Abstract
Monomeric fluorescent proteins of different colors are widely used to study behavior and targeting of proteins in living cells. Fluorescent proteins that irreversibly change their spectral properties in response to light irradiation of a specific wavelength, or photoactivate, have become increasingly popular to image intracellular dynamics and super-resolution protein localization. Until recently, however, no optimized monomeric red fluorescent proteins and red photoactivatable proteins have been available. Furthermore, monomeric fluorescent proteins, which change emission from blue to red simply with time, so-called fluorescent timers, were developed to study protein age and turnover. Understanding of chemical mechanisms of the chromophore maturation or photoactivation into a red form will further advance engineering of fluorescent timers and photoactivatable proteins with enhanced and novel properties.
Introduction
Since the discovery that green fluorescent protein (GFP) from jellyfish is encoded by a single gene and its fluorescence requires no enzymes or cofactors except molecular oxygen, the fluorescent proteins (FPs) became invaluable tools in biomedical sciences. Cloning of the first FP with red-shifted excitation and emission spectra, DsRed, led to the discovery of many new orange and red fluorescent proteins (RFPs) in non-bioluminescent organisms [1]. Further directed evolution of wild-type FPs allowed the creation of a wide palette of enhanced FPs, which span the visible spectrum from 420 nm to almost 650 nm [2]. Development of the monomeric RFPs allowed extending possibilities of a Förster resonance energy transfer (FRET) approach to three and four colors in a single cell [3].
The RFPs, whose chromophores are formed by induction with light, are known as the photoactivatable FPs (PA-RFPs). Two different groups of PA-RFPs are presently being distinguished. Members of the first group exhibit an irreversible photoconversion from the non-fluorescent or green fluorescent state to the red fluorescent state. Members of the second group undergo reversible photoswitching between the non-fluorescent and fluorescent states. Introduction of photoactivatable FPs into cell biology greatly extended the spatio-temporal limits of in vivo biological dynamics [4] and have become useful tools for the super-resolution microscopy approaches such as a photoactivation localization microscopy (PALM) [5]. Therefore, recently developed monomeric irreversible PA-RFPs are of a particular interest for tracking individual intracellular molecules.
In RFPs, the fluorescence shift toward the red is a result of the expansion in the π-system of the conventional GFP-like chromophore. Currently known RFPs share two types of chromophores, called a DsRed-like chromophore [6] and a Kaede-like chromophore [7] after the first proteins where they have been found. The DsRed-like chromophore may form either through autocatalytic post-translational modifications or via induction by irradiation with violet light. There are also some proteins containing derivatives of the DsRed-like structure, which form due to chemical modifications of the N-acylimine group in the DsRed-like chromophore [8,9,10]. The Kaede-like chromophore is characteristic for the green-to-red photoconvertible fluorescent proteins. Initially, the proteins of this group mature to a green-emitting state with the GFP-like chromophore. However, UV-violet light at approximately 350–450 nm efficiently converts them into the red fluorescent state.
Here, we provide a brief overview of RFPs and irreversible PA-RFPs published within the last few years.
Irreversibly photoactivatable red fluorescent proteins
Monomeric PA-RFP Dendra2 [11] has already found wide application for protein [12•] and cell tracking [13]. Dendra2 exhibits a high contrast photoconversion from the green to the red fluorescent state (Table 1). The unique feature of Dendra2 is that a low phototoxic 488 nm laser line can be used for its photoactivation. Furthermore, Dendra2 is simultaneously monomeric and efficiently matures at 37°C. Dendra2 performs well in sensitive fusions and possesses low cytotoxicity. The only disadvantage that should be mentioned is a relatively low pH stability of the activated red form.
Table 1.
