Synopsis
Frataxin is a highly conserved mitochondrial protein whose deficiency in humans results in Friedreich’s ataxia (FRDA), an autosomal recessive disorder characterized by progressive ataxia and cardiomyopathy. Although its cellular function is still not fully clear, the fact that frataxin plays a crucial role in Fe-S assembly on the scaffold protein Isu is well accepted. Here we report the characterisation of eight frataxin variants having alterations on two putative functional regions – the α1/β1 acidic ridge and the conserved β-sheet surface. We report that frataxin iron binding capacity is quite robust: even when five of the most conserved residues from the putative iron binding region are altered, at least 2 iron atoms per monomer can be bound, although with decreased affinity. Furthermore, we conclude that the acidic ridge is designed to favour function over stability. The negative charges have a functional role, but at the same time significantly impair frataxin’s stability. Removing five of those charges results in a thermal stabilization of ~24°C and reduces the inherent conformational plasticity. Alterations on the conserved β-sheet residues have only a modest impact on the protein stability, highlighting the functional importance of residues 122-124.
Keywords: yeast frataxin, protein folding, metallochaperone, binding affinities, protein plasticity
Introduction
Reduced expression of frataxin, or in some cases, expression of frataxin variants, leads to the development of Friedreich’s ataxia (FRDA), an autosomal recessive neurodegenerative and cardiodegenerative disease characterized by progressive ataxia and cardiomyopathy [1–3]. In spite of the availability of multi cellular models for the disease [4, 5], insights into possible therapeutic strategies [6, 7] and broader systematic clinical studies [8], the pathogenesis of Friedreich’s ataxia and its relationship with frataxin remains to be fully understood. Frataxin is a highly conserved protein localized in the mitochondria whose function is associated with iron homeostasis. Frataxin deficiency, as recently shown using conditional knockout mice, down regulates molecules involved in mitochondrial iron utilization pathways (FeS cluster and heme biosynthesis, and iron storage) while cellular iron uptake and its availability for mitochondrial uptake are increased [5]. This illustrates the complex interplay between frataxin deficiency and iron homeostasis in the context of FRDA [5].
The human and yeast frataxin (Yfh1) orthologs have a high degree of amino acid (65%) and structural identity. The phenotype observed as a consequence of frataxin’s deficiency is very similar in human and yeast cells [9, 10]. Hence, the yeast model system has been frequently used to study frataxin’s function, as well as the pathological events associated with FRDA. Although a detailed understanding of frataxin function is still missing, data suggests a role as an iron metallo-chaperone [11–14], delivering iron to its binding partners, promoting heme and Fe-S clusters biosynthesis, as well as Fe-S cluster repair [15–17]. While initially oligomerization was believed to be dispensable in vivo recently, it has been shown that oligomerization impairment is harmful under oxidative stress conditions and it leads to a decrease of yeast chronological lifespan [11, 18, 19]. Yfh1 is not essential, but its absence causes severe growth defects and a reduction in the activity of Fe-S cluster–containing enzymes, such as aconitase [9, 10, 19, 20]. Iron binding by frataxins, a requirement for a role as a metallochaperone, has been well documented by several laboratories [16, 21, 22]. Frataxins bind iron at micromolar binding affinities, with different stoichiometries: while monomeric human frataxin binds 6/7 iron ions (Kd ~ 12–55μM [16]), monomeric frataxin from Drosphila binds 1 ferrous iron (Kd ~6μM, [22]) and yeast frataxin binds 2 ferrous irons (Kd ~2.5μM, [21]). This variability suggests a certain plasticity of the frataxin fold in respect to binding iron. In fact, an NMR based investigation of the iron binding to Yfh1 at low stoichiometry (up to 2Fe:Yfh1) denoted changes in 10 distinct residues [23], which could indicate long range effects upon iron binding, or lower affinity sites. More complex effects arise at higher Fe:Yfh1 stoichiometries. Like human frataxin, Yfh1 undergoes iron-dependent oligomerization, above a threshold of 2 irons per monomer, according to the progression α → α3 → α6 → α12 → α24 → α48 [11, 18]. This property has been established as important for iron-induced oxidative stress protection, but not for Fe-S cluster biogenesis [19, 24].
The conserved frataxin fold consists of an α–β sandwich motif, with two α-helices (N- and C-terminal) that construct the helical plane and five antiparallel β-strands that form the β-sheet plane. A sixth (seventh in the human protein) β-strand intersects the helical and β-sheet planes [23, 25–28]. The first helix (α1) and the edge of the first β-strand (β1) form a semi-conserved acidic ridge that constitutes the putative iron binding region of the protein [21, 23, 28]. A set of conserved residues in the β-sheet surface constitutes the other putative functional region of the protein, believed to be involved in the binding of frataxin to its protein partners [23, 29]. Recently we have shown that mutations at the conserved β-sheet residues 122-124 affect neither iron binding capacity nor the oligomerization properties of frataxin, although this mutation does compromise the interaction with Isu [30].
To complement previous functional characterization of YFH1 mutants, here we address the structural and conformational effects of eight functional mutants in yeast frataxin that alter either the acid ridge or conserved residues on the β-sheet surface. We found that the negatively charged residues in the acid ridge that are important for iron binding reduce Yfh1’s stability, indicating a trade-off between functionality and stability. In addition, alterations in the conserved residues of the portion of the acid ridge found in the β-sheet, affected interaction with the scaffold protein Isu, but did not affect overall protein stability.
