methods for the study of the formation of high-affinity complexes by absorbance spectroscopy. An application to the studies of substrate binding in cytochrome P450. Biophys. Chem. 2006;123:95–101. doi: 10.1016/j.bpc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Sevrioukova I, Shaffer C, Ballou DP, Peterson JA. Equilibrium and transient state spectrophotometric studies of the mechanism of reduction of the flavoprotein domain of P450BM-3. Biochemistry. 1996;35:7058–7068. doi: 10.1021/bi960060a. [DOI] [PubMed] [Google Scholar]
- 52.Hanley SC, Ost TWB, Daff S. The unusual redox properties of flavocytochrome P450BM3 flavodoxin domain. Biochem. Biophys. Res. Commun. 2004;325:1418–1423. doi: 10.1016/j.bbrc.2004.10.189. [DOI] [PubMed] [Google Scholar]
- 53.Uvarov VY, Tretiakov VE, Archakov AI. Heme maintains catalytically active structure of cytochrome P-450. FEBS Lett. 1990;260:309–312. doi: 10.1016/0014-5793(90)80131-2. [DOI] [PubMed] [Google Scholar]
- 54.Pikuleva IA, Lapko AG, Chashchin VL. Functional reconstitution of cytochrome P-450scc with hemin activated with Woodward's reagent K. Formation of a hemeprotein cross-link. J. Biol. Chem. 1992;267:1438–1442. [PubMed] [Google Scholar]
- 55.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley-Interscience; 1975. [Google Scholar]
- 56.Hardt SL. Rates of diffusion controlled reactions in one, 2 and 3 dimensions. Biophys. Chem. 1979;10:239–243. doi: 10.1016/0301-4622(79)85012-7. [DOI] [PubMed] [Google Scholar]
- 57.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe J, Asaka Y, Kanai K, Kanamura S. Relation between cytochrome P-450 increase and endoplasmic-reticulum proliferation in hepatocytes of mice treated with phenobarbital - a microphotometric and morphometric study. J. Histochem. Cytochem. 1992;40:353–357. doi: 10.1177/40.3.1552174. [DOI] [PubMed] [Google Scholar]
- 59.Davydov DR, Knyushko TV, Kanaeva IP, Koen YM, Samenkova NF, Archakov AI, Hui Bon Hoa G. Interactions of cytochrome P450 2B4 with NADPH-cytochrome P450 reductase studied by a fluorescent probe. Biochimie. 1996;78:734–743. doi: 10.1016/s0300-9084(97)82531-x. [DOI] [PubMed] [Google Scholar]
- 60.Fantuzzi A, Meharenna YT, Briscoe PB, Guerlesquin F, Sadeghi SJ, Gilardi G. Characterisation of the electron transfer and complex formation between flavodoxin from D-vulgaris and the haem domain of cytochrome P450 BM3 from B-megaterium. Biochim. Biophys. Acta - Bioenergetics. 2009;1787:234–241. doi: 10.1016/j.bbabio.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Hamdane D, Xia CW, Im SC, Zhang HM, Kim JJP, Waskell L. Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450. J. Biol. Chem. 2009;284:11374–11384. doi: 10.1074/jbc.M807868200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aigrain L, Pompon D, Morera S, Truan G. Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase. EMBO Rep. 2009;10:742–747. doi: 10.1038/embor.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davydov DR, Davydova NY, Halpert JR. Allosteric transitions in cytochrome P450eryF explored with pressure-perturbation spectroscopy, lifetime FRET, and a novel fluorescent substrate, Fluorol-7GA. Biochemistry. 2008;47:11348–11359. doi: 10.1021/bi8011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sevrioukova IF, Li HY, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. USA. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lounnas V, Wade RC. Exceptionally stable salt bridges in cytochrome P450cam have functional roles. Biochemistry. 1997;36:5402–5417. doi: 10.1021/bi9622940. [DOI] [PubMed] [Google Scholar]
- 66.Davydov DR, Botchkareva AE, Kumar S, He YQ, Halpert JR. An electrostatically driven conformational transition is involved in the mechanisms of substrate binding and cooperativity in cytochrome P450eryF. Biochemistry. 2004;43:6475–6485. doi: 10.1021/bi036260l. [DOI] [PubMed] [Google Scholar]
- 67.Honeychurch M, Hill HA, Wong LL. The thermodynamics and kinetics of electron transfer in the cytochrome P450(cam) enzyme system. FEBS Lett. 1999;451:351–353. doi: 10.1016/s0014-5793(99)00610-9. [DOI] [PubMed] [Google Scholar]
- 68.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 69.Sligar SG. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry. 1976;15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 70.Fisher MT, Sligar SG. Control of heme protein redox potential and reduction rate-Linear free energy relation between potential and ferric spin state equilibrium. J. Am. Chem. Soc. 1985;107:5018–5019. [Google Scholar]
- 71.Das A, Grinkova YV, Sligar SG. Redox potential control by drug binding to cytochrome p450 3A4. J. Am. Chem. Soc. 2007;129:13778–13779. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Backes WL, Sligar SG, Schenkman JB. Kinetics of hepatic cytochrome P-450 reduction: correlation with spin state of the ferric heme. Biochemistry. 1982;21:1324–1330. doi: 10.1021/bi00535a034. [DOI] [PubMed] [Google Scholar]
- 73.Eyer CS, Backes WL. Relationship between the rate of reductase-cytochrome P450 complex formation and the rate of first electron transfer. Arch. Biochem. Biophys. 1992;293:231–240. doi: 10.1016/0003-9861(92)90390-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.