Properties of the new fluorescent proteins that become red fluorescent after light irradiation or with time.
| Protein | Oligomeric state | Excitation, nm | Emission, nm | Extinction coefficient, M−1cm−1 | Quantum yield | pKa | Maturation half-time at 37°C, h | Photostability, sec | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Dendra2 | monomer | 490 | 507 | 45,000 | 0.50 | 6.6 | ND | 45 | [11] |
| 553 | 573 | 35,000 | 0.55 | 6.9 | 378 | ||||
| IrisFP | tetramer | 488 | 516 | 52,200 | 0.43 | ND | ND | ND | [21] |
| 551 | 580 | 35,400 | 0.47 | ND | ND | ||||
| tdEosFP | dimer | 506 | 516 | 84,000 | 0.66 | 5.7 | ND | 47 | [18] |
| 569 | 581 | 33,000 | 0.60 | ND | 380 | ||||
| mEos2 | monomer | 506 | 519 | 56,000 | 0.84 | 5.6 | ND | 42 | [20] |
| 573 | 584 | 46,000 | 0.66 | 6.4 | 323 | ||||
| PA-mCherry1 | monomer | 404 | 466 | 6,500 | <0.001 | ND | 0.38 | ND | [17] |
| 564 | 594 | 18,000 | 0.46 | 6.3 | 18 | ||||
| mKikGR | monomer | 505 | 515 | 49,000 | 0.69 | ND | ND | 14 | [15] |
| 580 | 591 | 28,000 | 0.63 | ND | 21 | ||||
| Fast-FT | monomer | 403 | 466 | 49,700 | 0.30 | 2.8 | 0.25 | ND | [48] |
| 583 | 606 | 75,300 | 0.09 | 4.1 | 7.1 | ND | |||
| Medium-FT | monomer | 401 | 464 | 44,800 | 0.41 | 2.7 | 1.2 | ND | [48] |
| 579 | 600 | 73,100 | 0.08 | 4.7 | 3.9 | ND | |||
| Slow-FT | monomer | 402 | 465 | 33,400 | 0.35 | 2.6 | 9.8 | ND | [48] |
| 583 | 604 | 84,200 | 0.05 | 4.6 | 28 | ND |
ND, not determined.
In order to develop a monomeric version of KikGR [14], 21 amino acids mutations were introduced in 15 rounds of mutagenesis. mKikGR [15] has almost the same spectroscopic characteristics and kinetics of photoswitching as its parental protein KikGR [16]. High photostability and brightness of mKikGR activated red form allow for high resolution in photoactivation localization microscopy, as well as single-molecule tracking.
Several PA-mCherry variants, including PA-mCherry1, [17••] enable two-color diffraction-limited photoactivation imaging and super-resolution techniques, such as PALM. Irreversibly photoactivatable monomeric derivatives of mCherry, PA-mCherrys, are potentially less disruptive to tagged fusion partners. Before photoactivation these proteins have an absorbance maximum at about 400 nm and practically do not fluoresce. However, they can be easily photoactivated with a 405 nm laser line, achieving contrast up to 3,000–5,000-fold. In the photoactivated state, PA-mCherrys exhibit red fluorescence, which is stable in time and doesn’t relax back to the dark state. All variants have fast maturation time and perform excellent in protein fusions in live cells.
The green-to-red photoconvertible tandem dimeric tdEosFP protein [18] paired with the photoswitchable protein Dronpa was also applied to two-color PALM imaging [19], but tdEosFP still does not localize accurately in fusions, and its monomeric version mEosFP does not mature at 37°C [18]. Recently, McKinney et al. [20•] developed a true monomeric version of EosFP, called mEos2, which efficiently matures at 37°C. The spectral properties, brightness, pKa, photoconversion and contrast of the improved mEos2 are similar or better to those of tdEosFP, but there is a much better maturation at 37°C. Despite the dimerization tendency in vitro, mEos2 performs well even in difficult protein fusions in cells.