Experimental
Yeast strains, plasmids and media
Saccharomyces cerevisiae strains carrying YFH1 mutant or wild type genes on plasmids were created by sporulation and dissection of YFH1/yfh1Δ (W303 background) transformed with the plasmids. YFH1 mutants were generated by site-directed mutagenesis using pCM189-YFH1 as a template, having YFH1 under the control of the tetO promoter [31]. Constructs containing the following mutations were prepared: yfh1-D86A/E90A/E93A, yfh1-D101A/E103A, yfh1-D86A/E90A/E93A/D101A/E103A, yfh1-N122A, yfh1-N122K (corresponding to the human clinical mutation N146K), yfh1-K123T, yfh1-Q124A and yfh1-N122A/K123T/Q124A. Doxycycline was used at 1 μg/ml to reduce expression. Cells were grown in minimal synthetic media with –ura DO at 30°C for 2 days.
Aconitase activity in isolated mitochondria
Mitochondria were isolated, according to the method described in [32], from cells grown in synthetic minimal media (-ura), in the absence of doxycycline. Aconitase activity was measured in isolated mitochondria by monitoring the decrease in the absorbance of the substrate isocitrate at 240 nm as described in [33].
Gene Expression and Protein purification
The mature form of Yfh1 (amino acids 52-174) with an N-terminal His tag and a thrombin cleaving site in between was cloned into pET-3a vector (Novagen, Madison, WI, USA) [19]. Protein expression was induced over 3h at 37°C in BL21(DE3) E. coli by adding IPTG (isopropyl-b-galactopyranoside) at a final concentration of 0.5mM. With the exception of Yfh1-K123T, which was expressed at much lower levels (approximately 30%), all other mutant variants had expression levels identical to those of the wild type Yfh1. The reduced expression levels of Yfh1-K123T had already been described when expressing this mutant in yeast [30]. Under the tested conditions, wild type Yfh1 was exclusively expressed as a soluble protein, as were most of the mutants. The only exceptions are Yfh1-86/90/93A, Yfh1-N122A and Yfh1-N122A/K123/Q124A, in which ~15% of the expressed protein was found in the insoluble fraction. After harvesting the cells they were lysed on a French press. Cell lysate was subjected to His-binding resin (Amersham) chromatography and the protein was eluted with 500mM Imidazole in the binding buffer (50mM Tris-HCl Ph8.0, 200mM NaCl, 10mM Imidazole). The His tag was cleaved using biotinylated thrombin and thrombin was removed by streptavidin agarose (Amersham). When necessary (purity less then 90%) the protein was further purified by applying the sample on a Superdex 75. At the end the buffer was changed to 10mM HEPES, 50mM NaCl pH 7.0 using Centricons (Millipore). Protein concentration was determined using the extinction coefficient ε280nm = 15470 M−1.cm−1. The mature form of Isu1(amino acids 35-165) with an His tag and a TEV cleaving site in between was cloned into pKLD37 vector. Protein expression was induced over 4h at 30°C in C41 E. coli by adding IPTG (isopropyl-β-galactopyranoside) at a final concentration of 1mM the purification was performed as above described for the Yfh1 variants. Protein concentration was determined using the extinction coefficient ε280nm = 14161.4 M−1.cm−1
Spectroscopic methods
UV/Vis Spectra were recorded at room temperature in a Shimadzu UVPC-1601 spectrometer. Fluorescence spectroscopy was performed on a Cary Varian Eclipse instrument (λex =280nm, λem =340nm, slitex: 5 nm, slitem: 10 nm unless otherwise noted) equipped with cell stirring and Peltier temperature control. Far-UV CD spectra were recorded typically at 0.2 nm resolution on a Jasco J-715 spectropolarimeter fitted with a cell holder thermostated with a peltier.
Thermal denaturation
Thermal unfolding was followed by ellipticity (Δεmrw at 222nm) variations. In all experiments a heating rate of 1°C/min was used, and the temperature was changed from 10 to 90°C. All reactions were found to be reversible under the tested conditions, as inferred from obtaining identical far-UV CD spectra of the proteins before the thermal ramping and after cooling down the heated sample; a second temperature ramp of the same sample also yielded identical melting transitions. Protein aggregation was not observed during thermal unfolding. Data were analysed according to a two-state model and the thermodynamic data parameters were determined [34].
Trypsin limited proteolysis and LC-MS analysis
Frataxins were incubated with trypsin (bovine pancreas trypsin, sequencing grade, Sigma) at 37°C in 0.1 M Tris-HCl pH 8.5, in a 150 fold excess over the protease. Aliquots (300 μmol of protein) were sampled at different incubation periods and the reaction stopped by adding loading buffer with 5μM BSA followed by a 10 min incubation at 100°C. The products of the proteolysis reaction were analysed by SDS-PAGE [35].
