1. Introduction
The base excision repair (BER) pathway focuses mainly on base damage or loss and single-strand breaks (SSBs), and there are many complex variations within the overall BER process [1]. In our working model, an apurinic/apyrimidinic (AP) site formed by spontaneous base loss or enzymatic base removal is processed by AP endonuclease, yielding an intermediate with 3′-hydroxyl and 5′-deoxyribose phosphate (dRP) groups at the margins of a one-nucleotide gap. Alternatively, DNA glycosylase-mediated base removal is accompanied by AP lyase strand incision, yielding an intermediate with blocked 3′-hydroxyl and 5′-phosphate groups at the margins of a one-nucleotide gap. The 3′-hydroxyl block is removed by polynucleotide kinase or AP endonuclease, yielding the single-nucleotide (SN) gapped intermediate. BER then proceeds by two sub-pathways distinguished by the excision repair patch size, i.e., SN BER and long-patch (LP) BER. These sub-pathways appear to operate simultaneously in cells and cell extracts, unless a step is retarded or blocked, ultimately favoring one sub-pathway over the other. In SN BER, it is considered well established that DNA polymerase β (Pol β) fills the SN gap and removes the 5′-dRP group, creating a substrate for DNA ligase activity. In LP BER, where the repair patch size is two or more nucleotides long, there is a proliferating cell nuclear antigen (PCNA)-dependent branch, where replicative DNA polymerases and co-factors conduct “strand-displacement DNA synthesis,” producing the multi-nucleotide repair patch and a displaced flap that is removed by flap endonuclease 1 (FEN1), making way for DNA ligase activity [2–7]. In addition, LP BER also appears to occur by a PCNA-independent pathway involving gap-filling synthesis by Pol β [6–13]. However, this role for Pol β is not well documented in vivo, and little is known about the enzymology of the PCNA-independent branch of LP BER. There is a strong need for more experimental validation of the current working model for the PCNA-independent branch of LP BER. In the present study, we focused on use of a nucleotide excision repair (NER)-deficient cell background along with introduction of a strand break-containing BER intermediate with a bulky 5′-lesion where repair by SN BER is not possible.
In some lower eukaryotes, considered to be highly proficient in managing environmental UV exposure, a strand break 5′ to a UV-induced photoproduct is formed by the photoproduct-specific endonuclease [14] known as UV damage endonuclease (UVDE). This enzyme creates a nick 5′ to the cyclobutane pyrimidine dimer (CPD) or 6–4 photoproduct (6-4PP) leaving a 3′-hydroxyl group that is a substrate for excision repair strand displacement DNA synthesis [15] repair of the incised photoproduct-containing strand was shown to occur by the LP BER sub-pathway in Schizosaccharomyces pombe [16]. The repair involved removal of a 5′-CPD-containing flap by Rad27/FEN1 [17]. Interestingly, human FEN1 possesses a similar activity for removal of a 5′-CPD-containing flap [17]. Since human xeroderma pigmentosum complementation group A (XPA) cells lack NER, the CPDs and 6-4PPs formed by UV exposure in these cells accumulate. Yet, these lesions are removed in these human cells when they express UVDE, indicating that the cells have the constitutive capacity to repair such UV photoproducts once a nick adjacent to the photoproduct is formed. In the present study, we selected this NER-deficient system as an experimental approach to further investigate LP BER.
Briefly, an XPA cell line was transformed with a Neurospora crassa UVDE expression vector. The resulting cell line, stably expressing UVDE and termed “XPA-UVDE,” was shown to have CPD repair capacity after UV irradiation to the nucleus [18,19]. In vivo localization studies in the XPA-UVDE cell system using green fluorescent protein (GFP) fusions revealed that a known BER protein, i.e., x-ray repair cross-complementing group 1 (XRCC1), was recruited to sites of UV-induced DNA damage, consistent with involvement of LP BER in the repair of the UV-induced lesions [19]. Aphidicolin-sensitivity, a characteristic of replicative DNA polymerases, was observed for ~60% of the repair synthesis in the CPD LP BER process [18]. Yet, a significant level of aphidicolin-resistant DNA polymerase activity was also observed. Even though aphidicolin-resistance is characteristic of Pol β, the DNA polymerase responsible for this residual repair synthesis activity has not been investigated.
Here, using the XPA-UVDE cell system, we first examined in vivo repair of CPDs by immunostaining during a short repair period after UV irradiation. The results confirmed that CPD repair was strongly accelerated by expression of UVDE in these NER-deficient human cells. Therefore, these cells contained a constitutive repair system capable of repairing the UVDE-induced strand break adjacent to the UV photoproduct. Small interfering RNA (siRNA) knockdown in this system was used to demonstrate a Pol β role in the repair. In vivo localization studies demonstrated recruitment of GFP-fused Pol β and FEN1 to the sites of localized nuclear UV-induced damage. The involvement of Pol β in the LP BER of CPD-containing substrates in cell extracts was also demonstrated, and roles for purified Pol β and FEN1 in LP BER of CPD-containing substrates were examined. We found that the CPD-blocked 5′-end at the single-strand nick is a good substrate for Pol β strand-displacement DNA synthesis and also for FEN1 UV damaged strand excision.
2. Materials and Methods
2.1 Materials
[γ-32P]ATP (7,000 Ci/mmol) was from MP Biomedicals (Irvine, CA). [α-32P]dTTP, [α-32P]ddATP (3,000 Ci/mmol) and MicroSpin G-25 columns were from GE Healthcare (Piscataway, NJ). OptiKinase and terminal deoxynucleotidyl transferase were from USB Corporation (Irvine, CA) and Fermentas Inc. (Hanover, MD), respectively. AmpliTaq Gold DNA polymerase was from Perkin Elmer (Waltham, MA). S. pombe UVDE was from Trevigen, Inc. (Gaithersburg, MD). Protease inhibitor cocktail and Fugene 6 were from Roche Diagnostic Corp. (Indianapolis, IN). Glutamax, NuPAGE 4–12% Bis-Tris gels, NuPAGE LDS Sample Buffer, Alexa Fluor 594F(ab′)2 fragment of goat anti-mouse IgG (H+L) and ProLong Gold antifade reagent with DAPI were from Invitrogen (Carlsbad, CA). Triton X-100 was from Sigma-Aldrich Co. (St. Louis, MO). The anti-CPD monoclonal antibody, TDM-2, was from Cosmo Bio Co., Ltd. (Tokyo, Japan). Human Pol β and human FEN1 were purified as described [12]. The primary antibody, anti-mouse Pol β monoclonal antibody (18S), was prepared as described [20]. The anti-N. crassa UVDE polyclonal antibody was raised previously [21]. The anti-XRCC1 monoclonal antibody was a generous gift from Dr. Michael P. Thelen (Lawrence Livermore National Laboratory, Livermore, CA). The anti-FEN1, anti-LIG III and anti-LIG I monoclonal antibodies were from Genetex, Inc. (San Antonio, TX); the anti-human PARP-1 monoclonal antibody was from BD Biosciences (San Jose, CA); and the anti-PCNA monoclonal antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) antibody was from Alpha Diagnostics International Inc. (San Antonio, TX). The secondary antibodies, goat anti-mouse IgG (H+L)-horseradish peroxidase (HRP) conjugate and goat anti-rabbit IgG (H+L)-HRP conjugate, were from Bio-Rad Laboratories (Hercules, CA). SuperSignal West Pico Chemiluminescent substrate and Restore Western Blot Stripping Buffer were from Pierce Biotechnology Inc. (Rockford, IL). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), Dulbecco’s phosphate buffered saline (PBS) and Hanks’ balanced salt solution (HBSS) were from HyClone (Logan, UT). Polycarbonate isopore membrane filters with pores of 3 μm in diameter were from Millipore (Billerica, MA). Paraformaldehyde was from Electron Microscopy Sciences (Hatfield, PA). The pEGFP-C1 and DsRed-C1 (DR-C1) vectors were from Clontech Laboratories, Inc. (Mountain View, CA). 35-mm glass-bottom dishes coated by poly-L-lysine were from Matsunami Glass Ind., Ltd. (Osaka, Japan).