The only example of protein that displays both a reversible photoswitching and an irreversible photoactivation is the EosFP-derived protein, IrisFP [21], which has a single Phe181Ser amino acid substitution (here and below we use an amino acid numbering according to the alignment with wild-type GFP). IrisFP exhibits the irreversible green-to-red photoconversion under violet light, like its parental protein, and, in addition, both green and red fluorescent states can be turned off and on over again independently.
Novel monomeric red fluorescent proteins
Despite the growing role of photoactivatable FPs in advanced cell imaging approaches, common RFPs are still the proteins of choice for many standard biological applications. Although monomeric RFPs of the first generation are well-suited for protein labeling and exhibit efficient chromophore formation [2], they still have many drawbacks compared to common enhanced GFP (EGFP) and its derivatives.
Recently, several new RFPs with an enhanced brightness, rapid chromophore maturation and high photostability have been developed. Two wild-type RFPs, eqFP583 [22] and eqFP611 [23], were used to design a whole series of enhanced RFPs. Chudakov and coworkers subjected eqFP578 to a combination of site-specific and random mutagenesis to generate red and far-red monomeric FPs named TagRFP [22] and mKate [24], respectively. Crystallographic analysis of mKate [25] allowed for the substantial improvement of its pH-stability, brightness and photostability, resulting in mKate2 and tandem dimeric tdKatushka2 [26•].
The emission spectra of mKate2 and tdKatushka2 extend into a near-infrared “optical window” (650–900 nm), which is advantageous for light penetration in living tissues [27]. This feature makes them the RFPs of choice for visualizing fusion tags in tissues and whole animals. Further development of proteins with emission beyond 650 nm will possibly require extension of a conjugated π-electron system of the red chromophore or increasing Stokes shifts [28]. Alternatively, monomeric infrared FPs can be engineered on a basis of phytochromes, which incorporate an exogenous low-molecular weight chromophore [29].
Tsien and coworkers developed highly photostable FPs, named mOrange2 and TagRFP-T [30], which maintain most of the beneficial qualities of the original proteins and perform excellently for long-term imaging in fusion constructs. However, mOrange2 has a decreased brightness and chromophore maturation efficiency compared to parental mOrange. Tsutsui et al. generated a fast-maturating version of orange-emitting mKO, named mKOk, by introducing seven mutations [31•]. mKOk is 2-fold brighter than its precursor and fairly pH-resistant.
A monomeric RFP, mRuby, with emission maximum at 607 nm was recently shown to be a promising marker for peroxisomes in live mammalian cells [32]. In addition, Strack et al. [33,34] engineered several rapidly maturating tetrameric fluorescent proteins, called DsRed-Express2, DsRed-Max, E2-Orange and E2-Red/Green, with different spectral properties and low cytotoxicity. These proteins should facilitate production of transgenic organisms and stable cell lines.
Random mutagenesis of a chromoprotein derived from Montipora stony coral led to RFP named Keima, which exhibits a Stokes shift of 180 nm [35,36]. Structural and spectroscopic studies of mKeima revealed an excited-state proton transfer (ESPT) pathway from the chromophore hydroxyl to Asp165 acceptor, causing the large Stokes shift and pH-induced cis-trans chromophore isomerization [37]. Subach et al. [38•] showed that RFPs can be converted into the blue FPs using amino acids substitutions at limited number of positions. This strategy was applied to five RFPs, including mKeima, TagRFP, mCherry, HcRed1 and M355NA, which all were engineered into blue probes.