Iron-binding assays
Yeast frataxin oligomerizes in an iron-concentration dependent fashion, but at up to a stoichiometry of 2 irons per protein the protein remains in the monomer form [11, 18, 23, 36]. To avoid side effects from different oligomerization behaviours we have measured iron binding before oligomerization takes place, i.e. up to a iron:protein ratio of 2. Trp fluorescence emission spectroscopy was used to monitor iron binding, which quenches Trp emission as the iron-frataxin complex is being formed. As previously shown, this methods yield results identical to those obtained by isothermal titration calorimetry [16, 37]. For the purpose, the quenching of the Trp fluorescence (λexc=290nm; λem=340 nm) of a 50μM solution of apo frataxin in 100mM HEPES pH 7.0, 50mM NaCl was measured upon stepwise addition of an iron citrate solution, in 1 ml quartz cuvettes under continuous stirring [35, 37]. This data was used to calculate the fraction of binding sites occupied. The stoichiometry, p, and apparent dissociation constant, Kd, were then determined as described by Winzor and Sawyer [38].
Isu:Yfh1 interaction assay
Holo-Yfh1 was prepared using a stoichiometry of 2 irons per frataxin to avoid interference from oligomer formation. The formation of a Isu:holo-frataxin complex was followed monitoring the Trp fluorescence, taking advantage of the fact that yeast Isu does not contain any Trp residue. Thus, when Isu binds to holo-Yfh1, the Trp fluorescence emission (λexc=290nm; λem=340 nm) of the latter is quenched and this can be used to monitor binding. Holo-Yfh1 was prepared in 100 mM HEPES pH 7.0, 50mM NaCl. Aliquots of Isu were added to 2.5 mM holo frataxins under constant stirring. The stoichiometry, p, and apparent dissociation constant, Kd, were then obtained as described by Winzor and Sawyer [38].
Results and Discussion
Alterations in the conserved β-sheet surface result in functional effects
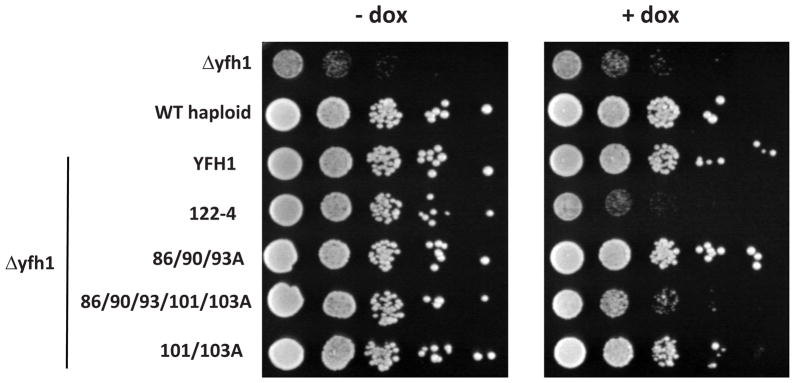
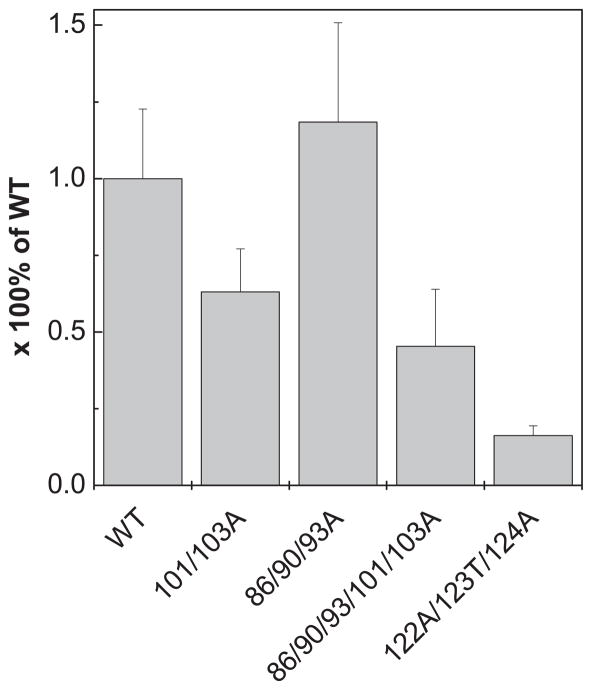
To gain a better understanding of the relationship between the structure and function of frataxin, we compared four Yfh1 variants. Three (Yfh1-D101/E103A, Yfh1-D86/E90/E93A and a combination of these two, Yfh1-86/90/93/101/103A) alter the putative iron binding region at the α1/β1 acidic ridge and oligomerization region (Fig. 1). The fourth, Yfh1-N122A/K123T/Q124A (hereafter referred to as Yfh1-122-4), alters the conserved β-sheet, the putative binding surface for interaction with Isu, the scaffold on which Fe-S clusters are built [30]. Since Yfh1 cellular levels can be significantly reduced before any growth defects are detected [39], wild type and mutant YFH1 genes were placed under the control of the tetO promoter allowing the modulation of protein expression using doxycycline. In this way, in vivo function could be tested under both normal and reduced levels of expression. At normal Yfh1 expression levels (minus doxycycline) all mutant gene were able to support growth as well as the wild type gene (Fig. 2). In addition, as we previously reported [19, 30], Yfh1-122-4 cells grew poorly when the variant was expressed at reduced levels (plus doxycycline), while cells expressing low levels of Yfh1-86/90/93A grew as well as wild type cells. However, Yfh1-101/103A was slightly compromised, and when these mutations were combined with those of Yfh1-86/90/93A, generating alterations in five of the charged residues in the acidic ridge, growth was severely compromised. Consistent with this enhanced growth phenotype, mitochondria isolated from Yfh1-101/103A had lower levels of activity of the Fe-S enzyme aconitase, even when the mutant protein was expressed at normal levels (Fig. 3). This affect was more extreme in Yfh1- 86/90/93/101/103A mitochondria, which had only 40% the aconitase activity of wild type mitochondria. Consistent with the affects on growth, Yfh1-122-4 mitochondria had only 20% the aconitase activity quantified for wild type mitochondria.
