Abstract
The DNA mismatch repair (MMR) system is highly conserved and vital for preserving genomic integrity. Current mechanistic models for MMR are mainly derived from in vitro assays including reconstitution of strand specific MMR and DNA binding assays using short oligonucleotides. However, fundamental questions regarding the mechanism and regulation in the context of cellular DNA replication remain. Using synchronized populations of HeLa cells we demonstrated that hMSH2, hMLH1 and PCNA localize to the chromatin during S-phase, and accumulate to a greater extent in cells treated with a DNA alkylating agent. In addition, using small interfering RNA to deplete hMSH2, we demonstrated that hMLH1 localization to the chromatin is hMSH2-dependent. hMSH2/hMLH1/PCNA proteins, when associated with the chromatin, form a complex that is greatly enhanced by DNA damage. The DNA damage caused by high doses of alkylating agents leads to a G2 arrest after only one round of replication. In these G2-arrested cells, an hMSH2/hMLH1 complex persists on chromatin, however, PCNA is no longer in the complex. Cells treated with a lower dose of alkylating agent require two rounds of replication before cells arrest in G2. In the first S-phase, the MMR proteins form a complex with PCNA, however, during the second S-phase PCNA is missing from that complex. The distinction between these complexes may suggest separate functions for the MMR proteins in damage repair and signaling. Additionally, using confocal immunofluorescence, we observed a population of hMSH6 that localized to the nucleolus. This population is significantly reduced after DNA damage suggesting that the protein is shuttled out of the nucleolus in response to damage. In contrast, hMLH1 is excluded from the nucleolus at all times. Thus, the nucleolus may act to segregate a population of hMSH2-hMSH6 from hMLH1-hPMS2 such that, in the absence of DNA damage, an inappropriate response is not invoked.
Keywords: DNA mismatch repair, hMSH2-hMSH6, chromatin, nucleolus, DNA replication, cell cycle
1. Introduction
The DNA mismatch repair (MMR) system plays an essential role in maintaining the fidelity of DNA replication by correcting single nucleotide mismatches and insertion/deletion (ID)-loops [1]. This system is further specified by the presence of three MutsS homologs (hMSH2, hMSH3, and hMSH6) that form two distinct heterodimers with hMSH2 as the common subunit. In addition, four MutL homologs have been identified (hMLH1, hMLH3, hPMS1, and hPMS2) that assemble into three functional heterodimers where the hMLH1-hPMS2 heterodimer is required for DNA MMR [2]. Loss of MMR is associated with a mutator phenotype and the hereditary cancer syndrome Hereditary Nonpolyposis Colorectal Cancer (HNPCC) [3-6].
The correction of single nucleotide mismatches has been reconstituted in vitro using mammalian cell extracts or purified recombinant proteins and a circular heteroduplex substrate [7-9]. From these studies the minimal factors required to support both 5′- and 3′-nick directed repair were elucidated and include hMSH2-hMSH6, hMLH1-hPMS2, hEXOI, RPA, RFC, PCNA, DNA polymerase δ and DNA ligase I. However, the question of how these protein players interact and function to execute the repair process still remains.
One model for MMR that has emerged based on the reconstitution data and experiments illuminating the biochemistry of the hMSH2-hMSH6 and hMLH1-hPMS2 heterodimers is the “molecular switch” model [10-12]. This model proposes that the MMR pathway is initiated by the hMSH2-hMSH6 heterodimer, which recognizes and binds to a single nucleotide mismatch. Exchange of ADP for ATP alters the conformation of the heterodimer such that it can freely diffuse in either direction along the DNA in an ATP hydrolysis-independent manner as a “sliding clamp”. Multiple hMSH2-hMSH6 heterodimers load onto the DNA in a similar fashion [13], and have been proposed to be important for the recruitment of hMLH1-hPMS2 to the mismatch [14]. Formation of hMLH1-hPMS2/hMSH2-hMSH6 protein complexes is thought to be involved in the recruitment of hEXOI and subsequent strand-specific excision. Resynthesis is carried out by replication factor C (RFC), proliferating cellular nuclear antigen (PCNA), and DNA polymerase δ.