2.2 Cell culture
The host cells of the human XPA-UVDE cell lines were derived from an XPA patient and the stable transfectants expressing UVDE were obtained as described previously [18]. Mouse XPA (−/−) primary fibroblasts were isolated from XPA knock-out mouse embryos, and XPA (−/−)/Pol β (−/−) primary fibroblasts were obtained from embryos produced by mating male and female XPA (−/−)/Pol β (+/− mice, developed by breeding heterozygous Pol β mice with the XPA knock-out mice [22,23]. Embryos were isolated and genotyped at embryonic day 14. Fibroblasts were cultured from each embryo separately following removal of the internal organs and the head. The remaining tissue from each embryo was incubated at 37°C in 5 ml 0.025% trypsin with vigorous shaking for 30–45 min. Isolated cells were suspended in 45 ml of DMEM supplemented with glutamax and 10% FBS, and a single-cell suspension was prepared by filtration through a 70-μm mesh filter. Cells were pelleted (3,000 rpm, 5 min), the medium discarded and the cells were again suspended in 10 ml growth medium and cultured in a 100-mm dish until confluent. Once confluent, the cells were isolated by trypsinization, suspended in 10 ml medium and seeded into 10 separate dishes, 1 ml per dish. At confluency, cells were isolated by trypsinization and either frozen immediately in liquid N2 (passage P1), or immortalized by expression of SV40 large T-Antigen. For immortalization, P1 cells were seeded into a 60-mm dish (5 × 105 cells/dish). After 24 h, they were transfected with 2 μg pSVTAg (SV40 large T-Antigen expression vector) using lipofectamine (10 μl), and cells were allowed to grow to confluence. The cells then were trypsinized and seeded into a 100-mm dish, grown to confluence and split 1 to 20. Once confluent, cells were again split 1 to 20 and this was repeated 5 times. Surviving cells were considered to be transformed and were maintained by passage 2–3 times per week. Cell genotypes were confirmed by Southern blotting and PCR.
2.3 Lentivirus preparation and cell transduction
Lentiviral particles for expression of GFP or co-expression of Pol β shRNA and GFP were prepared by transfection of plasmids [the control plasmid pLK0.1-Puro-tGFP or the human Pol β-specific shRNA plasmid pSIH1-H1-copGFP-h.pol β, plus pMD2.g(VSVG), pRSV-REV or pMDLg/pRRE] into 293-FT cells [24,25] using Fugene 6 transfection reagent. Culture medium from transfected cells was collected 48 h after transfection to isolate the viral particles, passed through 0.45-μm filters, used immediately or stored at −80°C in single-use aliquots. Transduction of XPA-vector or XPA-UVDE cells with control lentivirus (GFP expression only) and human Pol β-specific shRNA lentivirus was completed as follows: Briefly, 6 × 104 cells were seeded into 6-well plates and incubated for 24–30 h at 10% CO2 at 37°C. Cells were transduced for 18 h with virus at 32°C and cultured for 72 h at 37°C before isolation of the GFP-expressing population by fluorescence-activated cell sorting using the UPCI Flow Cytometry Facility. Single-cell clones were isolated by culturing GFP-expressing cells in 96-well plates at limiting dilution (0.3 cells/well). Cells were then cultured to expand each cell population and analyzed for expression of Pol β by immunoblot.
2.4 In vivo analysis of CPD repair after UV irradiation
The intracellular visualization technique for CPD was established previously [26]. Human XPA-vector and XPA-UVDE cells were grown in DMEM supplemented with glutamax and 10% FBS at 37°C, in a 10% CO2 incubator. Cells were plated at 1 × 105 cells/well in 6-well dishes and cultured overnight. Cells were then washed once with PBS, covered with a 3-μm membrane filter and irradiated with 30 J/m2 254 nm UVC light. The membrane filter was gently removed, and the cellular repair reaction was conducted at 37°C in complete DMEM. At the appropriate time, the cells were washed once with PBS and fixed in 4% paraformaldehyde for 10 min at room temperature. After washing twice with PBS, the cells were permeabilized in 0.5% Triton X-100 for 5 min on ice and cellular DNA was denatured by adding 2 M HCl for 30 min at room temperature. Cells were then washed 5 times with PBS and blocked with 20% FBS in PBS. CPD antibody, TDM-2, was then incubated with the cells at 1:1,500 dilution at 37°C for 30 min, and the visualization was achieved following incubation with Alexa Fluor 594F(ab′)2 fragment of goat anti-mouse IgG (H+L) at 1:100 dilution at 37°C for 30 min. The cells were mounted in ProLong Gold antifade reagent and cover-slipped. Images were obtained using an AxioVS50 Version 4.6.3.0 with an Olympus IX70 microscope and quantification analysis was done using MetaMorph Version 6.3r6 (Molecular Devices, Union City, CA). For each experimental time point, an image was obtained at 3 independent locations, and a randomly-selected region containing an average of 37 cells containing 27–323 spots was subjected to quantification. The background intensity was subtracted from each focus and the average focus intensity was calculated. Relative intensity was calculated from the average values of each region at 0 h after UV irradiation.
2.5 In vivo analysis of GFP- and DR-fused proteins after UV irradiation
Pol β, FEN1, and XRCC1 open reading frames were obtained following amplification of HeLa cDNA by PCR, and amplified fragments with additional SalI (Pol β and XRCC1) or XhoI (FEN1) sites at 5′ and NotI sites at 3′ were cloned into pEGFP-C1 vector (Pol β and FEN1) and DR-C1 vector (XRCC1), respectively. These plasmids with the GFP and DR tags at the N-terminus of the respective protein were introduced into XPA-vector and XPA-UVDE cells with Fugene 6 according to the manufacturer’s protocol. Cells were propagated in DMEM supplemented with 10% FBS at 37°C, 5% CO2. Cells were plated at 1 × 105 cells per 35-mm glass-bottom dish and cultured for two days. Cells were then washed twice with HBSS, covered with a polycarbonate isopore membrane filter with pores of 3-μm diameter, and irradiated with 100 J/m2 UVC light at 254 nm. Fluorescence images after irradiation were obtained and processed using a FV-500 confocal scanning laser microscopy system for live image (Olympus Corporation, Tokyo, Japan). The mean intensity of accumulation at a UV-irradiated site was obtained after subtraction of the background intensity in the irradiated cell (Fig. 4).
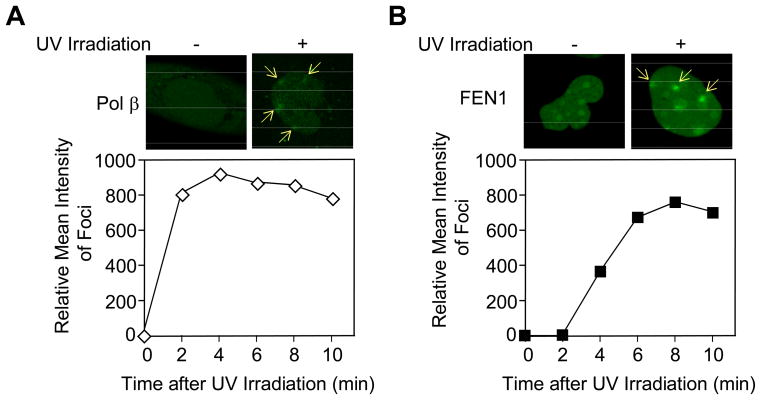
Fig. 4.