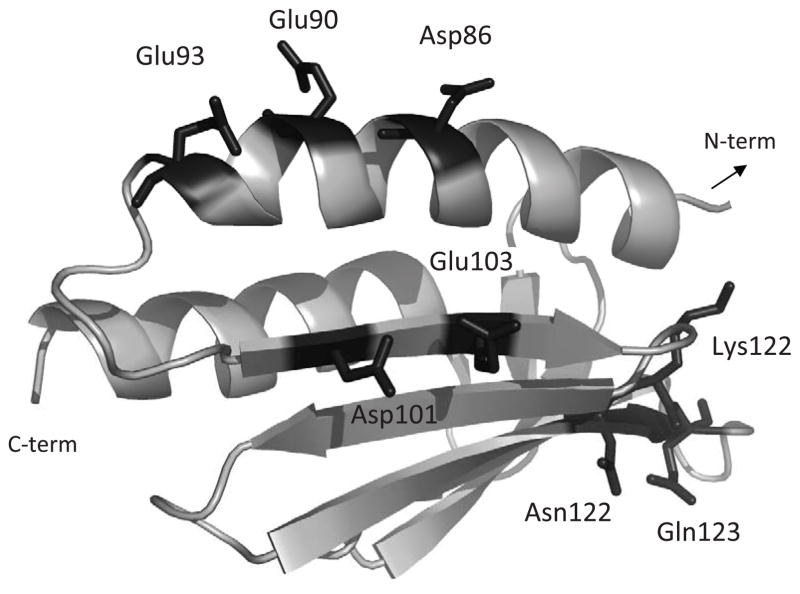
Figure 1.
Yfh1 ribbon structure generated using PyMOL (PDB accession #2ga5). Mutated residues are represented by sticks, labelled and highlighted in orange. The present study has focused on the structural and conformational characterization of the following variants: Yfh1-D86A/E90A/E93A, Yfh1-D101A/E103A, Yfh1-D86A/E90A/E93A/D101A/E103A, Yfh1-N122A, Yfh1-N122K (corresponding to the human clinical mutation N146K), Yfh1-K123T, Yfh1-Q124A and Yfh1-N122A/K123T/Q124A.
Figure 2. Yeast growth rescue of (Δyfh1) cells by Yfh1 variants.
A 10-fold dilutions of cell suspension of wild type, Δyfh1 and Δyfh1 transformed with plasmids harbouring the indicated YFH1 mutant genes under the control of the tet-O-regulatable wild type or mutant yfh1 were plated on minimal synthetic medium –ura DO containing (+) or lacking (−) doxycycline (dox). Plates were incubated at 30°C for 2 days.
Figure 3. Aconitase activity.
Extracts from mitochondria isolated from the indicated strains were prepared (150μg) and aconitase activity was measured. Data is presented as percentage of wild type aconitase activity (n=3).
None of the mutations abolish iron binding
Since iron binding is an essential functional feature of frataxin, we asked if iron-binding properties of the Yfh1 variants were affected. In order to exclude iron-induced frataxin oligomerization effects, we have investigated iron-binding at low stoichiometry (up to 2 Fe(II)/Yfh1), i.e. under conditions in which oligomerization does not take place. We have used Trp-fluorescence to monitor iron binding to Yfh1 variants, as Trp emission is a specific reporter for iron-binding to frataxin. Our measurements indicated that mutations altering the iron binding region, D86A, E93A, D101A and E103A, had no effect on the iron binding capacity at low stoichiometry, but did decrease binding affinity somewhat (Table 1). Conversely, iron must be able to bind to frataxin through other residues, as even the quintuple mutant Yfh1-86/90/93/101/103A retained the ability to bind ~2 Fe(II)/Yfh1. We hypothesise that iron is binding to secondary sites with lower affinity, as evidenced by the higher dissociation constants (~20 μM). In fact, a previous NMR study has shown that under identical conditions at low iron stoichiometry, iron binding to Yfh1 affected multiple sites: mainly interactions with carboxylate and nitrogen from acidic residues within the α1/β1 ridge (His83, Asp86, Glu93, His95, Asp101, Glu103), but other residues (Ala94, Leu104, Ser105 and Asn140) were also found to change their resonance positions upon binding of up to 2 irons per frataxin [23]. The lower binding affinity that we have determined in the Yfh1 variants (nevertheless still in the micromolar range comparable to that of human frataxins) could possibly indicate the recruitment of secondary positions, rather than unspecific binding. However, this it not the case, as the observed iron-binding is functional, as shown by the fact that yeast expressing these variants still have some aconitase activity (Fig. 3), which depends of frataxin-mediated iron transfer [15]. In addition, a previous study confirms our observations: it has been shown that single point mutations to alanine on residues 86, 90, 93, 101 and 103 reduce Yfh1 affinity to iron, but do not abolish iron binding [24]. Single point mutations in the Asn122-Gln124 segment seem to have an intermediate effect in respect to the binding affinity (~14–18 μM). Presumably, alterations in the protein-protein interaction region of Yfh1 result in long-range effects on the iron-binding acid ridge leading to decrease of the iron binding affinity.