Chromophore photochemistry in fluorescent proteins
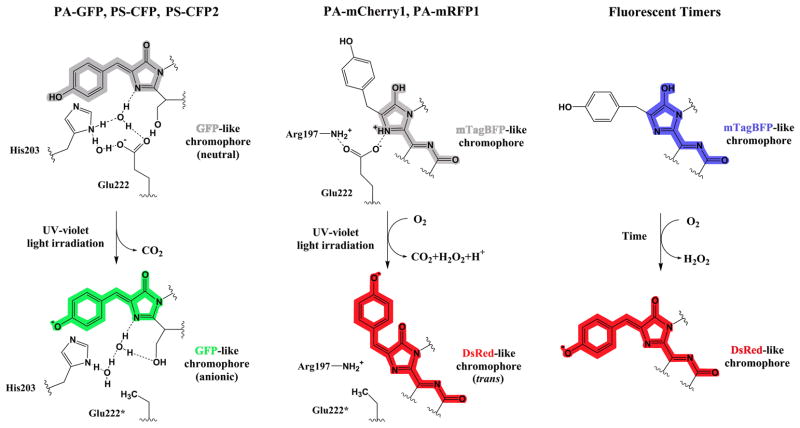
A structural basis for the photoactivation of PA-mCherrys, and the similar but rather dim PA-mRFP1 protein [39], remains unknown, while the mechanism of the green-to-red conversion for the Kaede-like proteins has been investigated thoroughly. However, it was suggested that the pre-activated PA-mCherry1 protein contains an mTagBFP-like chromophore, which absorbs violet light but does not fluoresce (FV Subach, VV Verkhusha, unpublished data). The PA-mCherry1 photoactivation possibly involves a decarboxylation of its Glu222 residue and subsequent oxidation of the mTagBFP-like chromophore that results in formation of the fluorescent DsRed-like chromophore but in a trans configuration (Figure 1). The quantitative decarboxylation of Glu222 via a Kolbe-like mechanism was detected after the PA-GFP photoactivation [40], and likely occurs in the course of the PS-CFP photoconversion [41] (Figure 1).
Figure 1.
Mechanisms of chromophore conversion from a neutral (protonated) to anionic (deprotonated) forms are illustrated for three key subgroups of fluorescent proteins such as PA-GFP, PS-CFP and PS-CFP2 (left), PA-mCherry1 and PA-mRFP1 (middle), and Fluorescent Timers (right). Chemical structures are shown for chromophores of the representative proteins before (top) and after (bottom) the conversion reactions. Colors of the chemical structures correspond to the spectral range of the chromophore emission except for the gray color, which indicates the non-fluorescent chromophore. UV-violet light-induced decarboxylation of the Glu222 residue is followed by the reorganization of the hydrogen bond network around the GFP-like chromophore that results in the chromophore deprotonation (left). Photoactivation by UV-violet light involves decarboxylation of the Glu222 residue and oxidation of the mTagBFP-like chromophore by molecular oxygen to the DsRed-like chromophore in the trans-configuration (middle). Slowed down oxidation of the mTagBFP-like chromophore by molecular oxygen without any light irradiation results in the formation of the DsRed-like chromophore in Fluorescent Timers (right).
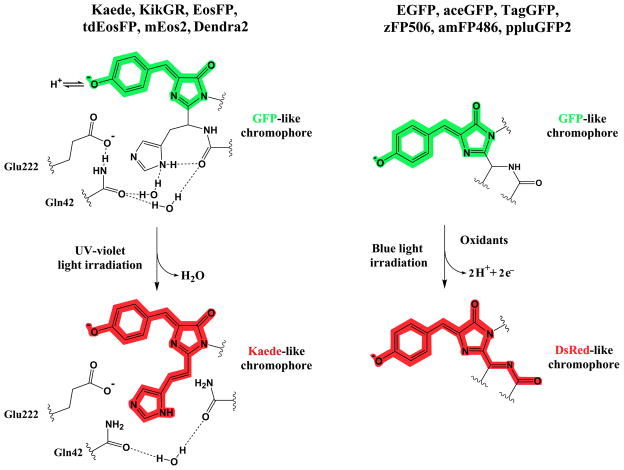
The Kaede-like proteins share the same chromophore-forming tripeptide His65-Tyr66-Gly67, which autocatalytically forms the green-emitting chromophore. X-ray analysis of the original green and photo-converted red chromophore forms revealed a light-induced extension of the chromophore π-electron system, known to result from backbone cleavage between the Nα and Cα atoms of His65 and formation of a double bond between the Cα and Cβ atoms in His65 [42,43] (Figure 2). This photo-induced process occurs only in the neutral state of the chromophore and requires no molecular oxygen. The structural basis for the β-elimination reaction is the unique positioning of His65 and the stereochemistry of the amino acid residues in chromophore environment. Glu222 is essential for the red chromophore formation, but it does not decarboxylate as it does in PA-mCherrys. Other Kaede-like proteins exhibit the similar chemical reaction during the photoconversion with minor variations, depending on the chromophore environment. In this way, a blue shift of absorption and emission peaks of Dendra2 can be explained by a local structural changes involving mainly Arg69 and neighboring water molecule [44]. It was shown that reversible photoswitching of IrisFP between the fluorescent and nonfluorescent states is based on the cis–trans chromophore isomerization, accompanied by protonation–deprotonation events [21].