Recruitment of Pol β and FEN1 in human XPA-UVDE cells before and after local UV irradiation at 254 nm. Accumulation of (A) GFP-fused Pol β and (B) GFP-fused FEN1 was measured before (left) and 5 min after local UV irradiation with 100 J/m2 (right). The sites of UV irradiation are indicated by arrows. Quantification of the time course of accumulation of Pol β (open diamonds) and FEN1 (solid squares) is shown below each image.
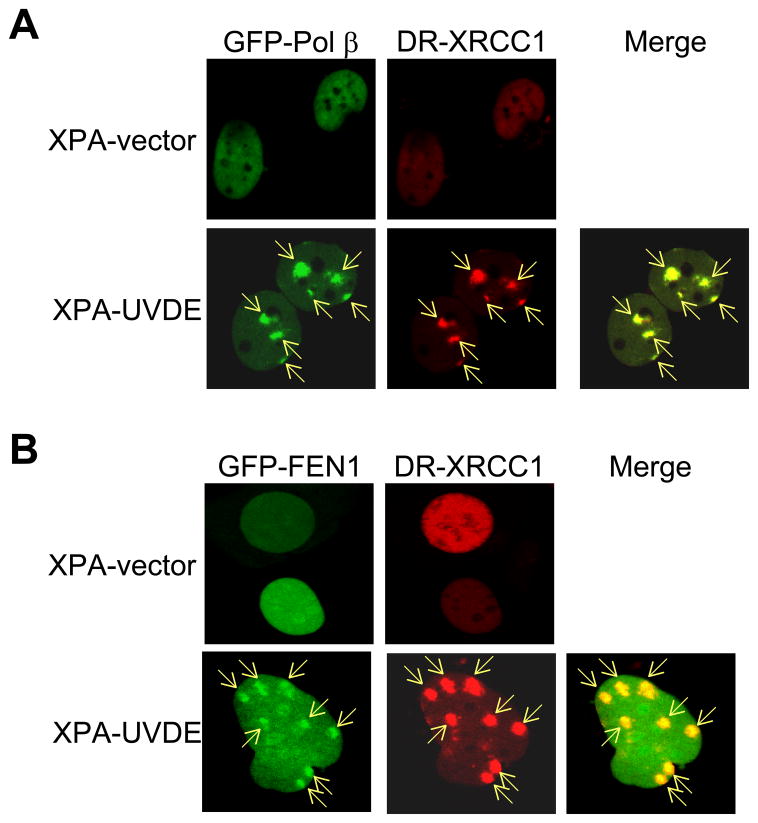
For the design of experiments on co-localization of Pol β and FEN1 (see Fig. 3), we used XRCC1 accumulation as a positive control for BER protein recruitment to the localized site with a single-strand break [19]. We then superimposed recruitment of GFP-tagged Pol β and FEN1, respectively, with DR-tagged XRCC1. In preliminary experiments to validate this approach, we first found that XRCC1 accumulated at virtually all of localized irradiation sites (> 99%). Next, we found that virtually all of the sites (>99%) of XRCC1 accumulation were positive for co-localization with Pol β and also for co-localization with FEN1.
Fig. 3.
Recruitment of Pol β, FEN1, and XRCC1 in human XPA-vector and XPA-UVDE cells after 254 nm local UV exposure using a polycarbonate isopore membrane filter as described in Section 2. (A) GFP-fused Pol β and DR-fused XRCC1 and (B) GFP-fused FEN1 and DR-fused XRCC1 were measured 5 min after localized UV irradiation with 100 J/m2 in XPA-vector (top) and XPA-UVDE (bottom) cells. The sites of localized UV irradiation are indicated by arrows. All sites of XRCC1 accumulation were positive for Pol β accumulation and FEN1 accumulation.
2.6 Cell extract preparation
Whole cell extracts (WCEs) were prepared as described [2]. Briefly, cells were washed twice with cold PBS, detached by scraping, collected by centrifugation, and resuspended in Buffer I (10 mM Tris-HCl, pH 7.8, 200 mM KCl and protease inhibitor cocktail). An equal volume of Buffer II (10 mM Tris-HCl, pH 7.8, 200 mM KCl, 2 mM EDTA, 40% glycerol, 0.2% Nonidet P-40, 2 mM dithiothreitol [DTT] and protease inhibitor cocktail) was added. The suspension was rotated for 1 h at 4°C, and the resulting extracts were clarified by centrifugation at 14,000 rpm at 4°C and frozen at −80°C until use in the in vitro BER assay. The protein concentration of the WCE was determined by Bio-Rad protein assay kit using bovine serum albumin (BSA) as standard.
2.7 Immunoblotting
Immunoblotting was performed essentially as described [27]. Briefly, WCE (20 μg) was separated by NuPAGE 4–12% Bis-Tris gel and transferred onto a nitrocellulose membrane. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBST) and then probed with monoclonal antibody to Pol β (18S), FEN1, XRCC1, PARP-1, LIG I, LIG III, PCNA, or G3PDH, or polyclonal antibody to UVDE, as indicated. After a 2-h incubation with primary antibody, the membranes were washed three times with TBST and incubated for 1 h with secondary antibody, either goat anti-mouse IgG (H+L)-HRP conjugate for anti-Pol β, anti-FEN1, anti-XRCC1, anti-PARP-1, anti-LIG I, anti-LIG III, anti-PCNA, or anti-G3PDH, or goat anti-rabbit IgG (H+L)-HRP conjugate for anti-UVDE. The HRP activity was detected by enhanced chemiluminescence system using SuperSignal West Pico Chemiluminescent substrate. The membranes were stripped with Restore Western Blot Stripping Buffer by incubating for 30 min at room temperature. After washing three times with TBST and blocking with 5% nonfat dry milk in TBST, the membranes were probed with another antibody.
2.8 PCR genotyping
Genomic DNA was isolated from the cells and amplified with AmpliTaq Gold DNA polymerase. The primers are listed in Table 1, where PolB5W and PolB3W were used for Pol β wild type allele, PolB5N and PolB3N were for Pol β null allele, XPA5 and XPA3W were for XPA wild type allele, and XPA5 and XPA3N were for XPA null allele. The PCR conditions for Pol β wild type allele were 95°C for 10 min, 35 cycles at 95°C for 0.5 min, 68°C for 1 min, 72°C for 1 min and 72°C for 10 min. The PCR conditions for Pol β null allele were 95°C for 10 min, 34 cycles at 95°C for 1.0 min, 68°C for 1 min, 72°C for 2 min and 72°C for 10 min. For XPA wild type allele, the PCR conditions were 95°C for 10 min, 34 cycles at 95°C for 1 min, 60°C for 1 min, 72°C for 1 min, and 72°C for 10 min. The PCR conditions for XPA null allele were 95°C for 10 min, 35 cycles at 95°C for 1 min, 60°C for 1 min, 72°C for 2 min, and 72°C for 10 min. The resulting PCR products were analyzed by 1.5% agarose gel electrophoresis, and were expected to be 0.49 kb for Pol β wild type allele, 0.42 kb for Pol β null allele, 214 bp for XPA wild type allele, and 132 bp for XPA null allele.
2.9 DNA substrates
The sequences of the synthetic ODNs used are shown in Table 1. The ODNs containing a CPD, 42D and 28D, were synthesized as described [28]. The ODN, 28D, was synthesized with a 5′-phosphate. Other ODNs were from Operon Biotechnologies Inc. (Huntsville, AL). The DNA substrate for the cell extract-based in vitro BER assay was a 42-mer CPD-containing duplex DNA and was prepared by annealing with the complementary strand. The DNA substrate thus contained a CPD at positions 15 and 16. Other DNA substrates for Pol β and FEN1 analyses were constructed by annealing various combinations of upstream and downstream ODNs with the complementary strand, 42COM, as shown in Table 1.