Interactions between the various components of the MMR pathway have been studied extensively using an array of in vitro techniques; however, the intricacies remain poorly understood. The current consensus is that formation of the hMLH1-hPMS2/hMSH2-hMSH6 ternary complex is ATP-dependent and enhanced in the presence of a heteroduplex substrate. However, both hMSH2-hMSH6 and hMLH1-hPMS2 heterodimers have some affinity for homoduplex substrates in vitro [15-18]. PCNA, the processivity factor for DNA polymerase has also been demonstrated to interact with MSH6, MSH3, and MLH1 via a conserved interaction motif called the PIP box [19-21]. Both MSH6 and MSH3 colocalize with PCNA at replication foci during S-phase [19]. The importance of this interaction is highlighted by the fact that a yeast Msh6 carrying mutations in the PIP box are unable to rescue defects conferred by an Msh6 deletion strain [21, 22]. PCNA-hMSH2, and PCNA-hEXOI interactions have also been demonstrated [15,21,23-25].
How these in vitro interaction data translate to the function of the MMR pathway in the cell during DNA replication remains an important question. Studies of the MMR pathway within the cell have focused primarily on its role in signaling a DNA damage response. MMR deficient cells have been shown to be much more resistant to the cytotoxic effects of several DNA damaging agents [26]. Most intensely studied are the DNA methylating compounds such as the SN1 type alkylating agent N-methyl-N′-nitro N-nitrosoguanidine (MNNG). MNNG generates a variety of DNA lesions including the cytotoxic O6-methyl-guanine (O6-meG) adduct. This lesion can be repaired by methylguanine methyltransferase (MGMT) in an irreversible one-step reaction whereby the methyl group is transferred from the donor guanine to MGMT [27]. However, if DNA replication occurs prior to repair, there is a propensity for O6-meG-thymine (O6-meG/T) mismatches to form which are recognized by the hMSH2-hMSH6 heterodimer [28]. MMR proficient cells induce a G2 arrest that, interestingly, requires two rounds of DNA replication following MNNG treatment [29,30]. The arrest is followed by induction of apoptosis as cells with intact MMR are approximately 100-fold more sensitive to alkylating agents than a MMR-deficient cell line [26,31]. Alternatively, as a recently published report suggests, these cells may not be arresting in G2, but rather are proceeding through mitosis into a third cycle in the absence of cell division, ultimately resulting in cell death [32].
Though a MMR-dependent response to alkylation damage has been characterized, the mechanistic role of the MMR proteins in activating this response is not clear. Using simple predictions based on MMR mechanistic models derived from in vitro studies, we attempted to characterize the interactions between the major MMR proteins and with DNA in cells treated with an alkylating agent. We analyzed the localization of MMR proteins in the nucleus as well as the formation of MMR protein complexes on and off the chromatin during an unperturbed cell cycle or in cells treated with MNNG.
2. Materials and Methods
2.1 Cell Lines and Small Interfering RNA (siRNA)
HeLa S3 cells were grown in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen). Hec59 cells were grown in DMEM:Nutrient Mixture F-12 (DMEM/F-12). Hec59+chromosome 2 cells were grown in DMEM/F-12 supplemented with 500 ug/mL G418 sulfate (Gibco). All medium contained 10% fetal bovine serum (FBS; Gibco). Cells were incubated at 37°C in a 5% CO2 humidified atmosphere. Transfections of siRNA were performed with Lipofectamine 2000 (Invitrogen). The hMSH2 target sequence was TCCAGGCATGCTTGTGTTGAA. The MLH1 target sequence was GTGGCTCATGTTACTATTACA. All siRNA oligonucleotides were purchased from QIAGEN.