Figure 2.
Mechanisms of chromophore photoconversion from an anionic (deprotonated) green to anionic (deprotonated) red forms are illustrated for two key subgroups of fluorescent proteins such as Kaede, KikGR, EosFP, tdEosFP, mEos2 and Dendra2 (left), and EGFP, aceGFP, TagGFP, zFP506, amFP486 and ppluGFP2 (right). Chemical structures are shown for chromophores of the representative proteins before (top) and after (bottom) the photoconversion reactions. Colors of the chemical structures correspond to the spectral range of the chromophore emission. The Glu222 residue stabilizes a transition state of the UV-violet light-induced polypeptide backbone cleavage by forming the hydrogen bond network with the Gln42 residue and chromophore forming His65 residue via water molecules; protonation-deprotonation equilibrium shown for the green chromophore is important for the photochemical behavior (left). The oxidative redding of the GFP-like chromophore is a one-photon process, which requires two equivalents of the oxidant per molecule of the fluorescent protein and possibly goes via formation of a radical of the chromophore, resulting in the DsRed-like chromophore (right).
Essentially distinctive green-to-red photocoversion mechanism has been recently revealed for EGFP and other green FPs, including aceGFP, TagGFP, zFP506, amFP486 and ppluGFP2 [45••]. In contrast to the previously described anaerobic EGFP redding, the authors found that a redding of green FPs also occurs in the presence of oxidants in common aerobic conditions. This oxidative redding occurred in solution as well as in live cells, when the protein was irradiated with a high-intensity 488 nm laser. A possible explanation is that the DsRed-like red chromophore is formed as the result of a two-electron oxidation (Figure 2). The same mechanism could also explain the photoconversion into a far-red state found in the mOrange variants [46•] and mKO [47]. Efficient photoconversion of mOrange occurred at 458 nm or 488 nm laser excitation. Excitation and emission maxima of the photoconverted state were approximately at 610 nm and 640 nm, respectively. In contrast to Kaede-like proteins, the mOrange photoconversion was not substantially affected by pH, however it required the 4–5-fold higher illumination power than that for Dendra2 photoconversion.
Interestingly, tdKatushka and mKate underwent a red-to-green photoconversion upon single-photon laser excitation at 405 and 561 nm, resulting in the green state with excitation and emission maxima at 495 nm and 518 nm, respectively [46•]. This red-to-green photoconversion suggests a reduction of the π-conjugated system of the DsRed-like chromophore. Monomeric state of mKate, low dependence of photoconversion from pH, and low phototoxicity of the converting blue light make mKate a promising template to design an efficient optical highlighter for live cell imaging. Furthermore, induction of a light-driven electron transfer in EGFP could be applied to monitor and manipulate with light the intracellular redox processes.