2.10 In vitro cell extract-based nicking analysis
The CPD-containing ODN, 42D, was 5′-end labeled by incubation with OptiKinase in the presence of [γ-32P]ATP for 30 min at 37°C. The labeled ODN was then annealed with the complementary ODN, 42COM, and the DNA was purified with a MicroSpin G-25 column. The 32P-labeled duplex 42D/42COM (50 nM) was incubated with 10 μg of WCE at 37°C for the indicated time period in a reaction mixture (10 μl) that contained 20 mM Tris-HCl, pH 7.0, 5 mM MgCl2, 100 mM NaCl, 5 mM DTT, 100 μg/ml BSA, and 10% glycerol. To prevent the intrinsic exonuclease activity of the cell extract, 10 mM dGMP was added to the reaction mixture. After the incubation, the reaction was terminated by addition of an equal volume (10 μl) of DNA gel-loading buffer, and the products were separated by 15% denaturing polyacrylamide gel electrophoresis (PAGE). The ODNs, 42D and 14US, were 5′-end labeled and used as a size marker for 42-mer and 14-mer, respectively. A Typhoon PhophorImager was used for gel scanning and imaging.
2.11 In vitro cell extract-based LP BER analysis
The ODN, 14US, was annealed with CPD-containing downstream oligonucleoitde, 28D, and the complementary strand, 42COM, to create nicked CPD-containing duplex ODN. The in vitro CPD LP BER assay was reconstituted in a reaction mixture (10 μl) that contained nicked CPD ODN (20 nM), 10 μg of WCE, 20 mM Tris-HCl, pH 7.0, 5 mM MgCl2, 20 mM NaCl, 1 mM DTT, 20 μg/ml BSA, 2% glycerol, 4 mM ATP and 10 mM dGMP. In some cases, the reaction included 90 μg of pre-immune or Pol β antibodies. The mixture was incubated at 37°C for the indicated time period in the presence of 20 μM each of dATP, dGTP, dCTP, and 2.3 μM [α-32P]dTTP. After the incubation, the reaction was terminated, and the products were analyzed as described above.
2.12 In vitro Pol β-mediated DNA synthesis
The upstream ODN strand was 5′-end labeled as described above. Several combinations of upstream and downstream strands were annealed with the complementary strand, 42COM. Briefly, 14US and 26GAP were used as upstream and downstream strands, respectively, to construct the 2-nucleotide gapped substrate; 14US and 28D were used to create the nicked CPD substrate; 16US and 28D for a 2-nucleotide- nicked CPD-flap substrate; and 19US and 28D to produce a 5-nucleotide nicked CPD-flap substrate. After purification, the resultant duplex oligomers (100 nM) were incubated at 37°C for the indicated time periods with 0.1 nM human Pol β in 20 mM Tris-HCl (pH 7.0), 5 mM MgCl2, 20 mM NaCl, 1 mM DTT, 20 μg/ml BSA and 2% glycerol in the presence of 20 μM each of dATP, dGTP, dCTP and dTTP. The reaction products were analyzed as described above. Kinetic parameters for Pol β were determined by quantification of reaction products using ImageQuant TL™ software (GE Healthcare).
2.13 In vitro FEN1 cleavage
The CPD-containing ODN, 28D, was 3′-end labeled by incubation with terminal deoxynucleotidyl transferase in the presence of [α-32P]ddATP for 30 min at 37°C. The labeled 28D was then annealed with the upstream ODN, 14US, and the complementary ODN, 42COM, to create nicked CPD-containing DNA substrate. After purification, the in vitro FEN1 reaction was conducted in a reaction mixture (10 μl) that contained nicked CPD-containing ODN (10 nM), 20 or 100 nM of human FEN1, 20 mM Tris-HCl (pH 7.0), 5 mM MgCl2, 20 mM NaCl, 1 mM DTT, 20 μg/ml BSA, 2% glycerol, and 4 mM ATP. The mixture was incubated at 37°C for the indicated time period in the presence of 20 μM each of dATP, dGTP, dCTP, and dTTP. The reaction products were analyzed as described above.
2.14 In vitro Pol β-mediated DNA synthesis and FEN1 cleavage analysis
An ODN duplex substrate labeled on both ends was employed. 5′-End labeled 14US and 3′-end labeled 28D were annealed with 42COM. The duplex substrate (20 nM) was incubated for 30 min at 37°C with human Pol β (1 nM) and human FEN1 (20 nM or 100 nM) in 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 20 mM NaCl and 0.5 mM DTT in the presence of 20 μM each of dATP, dGTP, dCTP and dTTP. The reaction products were analyzed as described above.
3. Results and Discussion
3.1 CPD repair after UV exposure in human XPA-UVDE cells
For in vivo repair analysis, we used the XPA-UVDE cell system developed earlier. This system enables expression of GFP-fusion proteins and localized UV exposure to the nucleus [19], and UVDE introduces an SSB immediately 5′ to the CPD and 6-4PP [14,15,29,30]. First, we verified the XPA-UVDE cell system by measuring repair of the CPD lesion using an anti-CPD monoclonal antibody [26]. For comparison, the XPA-vector cell line transfected with empty vector was used instead of the UVDE-expression vector. Immediately following UV irradiation (30 J/m2 at 254 nm), both cell types exhibited strong CPD signals (Fig. 1). After a 2-h repair period in growth medium, the CPD signal had almost disappeared in the XPA-UVDE cells, whereas the signal persisted in the XPA-vector cells (Fig. 1). These results indicated that SSBs introduced by UVDE triggered CPD repair by a constitutive repair process in these cells, presumably LP BER.
Fig. 1.
(A) In vivo repair of SSB-bearing CPDs in human XPA cells. The distribution of CPDs was detected by anti-CPD monoclonal antibody as described in Section 2. Disappearance of CPD signal was measured in human XPA-vector and XPA-UVDE cells for the indicated repair incubation times (0–2 h) after 254 nm UV exposure of 30 J/m2. Nuclei were visualized after staining with DAPI. The DAPI and CPD staining was superimposed in the panels marked “overlay.”
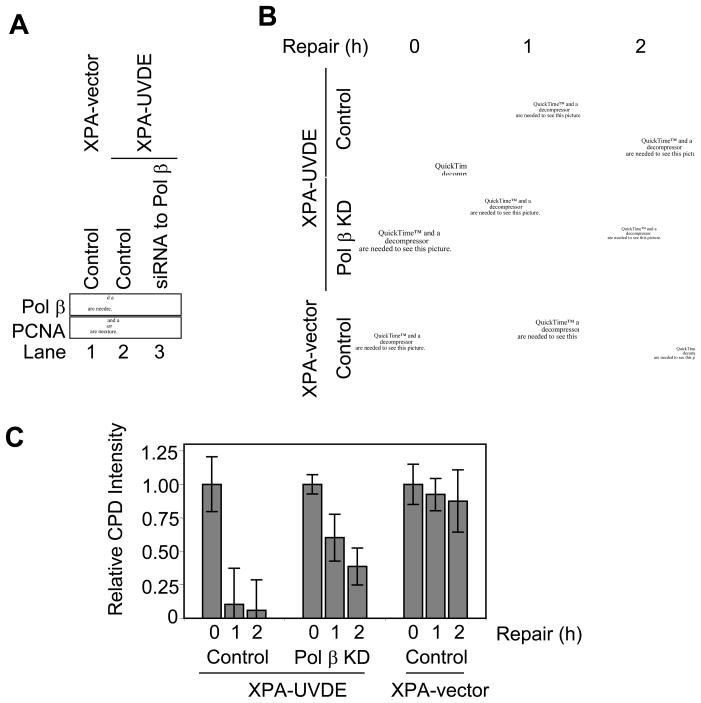
In light of the suggested role of Pol β in LP BER [6–13] and the chemical nature of the incised repair intermediate, i.e., 3′-hydroxyl group at one margin of a gap and a 5′-phosphate at the other margin [31,32], we considered Pol β as a good candidate to be a DNA polymerase involved in the repair patch synthesis step. This role also appeared feasible because an earlier study of repair synthesis in this system indicated a significant amount of aphidicolin-resistant synthesis activity, as would be the case for Pol β mediated repair [18]. To explore the possibility of a Pol β role in the CPD repair, we conducted siRNA knockdown experiments in the XPA-UVDE cell system that were directed to Pol β. A strong reduction in Pol β level was achieved, as can be seen from the results of immunoblotting analysis shown in Fig. 2A. The Pol β knockdown (Pol β KD) cells and the companion controls cells were subjected to CPD repair analysis as described above. Pol β knockdown resulted in a clear reduction in CPD repair as compared to the repair in the Pol β-expressing cells. However, a significant level of residual repair (~40%) was noted in the Pol β KD cells. These results are consistent with a role of Pol β in the repair of incised CPD lesions in the XPA-UVDE cells and also suggest that other polymerases can partially complement for a Pol β deficiency.