2.2 Chemicals, Antibodies, and Reagents
N-Methyl-N′-nitro-N-nitrosoguanidine (MNNG) (obtained from the National Cancer Institute Chemical Carcinogen Reference Standard Repository; CAS: 70-25-7) was dissolved in DMSO to a concentration of 10 mM and stored at −20°C until use. O6-Benzylguanine (O6-BG; CAS: 19916-73-5) was purchased from Sigma, dissolved in DMSO to a concentration of 25 mM and stored at −80°C until use. Thymidine was purchased from Sigma (T1895), dissolved in 1 N NaOH to a final concentration of 1 M and stored at room temperature until use. Antibodies against Nucleolin (SC-55486 mouse), PCNA (SC-56), and rabbit IgG (SC-2027) were purchased from Santa Cruz Biotechnology. Antibodies against Phospho-Chk1 S317 (#2344), Phospho-Chk2 Thr68 (#2661), Chk1 total (2G1D5), and Chk2 Total (#2662) were purchased from Cell Signaling. Antibodies against hMSH6 (A300-023A) and hMLH1 (A300-015A for nucleolar immunofluorescence) were purchased from Bethyl Laboratories. Antibodies against hMLH1 (550838 for western blot analysis and immunoprecipitations), hMLH1 (554073 for immunofluorescence studies), hMSH2 (556349 for western blot analysis) and Orc2 (551178) were purchased from BD Biosciences. Antibody against hMSH2 (NA27 for immunoprecipitation) was purchased from Oncogene. Antibody against Actin (A5060) was purchased from Sigma. Alexa Fluor-488, −594 anti-rabbit IgG or Alexa Fluor-488, −594 anti-mouse IgG were purchased from Molecular Probes.
2.3 Cell Synchronization
Synchronization at the G1/S boundary was performed by a double thymidine block (DTB) [33]. HeLa cells were grown for 18h in complete medium containing 2 mM thymidine, an additional 10h without thymidine, and then incubated again for 16h with 2 mM thymidine. At 14h into the second block, 25 μM O6-BG, a competitive inhibitor of MGMT, was added until the cells were released into serum free medium containing 0.2, 2, or 10 μM MNNG and 25 μM O6-BG. After 1h, cells were incubated in complete medium until harvested. For cell synchronization at G2/M, HeLa cells were grown in 300 ng/mL nocodazole for 16h at which time cells were detached by mechanical shake-off and replated in fresh complete medium without nocodazole. After 3h, 25 μM O6-BG was added and cells were incubated for an additional 2h. Serum free medium was then added containing 10 μM MNNG and 25 μM O6-BG. After 1h, cells were incubated in complete medium until harvested.
2.4 Cell Cycle Analyses
Cell cycle analyses were performed using propidium iodide (PI) staining for DNA content and subsequent detection by flow cytometry. Briefly, cells were collected at various time points post release from either double thymidine or nocodazole block and fixed in 70% ethanol at −20°C. Cells were then treated with 20 μg/mL PI and 200 μg/mL RNase A and incubated at 37°C for 1h, filtered, and analyzed with a FACS Calibur flow cytometer (BD Biosciences). The resulting data was analyzed by Modfit analysis software (Verity Software House). Time points were performed in duplicate.
2.5 Chromatin Localization
Cells were collected by centrifugation at 3400 rpm for 5 min and washed with phosphate-buffered saline (PBS) and pelleted as above. Cells were then washed once with buffer A (10 mM NaCl, 50 mM Tris-HCl pH 7.5, 2 mM MgCl2). Cells were resuspended in buffer B (buffer A plus 0.5 mM DTT, protease and phosphatase inhibitors, and 0.5% NP-40) and incubated on ice for 10 min. Subsequently, the cells were processed by a dounce homogenizer and centrifuged at 3400 rpm for 15 min. The soluble fraction was recovered and stored at −80°C until further use. The remaining nuclei were resupended in buffer C (420 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM DTT, 2 mM MgCl2, 5% glycerol, 0.75% Triton-X 100, 1 unit DNase I, protease and phosphatase inhibitors) and rocked for 35 min at 4°C. The samples were then centrifuged at 13,200 rpm for 15 min at 4°C. The recovered supernatant was used as the chromatin-enriched fraction. Protein concentrations were determined by a Bradford assay and equal protein concentrations were separated by denaturing SDS-PAGE and examined by Western blot.