Table 1.
Thermodynamic parameters for thermal denaturation of yeast frataxin variants. Effect of functional mutations on the protein thermal stability and iron binding affinity.
| Protein | ΔH (kJ.mol−1) | ΔGa (J.mol−1) | Tm (°C) | ΔTm (°C) | Δ(ΔG)b (J.mol−1) | Kd (μM)c |
|---|---|---|---|---|---|---|
| Wild Type | 118.0 ± 1.3 | 5946.7 | 40.4 ± 0.1 | - | - | 10.4 ± 2.1 |
| N122K | 145.6 ± 2.1 | 7772.2 | 44.7 ± 0.1 | + 4.3 ± 0.2 | 2286.6 | 11.0 ± 3.8 |
| N122A | 125.9 ± 1.7 | 4106.2 | 35.9 ± 0.1 | − 4.5 ± 0.2 | −2461.5 | 14.0 ± 1.0 |
| K123T | 120.1 ± 1.7 | 6197.8 | 41.0 ± 0.1 | + 0.6 ± 0.2 | 323.0 | 18.8 ± 2.4 |
| Q124A | 143.5 ± 1.3 | 5366.0 | 39.0 ± 0.1 | − 1.4 ± 0.2 | 758.1 | 13.2 ± 3.8 |
| N122A/K123T/Q124A | 90.8 ± 1.3 | 5863.5 | 40.2 ± 0.1 | − 0.2 ± 0.2 | 107.9 | 27.1 ± 1.0 |
| 101/103A | 230.5 ± 3.3 | 11001.4 | 52.0 ± 0.1 | + 11.6 ± 0.2 | 6030.4 | 20.9 ± 0.6 |
| 86/90/93A | 214.6 ± 3.8 | 11546.2 | 53.2 ± 0.1 | + 12.8 ± 0.2 | 6630.0 | 21.3 ± 1.0 |
| 86/90/93/101/103 A | 322.6 ± 6.3 | 16687.9 | 64.2 ± 0.1 | + 23.8 ± 0.2 | 11925.2 | 22.8 ± 0.9 |
ΔG at 25°C, considering an estimate for ΔCp of 50.2 J/mol/K.
Δ(ΔG) =[Δ(Tm)] × ΔSm = [Δ(Tm)] × (ΔHm/Tm), where ΔSm and ΔHm are values for the wild type.
(n=2)
Isu binding may also involve residues from the acidic ridge
Since low stoichiometry iron binding was not impaired in the mutants, we next evaluated whether the interaction between Yfh1 and Isu was compromised. This interaction is mediated by iron, as only holo-Yfh1 interacts with Isu [16]. Yfh1-86/90/93A was found to bind to Isu with a wild type like affinity (Kd ~5μM), while no interaction with Yfh1-122-4 was detected, consistent with previously published results [19] and [30], respectively. The alteration of residues 122, 123 and 124 individually (N122K, N122A, K123T or Q124A), severely affected the Isu interaction supporting the hypothesis that all three residues are important for this interaction.
No interaction between Isu and Yfh1-101/103A (or Yfh1-96/90/93/101/103A) was detected in our in vitro assay. This reduced interaction is somewhat surprising considering that Yfh1-101/103A could significantly rescue the growth defect of yfh1 cells, even when expressed at low levels (Fig. 2). Mutating these two residues has only been found to cause a growth defect when the alterations are to lysines and the medium is supplemented with high levels of iron [20]. In vivo other cellular factors may promote the interaction between Yfh1-101/103A and Isu, explaining the difference between the in vivo and in vitro results. Indeed frataxin was found to interact with Isd11 of the Nfs1/Isu complex and multiple mitochondrial chaperones [40]. Alternatively, significantly reduced affinity may be tolerated in vivo.
Charge-to-neutral alterations in the α1/β1 acidic ridge increase stability
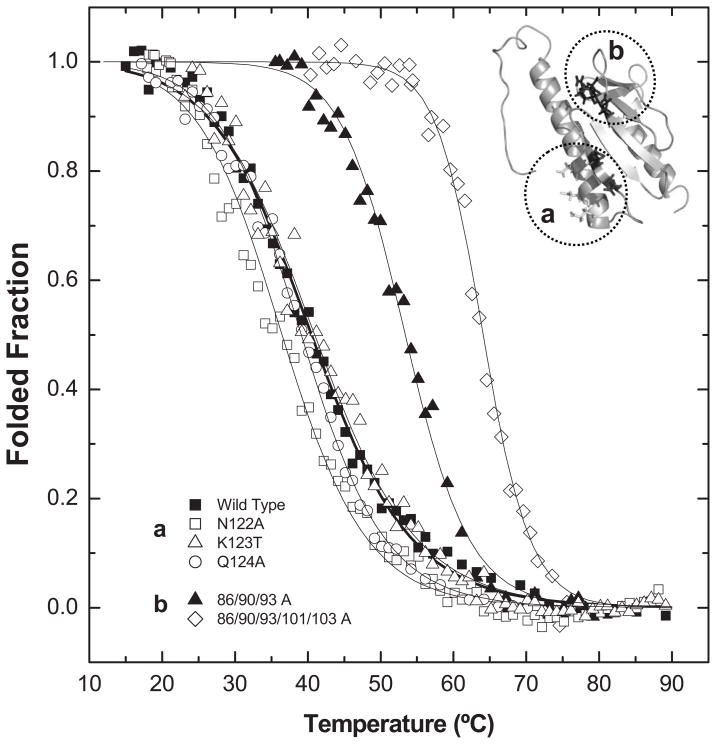
In order to evaluate if the functional impairment may result from decreased protein stability, the effect of the alterations on Yfh1 folding thermodynamics was analyzed by comparing the thermal stability of mutant variants to that of wild type. According to the analysis of the far-UV CD spectra at 20°C, before and after thermal denaturation, thermal unfolding was reversible for all protein variants studied and no aggregation was observed after thermal unfolding. The results showed that charge-to-neutral alterations in the acidic ridge result in an impressive stabilization of the protein fold: an increase of up to ~24°C was noted for the Yfh1-86/90/93/101/103A variant, while alterations in the β–sheet surface had almost no effect on protein thermal stability (Fig. 4, Table 1).