Fig. 2.
(A) Immunoblotting for Pol β in siRNA knockdown (KD) XPA-vector and XPA-UVDE cells. Cell extracts were prepared from human XPA-vector cells with control siRNA (lane 1) and human XPA-UVDE cells with control siRNA (lane 2) or siRNA to Pol β (lane 3). They were immunoblotted with antibody for Pol β and PCNA as a loading control. (B) In vivo repair of SSB-bearing CPDs in siRNA knockdown XPA-UVDE and XPA-vector cells. The experiment was conducted as described in Fig. 1. (C) Relative intensities of CPD signal during the indicated repair period after 254 nm local UV exposure of 30 J/m2 in control and Pol β KD XPA-UVDE cells and XPA-vector cells.
3.2 Recruitment of LP BER proteins at sites of UV irradiation in vivo
To obtain further insight into CPD repair in vivo, co-localization experiments at the sites of UV irradiation were conducted with GFP- and DR-fusion BER proteins. XRCC1 is known to be recruited to SSB intermediates during BER [19], and XRCC1 was used here as a positive control for concentration of a BER protein at sites of localized SSB formation. This recruitment was considered to be specific to XPA-UVDE cells because earlier experiments with XPA-vector control cells were negative for the recruitment of XRCC1 [19]. In our current experiments, recruitment of neither Pol β nor FEN1 was observed in the XPA-vector cells (Fig. 3). In experiments with XPA-UVDE cells, Pol β and XRCC1 co-localized at sites of localized UV irradiation (Fig. 3A); GFP-fused FEN1 also co-localized with DR-fused XRCC1 (Fig. 3B). In preliminary experiments with these cells, we found that virtually all of the sites of XRCC1 accumulation also were positive for Pol β and FEN1 accumulation.
To obtain further evidence for involvement of BER enzymes, we assessed the time course of recruitment of GFP-fusion proteins to the sites of UV irradiation. In the absence of UV exposure, GFP-fused Pol β was distributed throughout the nucleus in XPA-UVDE cells. After UV irradiation, Pol β was recruited to the sites of irradiation damage (Fig. 4A). GFP-fused FEN1 was distributed throughout the nucleus in XPA-UVDE cells, and after UV irradiation, recruitment of FEN1 to the sites of damage was observed (Fig. 4B). A time course of recruitment of GFP-fused Pol β and FEN1 to sites of UV irradiation in XPA-UVDE cells was obtained (Fig. 4). Near-maximal accumulation of Pol β was observed 2 min after UV irradiation, whereas initial recruitment of GFP-FEN1 was observed 4 min after irradiation, and recruitment reached a maximal level by 8 min. These rates of recruitment of the two BER enzymes appeared to be fast enough to accommodate the rate of repair observed in the experiments shown in Figs. 1 and 2. In summary, these in vivo localization experiments are consistent with a role for Pol β in repair of the incised CPD lesion in XPA-UVDE cells.
3.3 LP BER analysis in human XPA-UVDE cell extract
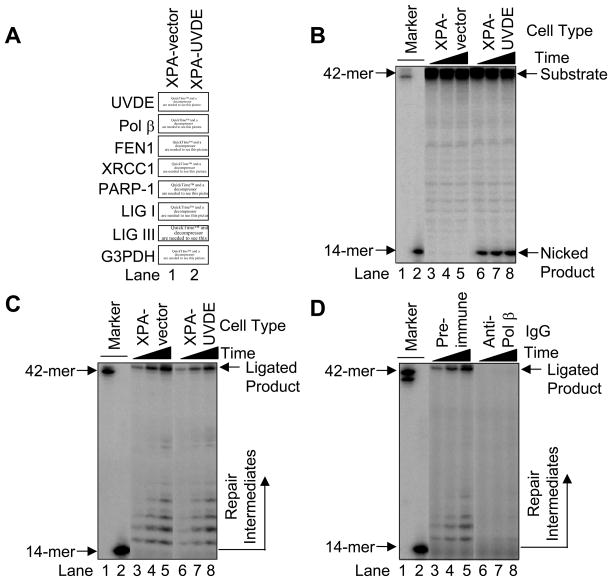
The results of the in vivo repair experiments and other results showing recruitment of LP BER enzymes to sites of CPD damage prompted us to establish an in vitro system for study of LP BER. First, the expression of UVDE in the XPA-UVDE cells and relevant potential LP BER proteins (i.e., Pol β, FEN1, XRCC1, PARP-1, LIG I, and LIG III) in the XPA-UVDE and XPA-vector cells (Fig. 5A) was confirmed by immunoblotting. Next, a 5′-end labeled duplex (42D/42COM) containing the CPD lesion was incubated with XPA-vector and XPA-UVDE cell extracts, respectively. The result showed that the XPA-vector cell extract did not provide any nicked product (Fig. 5B, lanes 3–5), whereas the XPA-UVDE cell extract produced a nicked product, indicating that the damage-containing strand was incised 5′ to the CPD lesion. However, we observed persistence of the intact substrate even with longer incubations (Fig. 5B, lanes 6–8).
Fig. 5.
(A) Immunoblotting for UVDE and known LP BER proteins. Cell extracts were prepared from human XPA-vector (lane 1) and XPA-UVDE (lane 2) cells and immunoblotted with antibody for N. crassa UVDE, Pol β, FEN1, XRCC1, PARP-1, LIG I, LIG III, and G3PDH as a loading control as described in Section 2. (B) Nicking activity of human XPA-vector and XPA-UVDE cell extract on CPD-containing ODN substrate. The in vitro cell extract-based nicking analysis was performed as described in Section 2. The incubation was conducted for 15 (lanes 3 and 6), 30 (lanes 4 and 7), and 60 min (lanes 5 and 8). The substrate (42-mer) and nicked product (14-mer) are shown. (C) Repair activity of human XPA-vector and XPA-UVDE cell extracts on the nicked CPD ODN substrate. The incubation was conducted for 5 (lanes 3 and 6), 15 (lanes 4 and 7), and 30 min (lanes 5 and 8). The position of repair intermediates and ligated product were shown. The mobility of the 42-mer marker was slightly faster than the ligated product due to the presence of 5′-phosphate. (D) Neutralization of Pol β in the in vitro cell extract-based LP BER. The in vitro repair reaction was conducted with pre-immune IgG (lanes 3–5) and anti-Pol β IgG (lanes 6–8) as described in Section 2. The incubation was conducted for 5 (lanes 3 and 6), 15 (lanes 4 and 7), and 30 min (lanes 5 and 8). The position of repair intermediates and ligated products are shown.
Next, we determined conditions necessary to observe in vitro CPD-initiated LP BER. To avoid possible complications from uncut substrate, we employed three ODNs to prepare the pre-nicked CPD-containing DNA substrate: the 5′ end-labeled upstream ODN (14US), CPD-containing downstream ODN (28D), and the complementary strand (42COM). This pre-nicked substrate was incubated with extract from XPA-vector and XPA-UVDE cells, respectively. Ligated BER products were observed with both XPA-vector and XPA-UVDE cell extracts (Fig. 5C), indicating that both cells had capacity for LP BER of the nicked CPD-containing substrate. A Pol β-neutralizing antibody was utilized to determine whether Pol β was responsible for the repair synthesis activity observed. The amount of ligated BER product formed was strongly reduced in the presence of the Pol β-neutralizing antibody, whereas the pre-immune IgG control had no effect (Fig. 5D, lanes 3–5 vs. 6–8). These results indicated a constitutive capacity in both cell types to repair the CPD-containing substrate by LP BER, and that Pol β was required for the repair.