2.6 Chromatin Immunoprecipitation Analyses
The soluble fraction and fragmented nuclei were recovered as described above. For analysis by chromatin immunoprecipitation, the fragmented nuclei were resuspended in buffer B, formaldehyde was added to a final concentration of 1.1%, and the nuclei were rocked at 4°C for 20 min. The reaction was quenched by rocking the nuclei at 4°C for 10 min in the presence of 125 mM Glycine pH 9.4. Nuclei were then washed twice in cold PBS and resuspended in buffer B for sonication. The cross-linked nuclei were sonicated seven times on ice for 10 sec with a 2 min rest in between each sonication. This yielded approximately 200-500 nt fragments. The samples were centrifuged at 13,200 rpm for 15min at 4°C. The recovered supernatant, (chromatin-enriched fraction) was used for immunoprecipitation. Soluble fractions were used directly for immunoprecipitation or, where noted, treated with formaldehyde as described for the nuclei preparations prior to immunoprecipitation.
2.7 Immunofluorescence
Cells were plated on cover slips 24 hours prior to DTB. Cells were harvested at 4 and 8 hours post-DTB release. For some experiments, soluble proteins were removed by extraction in CSK buffer (10 mM Hepes pH 7.4, 300 mM Sucrose, 100 mM NaCl, 3 mM MgCl2, 0.5% Triton-X 100) for 2 min at 4°C. Cells were fixed with 4% formaldehyde for 20 min at room temperature. For experiments using anti-PCNA antibody, cells were fixed with ice cold MeOH for 20 min at −20°C. Formaldehyde fixed cells were permeablized with phosphate buffered saline (PBS) containing 0.2% Triton X-100 for 15 min at room temperature. Cells were incubated overnight with primary antibodies as indicated at 4°C, then washed with PBS and incubated with secondary antibodies for 1 hour at room temperature. Cells were analyzed using a Zeiss LSM 510 meta-confocal microscope.
3. Results
3.1 Localization of MMR proteins to the chromatin during the cell cycle
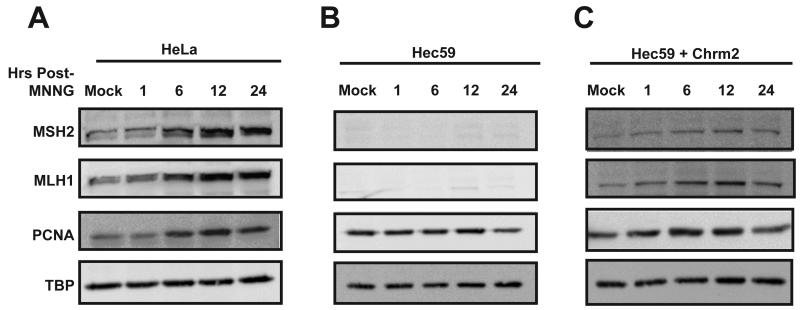
The goal of this study was to further understand the localization and formation of MMR protein complexes during cellular DNA replication and in response to alkylation damage. Based on in vitro models for MMR, an essential step in the pathway is the recruitment and association of MMR proteins on the DNA. To investigate the localization of hMSH2 and hMLH1 to chromatin in response to DNA damage, we first treated asynchronously growing HeLa cells with the alkylating agent MNNG. In this MMR-proficient background, there was a rapid increase in chromatin-associated hMSH2 and hMLH1 as early as 1 hour (h) after treatment (Fig. 1A). The accumulation of both MMR proteins was maximal 12h post-treatment with MNNG. In addition, we observed an increase in the levels of chromatin-associated PCNA. Similar to hMSH2 and hMLH1, PCNA levels peaked at 12h.
Fig. 1.