Figure 4. Thermal denaturation curves at pH 7.0 following Trp emission.
Impact of mutations on Yfh1 thermal stability. (■) Wild Type, (a) single point mutations of β-sheet surface residues (122-123). (□) N122A, (△) K123T, (○)Q124A, (b) Mutations on putative iron binding sites: (▲) D86A E90A E93A, (◇) 86/90/93/101/103 A. Lines represent fits to the two-state model [34]; for parameters see Table 2.
The protein stability decreased in the order: Yfh1-86/90/93/101/103 A > Yfh1-86/90/93 A > Yfh1-101/103 A > Yfh1-N122K > Yfh1-K123T > Wild Type > Yfh1-N122A/K123T/Q124A > Yfh1-Q124A > Yfh1-N122A. The exception for the effect on the β–sheet surface are the mutations in residue Asn-122: changing to an alanine (ΔTm = −4.5 °C) or to a lysine (ΔTm = +4.3 °C) had opposite effects, probably due to the effect these alterations would be expected to have on the β–hairpin between strands β3 and β4, which involves two hydrogen bonds (Asn122-Trp131 and Val120-A133). While the insertion of an Ala likely disrupts the hydrogen bond with Trp131, the positively charged Lys might strengthen it, stabilizing the protein. The 101/103A alterations stabilize the protein in spite of compromising 2 of the 3 hydrogen bonds involved in the β–hairpin connecting strands β1 and β2. This suggests that minimizing repulsive interactions overcomes the stabilisation obtained by the two hydrogen bonds.
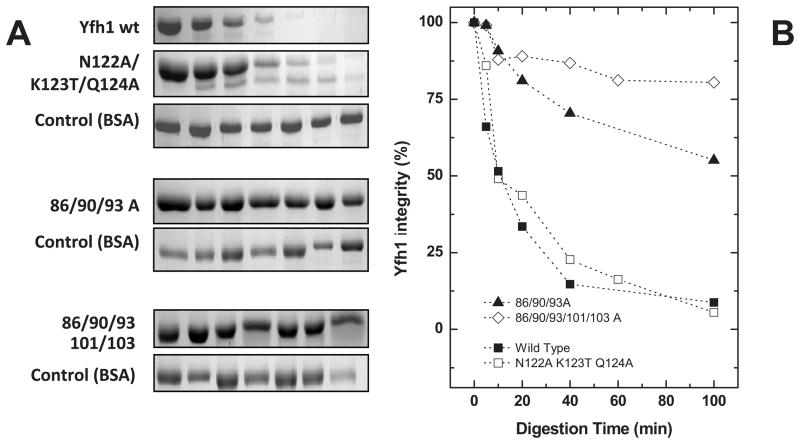
Since protein conformational plasticity affects both protein function and degradation rates. We next analyzed whether the functional mutations were also affecting frataxin flexibility, by performing limited proteolysis experiments using trypsin. Our underlying rationale was that mutations resulting in an increased structural flexibility would increase trypsin access to cleavage sites and consequently increase the degradation rate. The results show two distinct patterns, depending on the region in which the alteration is located (Fig. 5). Alterations on the acidic ridge had a pronounced effect on frataxin dynamics, making the protein more rigid and substantially less susceptible to proteolysis. In fact, the Yfh1-86/89/93/101/103A variant remains essentially intact under conditions in which wild type frataxin is essentially completely digested (80% versus 10% integrity after 100 min digestion). On the other hand, alterations on the β-sheet surface (Yfh1-N122K/K123T/Q124A) behave almost identically to the wild type, suggesting that modifications in this region have either a very small or no effect on the protein conformational plasticity.
Figure 5. Time course of trypsin limited proteolysis.
Comparison between wild type and functional mutants (A) SDS-PAGE analysis of the time course trypsin limited proteolysis experiments. (B) Evaluation/quantification of frataxin degradation during incubation with trypsin. Gels (A) densitometric analysis allowed the quantification of frataxin for the different incubation times with trypsin. (■) Wild Type, (□) N122A K123T Q124A, (▲) D86A E90A E93A, (◇) 86/90/93/101/103 A.