3.4 LP BER analysis in mouse cell extracts
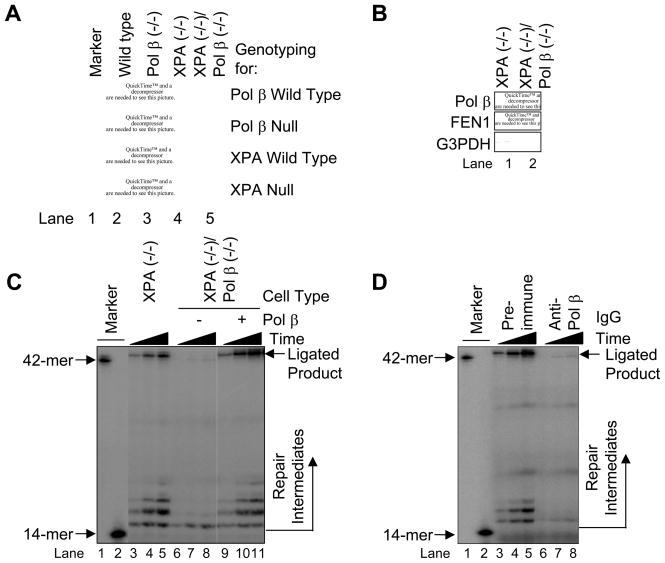
Since human XPA cells exhibited LP BER with the CPD-containing substrate, we decided to confirm and extend these results using a similar approach in mouse cell lines. With appropriate strain crosses, isogenic mouse cell lines were developed where the XPA gene was deleted and the Pol β gene was either deleted or wild type. The genotype for XPA and Pol β disruption was confirmed by PCR analysis (Fig. 6A), and the expression of Pol β and FEN1 was examined by immunoblotting using anti-Pol β and anti-FEN1 antibodies (Fig. 6B). As expected, the results revealed that Pol β expression was absent in the XPA (−/−)/Pol β (−/−) cells, whereas the FEN1 level was normal (Fig. 6B). Our attempts to create UVDE-expressing mouse cell lines were not successful.
Fig. 6.
(A) PCR genotyping for Pol β and XPA in mouse XPA/Pol β cell lines. PCR was conducted as described in Section 2 and the resulting product was analyzed by 1.5% agarose gel electrophoresis. (B) Immunoblotting of Pol β and FEN1 in mouse XPA null and XPA/Pol β double null cells. WCEs were prepared and immunoblotted as described in Section 2. The blots were probed with anti-G3PDH as a loading control. Repair activity of mouse XPA null (lanes 3–5) and XPA/Pol β double null (lanes 6–11) cell extracts on the nicked CPD ODN substrate. The in vitro cell extract-based LP BER assay was carried out as described in Section 2 using a nicked CPD substrate. The incubation was conducted for 5 (lanes 3, 6 and 9), 15 (lanes 4, 7 and 10), and 30 min (lanes 5, 8 and 11). Purified Pol β was added to the XPA/Pol β double null cell extract reaction mixture for complementation analysis (lanes 9–11). The positions of repair intermediate and ligated product are shown. The mobility of the 42-mer marker was slightly faster than the ligated product due to the presence of 5′-phosphate. (D) Repair activity of mouse XPA null cell extracts in the presence of pre-immune IgG (lanes 3–5) or anti-Pol β IgG (lanes 6–8) and incubated with [α-32P]dTTP. The incubation was conducted for 5 (lanes 3 and 6), 15 (lanes 4 and 7), and 30 min (lanes 5 and 8). The positions of repair intermediate and ligated product are shown.
Next, we examined involvement of Pol β in LP BER of the CPD-containing substrate using cell extracts. In the extract from Pol β-proficient cells, ligated BER products were observed (Fig. 6C, lanes 3–5). Such BER products were not observed when the repair reaction was performed with Pol β null cell extract (Fig. 6C, lanes 6–8). The addition of purified Pol β restored the BER reaction. This confirmed that the components of BER were able to function in the Pol β-deficient XPA cell extract (Fig. 6C, lanes 9–11). Similar results indicating a Pol β requirement were obtained by pre-incubation of the Pol β-proficient cell extract with the Pol β-neutralizing antibody (Fig. 6D, lanes 6–8). Pre-incubation of the extract with pre-immune IgG had no effect on the DNA repair reaction (Fig. 6D, lanes 3–5). Thus, LP BER in the mouse cell extracts exhibited a Pol β requirement.
3.5 Activity of purified Pol β on intermediates of LP BER
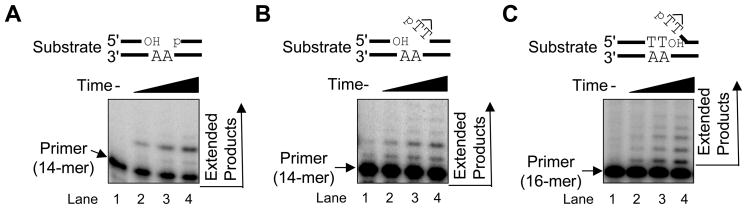
The activity of purified human Pol β was evaluated using substrates corresponding to intermediates produced during LP BER of a DNA molecule with an SSB adjacent to the CPD. These intermediates are illustrated in Fig. 7 and were as follows: nicked dimer representing the initial intermediate after UVDE incision (Fig. 7B); nicked dimer-flap representing the intermediate after 2-nucleotide gap-filling (Fig. 7C); and for comparison, a 2-nucleotide gapped substrate without the dimer-flap (Fig. 7A). With the 2-nucleotide gapped substrate, incorporation to fill the gap was observed (Fig. 7A), as expected from earlier work [33]. Further primer extension beyond 2-nucleotide gap filling was very weak, even after lengthening the incubation period. In the case of the nicked CPD substrate, a similar synthesis pattern was observed, but there was less synthesis and utilization of the primer substrate (Fig. 7B). Thus, the presence of the CPD appeared to perturb the weak strand displacement synthesis activity of Pol β. Finally, evaluation of synthesis with the nicked CPD-flap substrate revealed a different synthesis pattern; the pattern was consistent with strand displacement synthesis (Fig. 7C). These results confirmed that purified Pol β could utilize the nicked CPD-containing substrate for LP BER DNA synthesis.
Fig. 7.
In vitro Pol β synthesis with LP BER DNA intermediates. The Pol β synthesis reaction was carried out as described in Section 2 using (A) the 2-nucleotide gapped substrate, (B) the nicked CPD substrate, and (C) the nicked CPD-flap substrate for 0 (lane 1), 0.5 (lane 2), 1 (lane 3), and 1.5 (lane 4) min at 37°C. The positions of the primer band (14-mer for A and B; 16-mer for C) and extended product are shown.