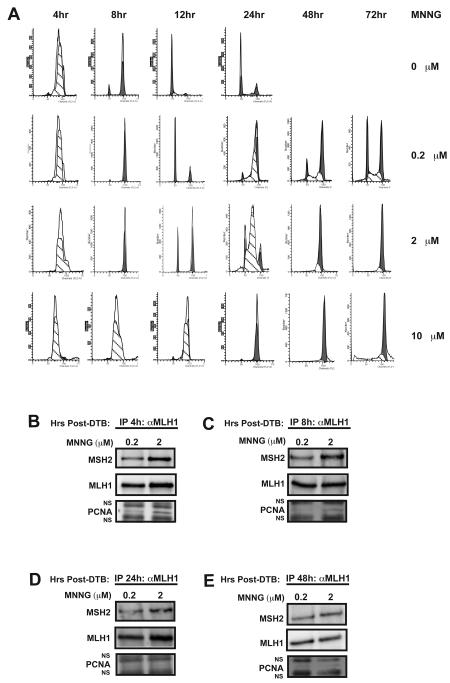
The MMR proteins localize to the chromatin in response to DNA damage. Asynchronously growing HeLa (A), Hec59 (B), or Hec59 + Chrm2 (C) cells were treated with 10 μM MNNG and O6-BG or DMSO for 1h. Chromatin-enriched (CE) fractions were prepared at the indicated time points post-treatment and examined by Western blotting with antibodies against hMSH2, hMLH1, and PCNA. Tata-binding protein (TBP) was used as the loading control. hMSH2, hMLH1 and PCNA localize to the chromatin rapidly following MNNG treatment, however, hMLH1 fails to localize to the chromatin in cells lacking hMSH2 expression.
Previous in vitro experiments using total internal reflectance to examine the interaction between E. coli MutS and MutL indicated that MutS binds to the mismatch first, but then must form an ATP-dependent sliding clamp in order to recruit MutL [14]. Similarly, using eukaryotic cell extracts and a heteroduplex oligonucleotide, it was demonstrated that hMSH2-hMSH6 is required for hMLH1-hPMS2 DNA binding and that protein complex formation is ATP-dependent [16,17]. To test whether the localization of hMLH1 to chromatin was hMSH2-dependent, we used an endometrial cancer cell line, Hec59, which harbors a biallelic mutation in hMSH2 that results in loss of a detectable protein product (Fig. 1B). In the absence of hMSH2, minimal hMLH1 is recruited to the chromatin in response to MNNG. However, chromatin-associated PCNA levels were similar to that observed using HeLa cells. To confirm that this was attributed to loss of hMSH2 and not an inherent difference between the two cell lines, we examined the localization of hMSH2 and hMLH1 using the Hec59 + chrm 2 cell line, in which hMSH2 expression has been restored by whole chromosome transfer [34]. In these cells with restored hMSH2 expression, both hMSH2 and hMLH1 localize to the chromatin following DNA damage (Fig. 1C).
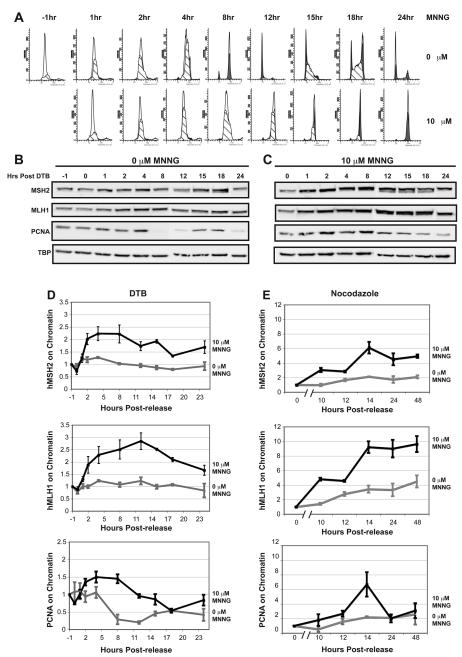
We next wished to further explore the recruitment of MMR proteins to the chromatin in response to DNA damage. Given that this DNA repair pathway recognizes lesions from MNNG treatment created during DNA replication [35], we first synchronized HeLa cells at the G1/S boundary using a double thymidine block (DTB). Two hours prior to release from the DTB, cells were treated with O6-BG to inhibit the repair activity of MGMT then released from the block into serum free media containing 10 μM MNNG and O6-BG or DMSO as a negative control for one hour. Cells were then returned to serum-containing normal growth media and harvested at various time points following treatment to analyze cell-cycle progression by flow cytometry. The DMSO-treated control cells completed the first S-phase by 8h and a complete cell cycle by 12h before entering into a second S-phase between 15h and 18h (Fig. 2A). By contrast, the MNNG-treated cells took approximately twice as long to progress through S-phase. Furthermore, after 24h the majority of these cells appeared to arrest in G2 (Fig. 2A).
Fig. 2.