Spectral properties of proteins could be also affected simply with time, without any light irradiation. The first monomeric fluorescent timers (FTs) [48•], which exhibit distinctive fast, medium, and slow blue-to-red chromophore maturation rates, were developed on the basis of mCherry (Table 1). It was suggested that a blue-emitting form of FTs contain the mTagBFP-like chromophore, which is converted after oxidation to the red-emitting DsRed-like chromophore (SV Pletnev, VV Verkhusha, unpublished data) (Figure 1). The blue and red forms of FTs are bright enough to use FTs either alone in protein fusions or together with green FPs for multicolor imaging. FTs exhibit the similar timing behavior in bacteria, insect and mammalian cells. The predictable time course of changing fluorescent colors allows a quantitative analysis of temporal and spatial molecular events based on the ratio between the blue and red fluorescence intensities. Availability of three FTs with distinctive blue-to-red maturation times will be useful for studies of intracellular processes with substantially different time scales.
Conclusions
Recently developed monomeric RFPs and PA-RFPs extend the range of available probes and provide exciting new options in biotechnology, developmental and cell biology. We expect further broadening of the applications of these proteins in intact tissues and transgenic animals, using spatial-restricted deeper multi-photon laser excitation. Further studies generating the novel far-red and infra-red genetically encoded fluorescent markers and photoactivatable probes are of great practical interest because of high transparency of animal tissues in the 650–900 nm region.
Acknowledgments
This work was supported by a grant from the National Institutes of Health, GM073913 to V.V.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 2.Stepanenko OV, Verkhusha VV, Kuznetsova IM, Uversky VN, Turoverov KK. Fluorescent proteins as biomarkers and biosensors: throwing color lights on molecular and cellular processes. Curr Protein Pept Sci. 2008;9:338–369. doi: 10.2174/138920308785132668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shcherbo D, Souslova EA, Goedhart J, Chepurnykh TV, Gaintzeva A, Shemiakina II, Gadella TW, Lukyanov S, Chudakov DM. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol. 2009;9:24. doi: 10.1186/1472-6750-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 5.Gould TJ, Verkhusha VV, Hess ST. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat Protoc. 2009;4:291–308. doi: 10.1038/nprot.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuno H, Mal TK, Tong KI, Ando R, Furuta T, Ikura M, Miyawaki A. Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 8.Quillin ML, Anstrom DM, Shu X, O’Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38 A resolution. Biochemistry. 2005;44:5774–5787. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 9.Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA. zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore. Biochemistry. 2005;44:202–212. doi: 10.1021/bi048383r. [DOI] [PubMed] [Google Scholar]
- 10.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 11.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- •12.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. Detailed protocols for the intracellular photoactivation and tracking of photoactivatable red Dendra2 and green PS-CFP2 are presented. The protocols include design of genetic fusion constructs, transfection of mammalian cells, primary visualization of the fusion proteins, their photoactivation, tracking of the fusion proteins, and quantification of the imaged dynamics. The paper has recommendations regarding the optimal filter sets and settings of several confocal microscopes. [DOI] [PubMed] [Google Scholar]
- 13.Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habuchi S, Tsutsui H, Kochaniak AB, Miyawaki A, van Oijen AM. mKikGR, a monomeric photoswitchable fluorescent protein. PLoS One. 2008;3:e3944. doi: 10.1371/journal.pone.0003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark DA, Kulesa PM. An in vivo comparison of photoactivatable fluorescent proteins in an avian embryo model. Dev Dyn. 2007;236:1583–1594. doi: 10.1002/dvdy.21174. [DOI] [PubMed] [Google Scholar]