3.6 Activity of purified FEN1 on intermediates of LP BER
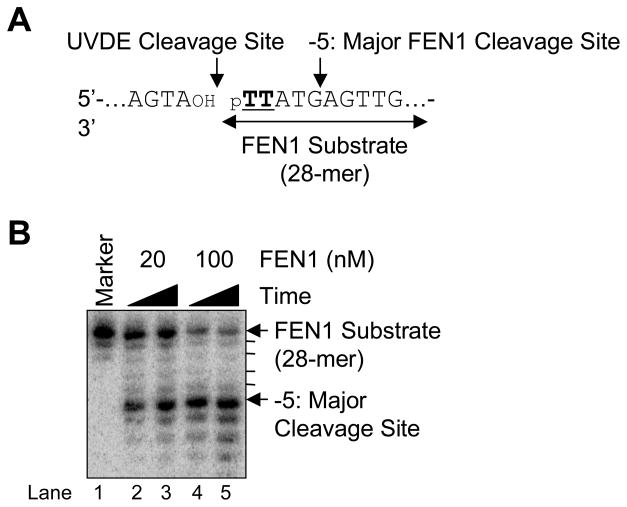
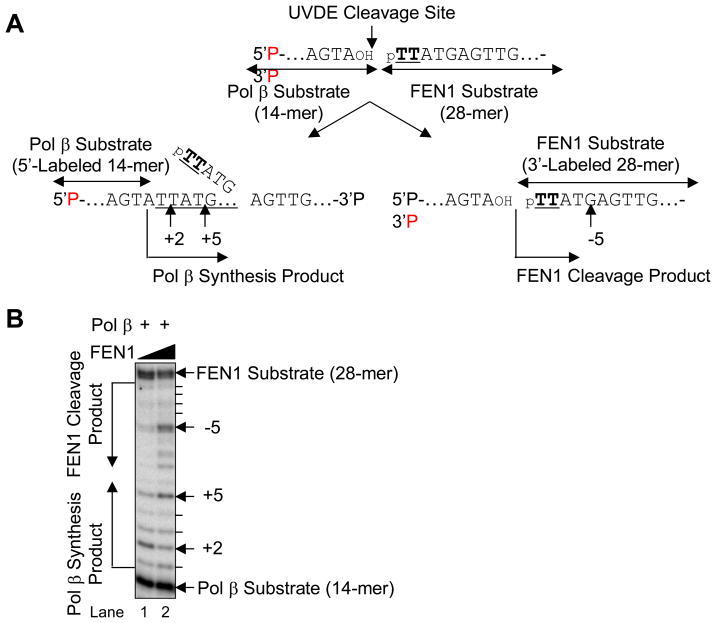
To examine the specificity of FEN1 during LP BER, the CPD-containing downstream strand was 3′-end labeled and annealed with upstream and the complementary strand (Fig. 8A). FEN1 specificity was examined by incubation with purified human FEN1. The results indicated that FEN1 cleaved the nicked CPD-containing LP BER intermediates, and the major FEN1 cleavage site was 5 bases downstream from the 5′ CPD lesion (Fig. 8B).
Fig. 8.
FEN1 cleavage on the nicked CPD ODN substrate. (A) Schematic diagram of the FEN1 cleavage reaction where TT indicates a CPD. The UVDE cleavage site and the major FEN1 cleavage site are indicated along with FEN1 substrate. (B) The in vitro FEN1 cleavage reaction was carried out as described in Section 2 using human FEN1 (20 nM and 100 nM) for 5 min (lanes 2 and 4) and 15 min (lanes 3 and 5). The positions of DNA substrate and product are shown.
3.7 Balance between Pol β and FEN1 activities in a reconstituted LP BER system
In the in vitro LP BER assays described so far, the enzymatic activities of Pol β and FEN1 were measured individually on the nicked CPD-containing LP BER intermediates. To examine the influence of FEN1 on Pol β gap-filling synthesis on the LP BER intermediates, both ends of the nicked CPD-containing strand were labeled to enable measurements of both enzymatic activities in the same reaction mixture (Fig. 9A). In this experiment, Pol β utilized the 5′-end labeled upstream strand (14-mer) and provided synthesis products longer than the 14-mer, whereas FEN1 utilized the 3′-end labeled damage-containing strand (28-mer) and provided digestion products shorter than the 28-mer (Fig. 9A). We compared results with a lower and higher level of FEN1, 20 and 100 nM, respectively (Fig. 9B). The expected results from the experiment were as follows: based on the primary site of FEN1 cleavage 5 nucleotides from the CPD, the repair synthesis product was predicted to be 5 nucleotides long if FEN1 acted before Pol β. On the other hand, since Pol β incorporates two nucleotides at this nick, the repair intermediate was predicted to be two nucleotides long if Pol β acted before FEN1. At the lower FEN1 concentration, a mixture of Pol β products was observed, with the 2-nucleotide product the most abundant (Fig. 9B, lane 1). With the higher concentration of FEN1, the Pol β synthesis pattern was different, such that the predominant synthesis product was 5 nucleotides long. In addition, the 5-nucleotides long FEN1 product also accumulated (Fig. 9B, lane 2). Thus, under these latter conditions (100 nM FEN1), the size of the Pol β synthesis product was determined by FEN1 cleavage.
Fig. 9.
Coordination of Pol β and FEN1 on the nicked CPD ODN substrate. (A) Schematic diagram of the reaction. The Pol β substrate was the 5′-end labeled 14-mer upstream strand and the product was expected to be observed beyond 14-mer (left side scheme). The FEN1 substrate was the 3′-end labeled 28-mer downstream strand containing CPD lesion (TT) at the 5′-end. The observed FEN1 cleavage product was expected to less than 28-mer (right side scheme). (B) Typical result of Pol β and FEN1 coordination reaction is shown. The in vitro Pol β synthesis and FEN1 cleavage reaction was simultaneously conducted using Pol β (1 nM) and FEN1 [20 nM (lane 1) and 100 nM (lane 2)]. The positions of Pol β synthesis product and FEN1 cleavage product are shown.
To further evaluate LP BER substrate requirements for Pol β and FEN1, a kinetic measurement of enzymatic activities was conducted using three different model BER intermediates. These were the “nicked CPD” reflecting the substrate immediately after nick introduction, the “2-nucleotide nicked CPD-flap” reflecting the substrate after 2 nucleotide incorporations by Pol β, and the “5-nucleotide nicked CPD-flap” reflecting the substrate after 5 nucleotide incorporations by Pol β (Table 2). Assays of the steady-state activity of Pol β showed that its repair synthesis activity was similar on the nicked CPD substrate and 2-nucleotide nicked CPD-flap substrate. FEN1 activity also was similar on these two substrates. However, FEN1 activity was higher (~20-fold) on the 5-nucleotide nicked CPD-flap substrate. The results suggest that FEN1 could limit the size of a repair patch by virtue of its strong activity on the CPD-containing 5-base flap.
4. Concluding Remarks
We proposed that the role of Pol β in mammalian LP BER could be examined in an experimental system where a strand incision 5′ to a CPD lesion forces repair by the constitutively-expressed LP BER sub-pathway. This approach, which involved NER-deficient XPA cells, was used earlier to examine recruitment of some BER co-factors to the sites of UV irradiation [19]. In addition to analysis of living cells, in vitro biochemical studies may help define mechanistic aspects of the repair via LP BER. In this system, involvement of aphidicolin-sensitive DNA polymerases was observed earlier, e.g., Pol δ and/or Pol ε [18]; yet, repair synthesis was only partially inhibited by aphidicolin, consistent with the idea that other DNA polymerase(s) also could be involved. With regard to in vivo LP BER analyses, a damage containing EGFP plasmid was utilized to measure the size of the LP BER excision patch in living cells [34]. However, this study did not address to DNA polymerase responsible for repair patch synthesis in LP BER. Here, the results of in vivo localization studies were consistent with a role of Pol β in the repair, and this is the first evidence for Pol β recruitment to sites of LP BER in vivo. Involvement of Pol β also was demonstrated in XPA-UVDE cell extract-mediated LP BER of the CPD-containing DNA substrate. The extract-based studies in mouse cell lines are consistent with Pol β involvement in LP BER (Fig. 6). These observations are in agreement with those of earlier in vitro biochemical studies suggesting a role for Pol β in LP BER DNA synthesis [6,10–13]. The observations here also are consistent with an earlier in vivo study in mouse cell lines that showed a protective effect of Pol β against alkylating agent treatment under conditions where SN BER was blocked [9].