The MMR proteins localize to the chromatin during the S-phase and accumulate in response to DNA damage. (A) HeLa cells were synchronized to the G1/S boundary by a double thymidine block (DTB). In the last 2h of the DTB, the cells were treated with O6-BG or DMSO before being released into serum free media containing either 10 μM MNNG and fresh O6-BG or DMSO (T= −1h). After 1h, (T=0h) normal growth media was supplied and the cells were incubated until the indicated time points. Cell cycle progression was monitored by flow cytometry. Mock-treated cells enter S-phase immediately following release and complete S-phase within 8 hours. MNNG-treated cells show a delayed progression through S-phase, finally exiting between 12 and 15 hours. Each time point was performed in duplicate. (B and C) Chromatin enriched fractions were prepared from mock (B) and MNNG-treated (C) cells at the various time points post-DTB and examined by Western blotting against hMSH2, hMLH1, PCNA, and TBP. (D) Quantification of chromatin-associated protein levels from immunoblots in B using ImageJ software (NIH) and graphed using Excel. PCNA localizes to the chromatin during S-phase in both mock and treated cells. hMSH2 and hMLH1 show a slight increase in localization to the chromatin during S-phase in mock-treated cells, however, in MNNG-treated cells, the chromatin localization is increased 2- to 2.5-fold. Localization experiments were performed at least in triplicate. (E) Quantification of chromatin-associated protein levels from immunoblots of mock or MNNG-treated HeLa cells synchronized by nocodazole block. hMSH2 and hMLH1 levels are increased 2-4 fold during S-phase in mock-treated cells compared to cells arrested in mitosis (0h time point). MNNG treatment results in 6-10 fold increases in hMSH2 and hMLH1 chromatin levels during S-phase. Localization was performed in duplicate.
To monitor the localization of MMR proteins during S-phase, cells were released from the DTB into media with or without MNNG as described above, then harvested at various time points following treatment. Chromatin-enriched and soluble protein fractions were prepared and hMSH2, hMLH1 and PCNA levels were examined in each fraction by Western blot. Chromatin-bound proteins were quantitated and normalized to the level of chromatin-bound protein observed just prior to release from the DTB (−1 hour). In MNNG-treated cells, a rapid increase in the levels of hMSH2, hMLH1, and PCNA on the chromatin was observed by 1h, reaching peak chromatin binding levels by 8h and 12h for hMSH2 and hMLH1, respectively (Fig. 2C, and 2D). Though PCNA levels diminished as cells exited S-phase, populations of both hMSH2 and hMLH1 remained associated with the chromatin throughout S-phase and into G2. Interestingly, in mock treated cells we observed a subtle accumulation of both hMSH2 and hMLH1 on the chromatin during the first round of DNA replication. PCNA levels remained tightly coupled to the cell cycle, associating with the chromatin only during S-phase (Fig. 2B and 2D). We did not observe the same increase in chromatin-bound hMSH2 or hMLH1 levels during the second round of replication between 15h and 18h post-release from the DTB. This could be attributed to the fact that after a complete cell cycle the cells became more asynchronous, which may hinder our ability to detect the subtle increase. Furthermore, the accumulation of chromatin-bound MMR proteins may be understated in these experiments as we already detect some MMR proteins bound to the chromatin at the -1 hour time point (our normalization time point). As the DTB synchronizes cells at the G1/S boundary, some MMR proteins may accumulate on the chromatin in preparation for entry into S-phase. Alternatively, there may be a low-level accumulation of MMR proteins on the chromatin in response to DNA damage caused by the DTB treatment. Therefore, to further investigate the accumulation of MMR proteins on chromatin during S-phase, we synchronized HeLa cells using a nocodazole block. Nocodazole inhibits mitosis and causes cells to arrest with a G2/M DNA content when analyzed by flow cytometry (Fig. S1A). In the absence of MNNG, enrichment of cells in S-phase occurs between 12h and 14h post-release. We observed a 2-4 fold accumulation of chromatin-associated hMSH2 and hMLH1 during S-phase in our mock-treated cells (Fig. 2E and Fig. S1B). This increase was further enhanced in cells treated with MNNG (Fig 2E and Fig. S1C). Similar to the experiments performed using the DTB, treatment of nocodazole-synchronized cells with MNNG resulted in an extended S-phase progression followed by a pronounced G2 arrest after a single round of replication (Fig. S1A). Furthermore, chromatin-associated hMSH2 and hMLH1 persisted throughout S-phase and into G2 in MNNG-treated cells (Fig. 2E and Fig. S1C), consistent with the results performed using the DTB (Fig. 2C and 2D). Taken together, these data reveal that the MMR proteins hMSH2 and hMLH1 localize to the chromatin as a consequence of normal DNA replication. In response to DNA damage there is an increased localization of both hMSH2 and hMLH1 to the chromatin that persists throughout S-phase and into G2.