Overall, alterations in the acidic ridge that prevent iron binding at the primary sites increased substantially the protein stability and decreased its flexibility decreases in the structural flexibility may prevent conformational changes necessary to allow the interaction with protein partners. This increase in both thermal stability and resistance to proteolytic degradation suggests that the iron binding region is particularly susceptible to an activity-stability trade off.
Conclusions
Here we present a detailed characterization of eight yeast frataxin functional variants, that either alter the acidic ridge between α-helix 1 and β-sheet 1, or the conserved β-sheet surface between strands 3 and 4. Changing the conserved β-sheet residues Asn122-Lys-123-Gln124 had almost no effect on Yfh1 stability and plasticity, indicating that changes in this region did not disrupt overall conformation, but are relevant for the Yfh1-Isu interaction. Alteration of up to five residues in the acidic ridge region, four of which had been identified as iron binding sites [23], significantly increase frataxin stability. Thus illustrates a rather interesting trade off between activity and stability in this region. In addition, our study suggests that residues Asp101 and Glu103 are involved in the iron-mediated interaction between Isu and Yfh1, but their alteration does not abrogate the interaction, as evidenced by the rescue of the Δyfh1 phenotype.
Acknowledgments
This work is partly financed by grants from the Fundação para a Ciência e Tecnologia (PTDC/QUI/70101) and National Ataxia Foundation (to CMG) and grants from the National Institutes of Health Grant (GM27870) and Muscular Dystrophy Association (to EAC.). A.R.C and T.W were recipients of FCT PhD Fellowship (SFRHBD/24949/2005) and an American Heart Association Postdoctoral Fellowship 0525728Z, respectively.
References
- 1.Pandolfo M. Friedreich ataxia: the clinical picture. Journal of neurology. 2009;256(Suppl 1):3–8. doi: 10.1007/s00415-009-1002-3. [DOI] [PubMed] [Google Scholar]
- 2.Becker E, Richardson DR. Frataxin: its role in iron metabolism and the pathogenesis of Friedreich’s ataxia. The international journal of biochemistry & cell biology. 2001;33:1–10. doi: 10.1016/s1357-2725(00)00067-4. [DOI] [PubMed] [Google Scholar]
- 3.Durr A, Brice A. Clinical and genetic aspects of spinocerebellar degeneration. Current opinion in neurology. 2000;13:407–413. doi: 10.1097/00019052-200008000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Puccio H. Multicellular models of Friedreich ataxia. Journal of neurology. 2009;256(Suppl 1):18–24. doi: 10.1007/s00415-009-1004-1. [DOI] [PubMed] [Google Scholar]
- 5.Huang ML, Becker EM, Whitnall M, Rahmanto YS, Ponka P, Richardson DR. Elucidation of the mechanism of mitochondrial iron loading in Friedreich’s ataxia by analysis of a mouse mutant. Proc Natl Acad Sci U S A. 2009;106:16381–16386. doi: 10.1073/pnas.0906784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson DR. Friedreich’s ataxia: iron chelators that target the mitochondrion as a therapeutic strategy? Expert opinion on investigational drugs. 2003;12:235–245. doi: 10.1517/13543784.12.2.235. [DOI] [PubMed] [Google Scholar]
- 7.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nature chemical biology. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 8.Schulz JB, Boesch S, Burk K, Durr A, Giunti P, Mariotti C, Pousset F, Schols L, Vankan P, Pandolfo M. Diagnosis and treatment of Friedreich ataxia: a European perspective. Nature reviews. 2009;5:222–234. doi: 10.1038/nrneurol.2009.26. [DOI] [PubMed] [Google Scholar]
- 9.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 10.Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 11.Gakh O, Adamec J, Gacy AM, Twesten RD, Owen WG, Isaya G. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry. 2002;41:6798–6804. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 12.Schulz JB, Dehmer T, Schols L, Mende H, Hardt C, Vorgerd M, Burk K, Matson W, Dichgans J, Beal MF, Bogdanov MB. Oxidative stress in patients with Friedreich ataxia. Neurology. 2000;55:1719–1721. doi: 10.1212/wnl.55.11.1719. [DOI] [PubMed] [Google Scholar]
- 13.Thierbach R, Schulz TJ, Isken F, Voigt A, Mietzner B, Drewes G, von Kleist-Retzow JC, Wiesner RJ, Magnuson MA, Puccio H, Pfeiffer AF, Steinberg P, Ristow M. Targeted disruption of hepatic frataxin expression causes impaired mitochondrial function, decreased life span and tumor growth in mice. Hum Mol Genet. 2005;14:3857–3864. doi: 10.1093/hmg/ddi410. [DOI] [PubMed] [Google Scholar]
- 14.Vazquez-Manrique RP, Gonzalez-Cabo P, Ros S, Aziz H, Baylis HA, Palau F. Reduction of Caenorhabditis elegans frataxin increases sensitivity to oxidative stress, reduces lifespan, and causes lethality in a mitochondrial complex II mutant. Faseb J. 2006;20:172–174. doi: 10.1096/fj.05-4212fje. [DOI] [PubMed] [Google Scholar]
- 15.Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 16.Yoon T, Cowan JA. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J Am Chem Soc. 2003;125:6078–6084. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 17.Yoon T, Cowan JA. Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J Biol Chem. 2004;279:25943–25946. doi: 10.1074/jbc.C400107200. [DOI] [PubMed] [Google Scholar]