- ••17.Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. Using ensemble and single-molecule characteristics as screening criteria, monomeric PA-RFP PAmCherry1 was developed. Compared to other PA-RFPs, it has faster maturation, better pH stability, faster photoactivation, higher photoactivation contrast, and better photostability. Lack of green fluorescence in original state and its single-molecule behavior makes monomeric PAmCherry1 a preferred tag for two-color diffraction-limited photoactivation imaging and for super-resolution techniques such as a PALM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Röcker C, Salih A, Spindler KD, Nienhaus GU. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci USA. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •20.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. The paper describes an improved and true monomeric variant of EosFP, called mEos2. Authors characterize mEos2 in several fusion constructs and demonstrate its good incorporation into endogenous intracellular structures. An excellent photostability of the mEos2 photoconverted species enabled the long-term cell imaging. They also compare characteristics of mEos2 with those of other PA-RFPs both in purified samples and in cellular PALM experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adam V, Lelimousin M, Boehme S, Desfonds G, Nienhaus K, Field MJ, Wiedenmann J, McSweeney S, Nienhaus GU, Bourgeois D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc Natl Acad Sci USA. 2008;105:18343–18348. doi: 10.1073/pnas.0805949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, Chudakov DM. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 23.Nienhaus GU, Wiedenmann J. Structure, dynamics and optical properties of fluorescent proteins: perspectives for marker development. Chemphyschem. 2009;10:1369–79. doi: 10.1002/cphc.200800839. [DOI] [PubMed] [Google Scholar]
- 24.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–6. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 25.Pletnev S, Shcherbo D, Chudakov DM, Pletneva N, Merzlyak EM, Wlodawer A, Dauter Z, Pletnev V. A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis-trans isomerization of the chromophore. J Biol Chem. 2008;283:28980–7. doi: 10.1074/jbc.M800599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •26.Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Haelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–74. doi: 10.1042/BJ20081949. The paper reports on mKate2, a monomeric far-red uorescent protein that is 3-fold brighter than the original mKate and 4-fold brighter than mPlum. The authors also present tdKatushka2, a tandem far-red tag that performs well in fusions, provides 4-fold brighter near-IR uorescence compared with mRaspberry or mCherry, and is 20-fold brighter than mPlum. Currently, tdKatushka2 is the best far-red fluorescent protein for a whole body imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.König K. Multiphoton microscopy in life sciences. J Microsc. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 28.Shu X, Wang L, Colip L, Kallio K, Remington SJ. Unique interactions between the chromophore and glutamate 16 lead to far-red emission in a red fluorescent protein. Protein Sci. 2009;18:460–466. doi: 10.1002/pro.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–5. doi: 10.1038/nmeth.1235. The paper describes an improved orange-emitting monomeric fluorescent protein, named mKOk. The author used mKOk to construct a voltage sensor based on FRET from the coral-derived green fluorescent protein, named mUKG. The FRET sensor allowed visualization of the electrical activities in cultured cells and had the fast on-off switching rate. [DOI] [PubMed] [Google Scholar]
- 32.Kredel S, Oswald F, Nienhaus K, Deuschle K, Röcker C, Wolff M, Heilker R, Nienhaus GU, Wiedenmann J. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, Keenan RJ, Glick BS. A noncytotoxic DsRed variant for whole-cell labeling. Nat Methods. 2008;5:955–7. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strack RL, Bhattacharyya D, Glick BS, Keenan RJ. Noncytotoxic orange and red/green derivatives of DsRed-Express2 for whole-cell labeling. BMC Biotechnol. 2009;9:32. doi: 10.1186/1472-6750-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kogure T, Karasawa S, Araki T, Saito K, Kinjo M, Miyawaki A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 36.Kogure T, Kawano H, Abe Y, Miyawaki A. Fluorescence imaging using a fluorescent protein with a large Stokes shift. Methods. 2008;45:223–6. doi: 10.1016/j.ymeth.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Violot S, Carpentier P, Blanchoin L, Bourgeois D. Reverse pH-dependence of chromophore protonation explains the large Stokes shift of the red fluorescent protein mKeima. J Am Chem Soc. 2009 doi: 10.1021/ja903695n. in press (published on-line on July 10). [DOI] [PubMed] [Google Scholar]