In addition to DNA repair, the MMR pathway has a role in eliciting a cell cycle checkpoint response to alkylation damage [1,26,36]. In cells treated with MNNG, Chk1, an essential kinase in MMR-dependent checkpoint activation, was activated (demonstrated by phosphorylation of Serine 317) as early as 1h post-treatment (Fig. S2B). Phosphorylated Chk1 persisted throughout S-phase and declined slightly as cells entered G2. In the absence of MNNG Chk1 was also activated during S-phase; however, there was a substantial reduction in this population as cells cycled into G2 (Fig. S2A). Furthermore, we observed an additional increase in Chk1-pS317 at 15h post-release from the DTB, which corresponded to a second round of DNA replication (Fig. S2A). We also observed activation of Chk1 in the absence of DNA damage during S-phase after release from a nocodozole block (Fig. S1D, see lanes 3 and 4). This activation was further enhanced by treatment with MNNG (Fig. S1E, see lanes 2-5).
3.2 Identification of an hMSH2 /hMLH1 complex in MMR-proficient HeLa cells
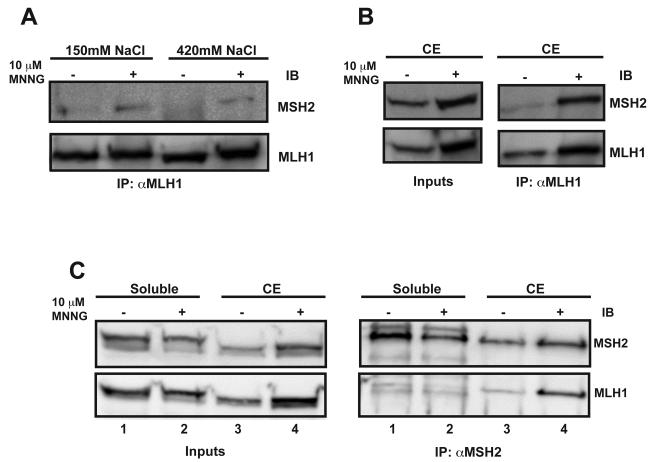
In vitro evidence suggests that an essential step in the MMR pathway is the formation of the hMSH2-hMSH6/hMLH1-hPMS2 ternary complex [17]. Therefore, we reasoned that we should be able to identify an interaction between hMSH2 and hMLH1 within a MMR-proficient cell line. Asynchronously growing HeLa cells were treated with 10 μM MNNG and incubated for 24h. We next extracted the chromatin-associated proteins with high and low salt concentrations and performed immunoprecipitation (IP) against hMLH1. Co-IP of hMSH2 with hMLH1 was only observed in MNNG-treated cells (Fig. 3A). However, given the amount of the target protein immunoprecipitated, the interaction appeared relatively weak. As the “molecular switch” model for MMR suggests that hMLH1 interacts with DNA-bound hMSH2-hMSH6 sliding clamps [14], we questioned whether the extraction of the proteins from the chromatin led to a disassembly of the complex. To stabilize and capture the MMR complex associated with the chromatin, we first cross-linked proteins to the chromosomal DNA using formaldehyde fixation of the nuclei, followed by sonication of the genomic DNA into 200-500nt fragments and IP against hMLH1 similar to a chromatin immunoprecipitation assay (ChIP). Using this approach, we observed the co-IP of hMSH2 with hMLH1 in a DNA-damage dependent manner (Fig. 3B). The reciprocal IP against hMSH2 also demonstrated the formation of an hMSH2-hMLH1 complex in MNNG-treated cells (Fig. 3C, right panels compare lanes 3 and 4). To test whether formaldehyde cross-linking resulted in an interaction between the non-chromatin associated hMSH2 and hMLH1 proteins, we treated the soluble fractions of MNNG or DMSO-treated cells with formaldehyde, similar to the fixation of nuclei, and performed the same IP against hMSH2. No specific interaction was observed in the soluble fraction between hMSH2 and hMLH1 (Fig. 3C, right panels, lanes 1 and 2).