- 18.Adamec J, Rusnak F, Owen WG, Naylor S, Benson LM, Gacy AM, Isaya G. Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich ataxia. Am J Hum Genet. 2000;67:549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aloria K, Schilke B, Andrew A, Craig EA. Iron-induced oligomerization of yeast frataxin homologue Yfh1 is dispensable in vivo. EMBO Rep. 2004;5:1096–1101. doi: 10.1038/sj.embor.7400272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foury F, Pastore A, Trincal M. Acidic residues of yeast frataxin have an essential role in Fe-S cluster assembly. EMBO Rep. 2007;8:194–199. doi: 10.1038/sj.embor.7400881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR, Stemmler TL. Monomeric yeast frataxin is an iron-binding protein. Biochemistry. 2006;45:7767–7777. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondapalli KC, Kok NM, Dancis A, Stemmler TL. Drosophila frataxin: an iron chaperone during cellular Fe-S cluster bioassembly. Biochemistry. 2008;47:6917–6927. doi: 10.1021/bi800366d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Alam SL, Proteasa SV, Zhang Y, Lesuisse E, Dancis A, Stemmler TL. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43:16254–16262. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gakh O, Park S, Liu G, Macomber L, Imlay JA, Ferreira GC, Isaya G. Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Hum Mol Genet. 2006;15:467–479. doi: 10.1093/hmg/ddi461. [DOI] [PubMed] [Google Scholar]
- 25.Cho SJ, Lee MG, Yang JK, Lee JY, Song HK, Suh SW. Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proc Natl Acad Sci U S A. 2000;97:8932–8937. doi: 10.1073/pnas.160270897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhe-Paganon S, Shigeta R, Chi YI, Ristow M, Shoelson SE. Crystal structure of human frataxin. J Biol Chem. 2000;275:30753–30756. doi: 10.1074/jbc.C000407200. [DOI] [PubMed] [Google Scholar]
- 27.Musco G, de Tommasi T, Stier G, Kolmerer B, Bottomley M, Adinolfi S, Muskett FW, Gibson TJ, Frenkiel TA, Pastore A. Assignment of the 1H, 15N, and 13C resonances of the C-terminal domain of frataxin, the protein responsible for Friedreich ataxia. J Biomol NMR. 1999;15:87–88. doi: 10.1023/a:1008398832619. [DOI] [PubMed] [Google Scholar]
- 28.Nair M, Adinolfi S, Pastore C, Kelly G, Temussi P, Pastore A. Solution structure of the bacterial frataxin ortholog, CyaY: mapping the iron binding sites. Structure (Camb) 2004;12:2037–2048. doi: 10.1016/j.str.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Musco G, Stier G, Kolmerer B, Adinolfi S, Martin S, Frenkiel T, Gibson T, Pastore A. Towards a structural understanding of Friedreich’s ataxia: the solution structure of frataxin. Structure Fold Des. 2000;8:695–707. doi: 10.1016/s0969-2126(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang T, Craig EA. Binding of yeast frataxin to the scaffold for FeS cluster biogenesis, Isu. J Biol Chem. 2008;283:12674–12679. doi: 10.1074/jbc.M800399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Kogan M, Knight SA, Pain D, Dancis A. Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem. 1999;274:33025–33034. doi: 10.1074/jbc.274.46.33025. [DOI] [PubMed] [Google Scholar]
- 33.Murakami H, Pain D, Blobel G. 70-kD heat shock-related protein is one of at least two distinct cytosolic factors stimulating protein import into mitochondria. J Cell Biol. 1988;107:2051–2057. doi: 10.1083/jcb.107.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pace CN, Hebert EJ, Shaw KL, Schell D, Both V, Krajcikova D, Sevcik J, Wilson KS, Dauter Z, Hartley RW, Grimsley GR. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J Mol Biol. 1998;279:271–286. doi: 10.1006/jmbi.1998.1760. [DOI] [PubMed] [Google Scholar]
- 35.Correia AR, Pastore C, Adinolfi S, Pastore A, Gomes CM. Dynamics, stability and iron-binding activity of frataxin clinical mutants. Febs J. 2008;275:3680–3690. doi: 10.1111/j.1742-4658.2008.06512.x. [DOI] [PubMed] [Google Scholar]
- 36.Bencze KZ, Kondapalli KC, Cook JD, McMahon S, Millan-Pacheco C, Pastor N, Stemmler TL. The structure and function of frataxin. Crit Rev Biochem Mol Biol. 2006;41:269–291. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Correia AR, Adinolfi S, Pastore A, Gomes CM. Conformational stability of human frataxin and effect of Friedreich’s ataxia-related mutations on protein folding. Biochem J. 2006;398:605–611. doi: 10.1042/BJ20060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winzor DJ, Sawyer WH. Quantitative characterisation of ligand binding. Wiley-Liss; New York: 1995. [Google Scholar]
- 39.Karthikeyan G, Santos JH, Graziewicz MA, Copeland WC, Isaya G, Van Houten B, Resnick MA. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Hum Mol Genet. 2003;12:3331–3342. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- 40.Shan Y, Napoli E, Cortopassi G. Mitochondrial frataxin interacts with ISD11 of the Nfs1/ISCU complex and multiple mitochondrial chaperones. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm038. [DOI] [PubMed] [Google Scholar]