- •38.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Grüenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. The paper describes a structure-based rational design strategy to develop blue fluorescent proteins. The strategy was applied to RFPs of the different genetic background. Further improvement of a TagRFP’s blue variant using random mutagenesis resulted in an enhanced monomeric protein, mTagBFP, characterized by higher brightness, pH-stability and faster chromophore formation than common blue fluorescence proteins with histidine in the chromophore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verkhusha VV, Sorkin A. Conversion of the monomeric red fluorescent protein into a photoactivatable probe. Chem Biol. 2005;12:279–285. doi: 10.1016/j.chembiol.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Henderson JN, Gepshtein R, Heenan JR, Kallio K, Huppert D, Remington SJ. Structure and mechanism of the photoactivatable green fluorescent protein. J Am Chem Soc. 2009;131:4176–4177. doi: 10.1021/ja808851n. [DOI] [PubMed] [Google Scholar]
- 41.Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, Lukyanov S, Lukyanov KA. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 42.Nienhaus K, Nienhaus GU, Wiedenmann J, Nar H. Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc Natl Acad Sci USA. 2005;102:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi I, Mizuno H, Tong KI, Furuta T, Tanaka F, Yoshimura M, Miyawaki A, Ikura M. Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede. J Mol Biol. 2007;372:918–926. doi: 10.1016/j.jmb.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 44.Adam V, Nienhaus K, Bourgeois D, Nienhaus GU. Structural basis of enhanced photoconversion yield in green fluorescent protein-like protein Dendra2. Biochemistry. 2009;48:4905–4915. doi: 10.1021/bi900383a. [DOI] [PubMed] [Google Scholar]
- ••45.Bogdanov AM, Mishin AS, Yampolsky IV, Belousov VV, Chudakov DM, Subach FV, Verkhusha VV, Lukyanov S, Lukyanov KA. Green fluorescent proteins are light-induced electron donors. Nat Chem Biol. 2009;5:459–461. doi: 10.1038/nchembio.174. The authors discovered a novel feature of green FPs, such as EGFP, aceGFP, TagGFP, zFP506, amFP486 and ppluGFP2, to act as the light-induced electron donors. It was found that in common aerobic conditions, in the presence of oxidants, blue light induces the efficient green-to-red photoconversion, named an oxidative redding. Various electron acceptors such as potassium ferricyanide, benzoquinone, flavines, and some redox-active proteins substantially facilitated the oxidative redding. The green-to-red photoconversion was also observed in living cells without additional treatment. This finding changes the general view on the green FPs as the passive light absorbers-emitters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •46.Kremers GJ, Hazelwood KL, Murphy CS, Davidson MW, Piston DW. Photoconversion in orange and red fluorescent proteins. Nat Methods. 2009;6:355–358. doi: 10.1038/nmeth.1319. The paper describes a surprising photoactivation of orange-emitting mOrange and mOrange2, and far-red Katushka, mKate and HcRed1 fluorescent proteins, when they are irradiated with an intense light of the specific wavelengths. Interestingly, the orange RFPs photoconvert into a farred form while the far-red RFPs photoconvert into a green form. For all RFPs, the photoconversion efficiency nonlinearly depended on the light intensity suggesting a multiphoton phenomenon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW. Sensitive detection of p65 homodimers using red-shifted and fluorescent protein-based FRET couples. PLoS One. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •48.Subach FV, Subach OM, Gundorov IS, Morozova KS, Piatkevich KD, Cuervo AM, Verkhusha VV. Monomeric fluorescent timers that change color from blue to red report on cellular trafficking. Nat Chem Biol. 2009;5:118–126. doi: 10.1038/nchembio.138. The authors developed three monomeric fluorescent timers (FTs), which change their fluorescence from blue to red over time. FTs provide reliable way to analyze the spatial and temporal history of the FT-fused proteins and to detect activation and repression of their expression. FTs will also allow identification of protein recycling events among compartments, temporal tracking of molecules before and after a cellular event, and timing of particular intracellular post-translational modifications. [DOI] [PMC free article] [PubMed] [Google Scholar]