For repair of a SSB adjacent to a CPD, S. pombe extract mediated repair by LP BER [16] and this involved removal of the CPD-containing flap by Rad27/FEN1 [17]. Human FEN1 possesses a similar capability for removal of a 5′ CPD-containing flap (Fig. 8). We demonstrated recruitment of FEN1 to sites of UV irradiation consistent with the proposed role of FEN1 in LP BER (Figs. 3 and 4). The enzymatic activity of FEN1 appears to be vital for processing the excision patch in LP BER, and FEN1 could influence the Pol β gap-filling step as shown in Fig. 9.
In summary, we used protein accumulation at site of localized UV damage in human XPA-UVDE cells to obtain evidence for a role of Pol β in LP BER. Results pointing to a similar conclusion regarding a role for Pol β in LP BER were obtained using siRNA knockdown in XPA-UVDE cells, to create a Pol β deficiency in the XPA null background. A role of Pol β in LP BER also was demonstrated in extract-based experiments using Pol β-neutralizing antibody. Kinetic analyses confirmed substrate specificities of purified Pol β and FEN1 for their proposed roles in processing LP BER intermediates.
Acknowledgments
We thank William A. Beard for critical reading of the manuscript and Bonnie E. Mesmer and Jennifer P. Myers for editorial assistance. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES050158 & Z01-ES050159). This work also was supported in part by a Research Scholar grant (RSG-05-246-01-GMC) from the American Cancer Society and grants from NIH (1 R01 AG24364-01; 1 P20 CA132385-01) to R.W.S., by grants from NIH (CA40463) to J.S.T., and by a genome network project grant from the Ministry of Education, Science, Sports and Culture of Japan to A.Y.
Footnotes
Conflict of interest statement
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, DC: 2006. [Google Scholar]
- Biade S, Sobol RW, Wilson SH, Matsumoto Y. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. Journal of Biological Chemistry. 1998;273:898–902. doi: 10.1074/jbc.273.2.898. [DOI] [PubMed] [Google Scholar]
- Blank A, Kim B, Loeb LA. DNA polymerase δ is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, USA. 1994;91:9047–9051. doi: 10.1073/pnas.91.19.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37:3575–3580. doi: 10.1021/bi972999h. [DOI] [PubMed] [Google Scholar]
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. Journal of Biological Chemistry. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO Journal. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlanti E, Pascucci B, Terrados G, Blanco L, Dogliotti E. Aphidicolin-resistant and -sensitive base excision repair in wild-type and DNA polymerase beta-defective mouse cells. DNA Repair (Amst) 2004;3:703–710. doi: 10.1016/j.dnarep.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA polymerase β in the excision step of long patch mammalian base excision repair. Journal of Biological Chemistry. 1999;274:13741–13743. doi: 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- Horton JK, Prasad R, Hou E, Wilson SH. Protection against methylation-induced cytotoxicity by DNA polymerase β-dependent long patch base excision repair. Journal of Biological Chemistry. 2000;275:2211–2218. doi: 10.1074/jbc.275.3.2211. [DOI] [PubMed] [Google Scholar]
- Liu Y, Beard WA, Shock DD, Prasad R, Hou EW, Wilson SH. DNA polymerase β and flap endonuclease 1 enzymatic specificities sustain DNA synthesis for long patch base excision repair. Journal of Biological Chemistry. 2005;280:3665–3674. doi: 10.1074/jbc.M412922200. [DOI] [PubMed] [Google Scholar]
- Podlutsky AJ, Dianova, Podust VN, Bohr VA, Dianov GL. Human DNA polymerase β initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO Journal. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. Journal of Biological Chemistry. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase β-mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. Journal of Biological Chemistry. 2001;276:32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- Takao M, Yonemasu R, Yamamoto K, Yasui A. Characterization of a UV endonuclease gene from the fission yeast Schizosaccharomyces pombe and its bacterial homolog. Nucleic Acids Research. 1996;24:1267–1271. doi: 10.1093/nar/24.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman KK, Sidik K, Smith CA, Taylor JS, Doetsch PW, Freyer GA. A new ATP-independent DNA endonuclease from Schizosaccharomyces pombe that recognizes cyclobutane pyrimidine dimers and 6-4 photoproducts. Nucleic Acids Research. 1994;22:3026–3032. doi: 10.1093/nar/22.15.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleva JL, Zuo S, Hurwitz J, Doetsch PW. In vitro reconstitution of the Schizosaccharomyces pombe alternative excision repair pathway. Biochemistry. 2000;39:2659–2666. doi: 10.1021/bi992751n. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Swiderski PM, Kaplan BE, Takao M, Yasui A, Shen B, Pfeifer GP. Processing of UV damage in vitro by FEN-1 proteins as part of an alternative DNA excision repair pathway. Biochemistry. 1999;38:4809–4817. doi: 10.1021/bi990105i. [DOI] [PubMed] [Google Scholar]
- Okano S, Kanno S, Nakajima S, Yasui A. Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. Journal of Biological Chemistry. 2000;275:32635–32641. doi: 10.1074/jbc.M004085200. [DOI] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Molecular and Cellular Biology. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal RK, Prasad R, Wilson SH. DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. Journal of Biological Chemistry. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- Kanno S, Iwai S, Takao M, Yasui A. Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Research. 1999;27:3096–3103. doi: 10.1093/nar/27.15.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A, van Steeg H. Xpa knockout mice. Seminars in Cancer Biology. 1996;7:229–240. doi: 10.1006/scbi.1996.0031. [DOI] [PubMed] [Google Scholar]
- Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- Poeschla EM, Wong-Staal F, Looney DJ. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Katsumi S, Kobayashi N, Imoto K, Nakagawa A, Yamashina Y, Muramatsu T, Shirai T, Miyagawa S, Sugiura S, Hanaoka F, Matsunaga T, Nikaido O, Mori T. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. Journal of Investigative Dermatology. 2001;117:1156–1161. doi: 10.1046/j.0022-202x.2001.01540.x. [DOI] [PubMed] [Google Scholar]
- Kedar PS, Kim SJ, Robertson A, Hou E, Prasad R, Horton JK, Wilson SH. Direct interaction between mammalian DNA polymerase β and proliferating cell nuclear antigen. Journal of Biological Chemistry. 2002;277:31115–31123. doi: 10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Brockie IR, O’Day CL. A building block for the sequence-specific introduction of cis-syn thymine dimers into oligonucleotides. Solid-phase synthesis of TpT[c,s]pTpT. Journal of the American Chemical Society. 1987;109:6735–6742. [Google Scholar]
- Freyer GA, Davey S, Ferrer JV, Martin AM, Beach D, Doetsch PW. An alternative eukaryotic DNA excision repair pathway. Molecular and Cellular Biology. 1995;15:4572–4577. doi: 10.1128/mcb.15.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima H, Takao M, Yasuhira S, Zhao JH, Ishii C, Inoue H, Yasui A. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO Journal. 1995;14:1267–1271. doi: 10.1002/j.1460-2075.1995.tb07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogliotti E, Fortini P, Pascucci B, Parlanti E. The mechanism of switching among multiple BER pathways. Progress in Nucleic Acid Research and Molecular Biology. 2001;68:3–27. doi: 10.1016/s0079-6603(01)68086-3. [DOI] [PubMed] [Google Scholar]
- Srivastava DK, Vande Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. Journal of Biological Chemistry. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- Prasad R, Beard WA, Wilson SH. Studies of gapped DNA substrate binding by mammalian DNA polymerase β. Dependence on 5′-phosphate group. Journal of Biological Chemistry. 1994;269:18096–18101. [PubMed] [Google Scholar]
- Sattler U, Frit P, Salles B, Calsou P. Long-patch DNA repair synthesis during base excision repair in mammalian cells. EMBO Rep. 2003;4:363–367. doi: 10.1038/sj.embor.embor796. [DOI] [PMC free article] [PubMed] [Google Scholar]