Fig. 3.
hMSH2 and hMLH1 interact on chromatin in response to DNA damage. (A) Extracts were prepared from nuclei of mock and MNNG-treated asynchronously growing HeLa cells using 150mM or 420mM NaCl as indicated. The extracts were immunoprecipitated against hMLH1 and examined by Western blotting with antibodies against hMSH2 and hMLH1. A weak interaction is detected between hMLH1 and hMSH2 only in extracts from MNNG treated cells. (B and C) ChIP was performed from chromatin-enriched (CE) extracts from mock and MNNG-treated asynchronously growing HeLa cells using an antibody against hMLH1 (B) or hMSH2 (C). A strong interaction was detected between hMLH1 and hMSH2 in both ChIP assays from MNNG-treated cells. No interaction was detected between hMSH2 and hMLH1 in formaldehyde-fixed soluble fractions from mock or MNNG-treated HeLa cells.
3.3 Formation of MMR protein complexes occurs on the chromatin during DNA replication, is hMSH2-dependent, and is enhanced by treatment with MNNG
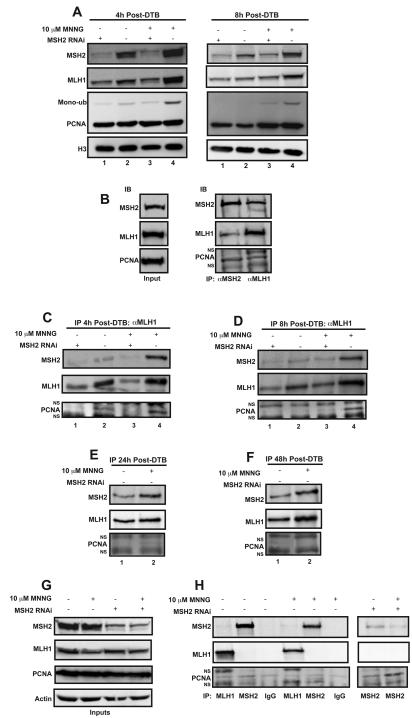
We next wished to further test that the recruitment of hMLH1 to the chromatin is dependent on hMSH2 by depleting hMSH2 levels with siRNA. In hMSH2 siRNA treated cells, the damage-induced chromatin localization of hMLH1 was severely diminished (Fig. 4A, left panels compare lanes 3 and 4). In addition, recruitment of hMLH1 to the chromatin during an unperturbed S-phase was also decreased in the absence of hMSH2 (Fig. 4A, left panels compare lanes 1 and 2). Similar results were observed at 8h post-treatment, whereby localization of hMLH1 to the chromatin was also hMSH2-dependent (Fig. 4A, right panels). PCNA localization to the chromatin was not affected in hMSH2 knockdown cells. However, an additional damage-dependent band migrating above the unmodified PCNA band in the PCNA immunoblot is dramatically reduced in hMSH2 knockdown cells (Fig. 4A, left panels compare lanes 3 and 4). This additional band has been reported to be a mono-ubiquitinated form of PCNA, which is required for association with the translesion synthesis polymerases [37]. A similar hMSH2- and damage-dependent increase in the mono-ubiquitinated form of PCNA was detectable at 8h as well (Fig. 4A, right panels compare lanes 3 and 4).
Fig. 4.
hMSH2, hMLH1 and PCNA form a complex on chromatin during S-phase. HeLa cells were transfected with siRNA targeting hMSH2 or a non-targeting control, synchronized by a DTB then treated with 10 μM MNNG and O6-BG or DMSO. ChIP assays were performed on cells harvested 4, 8, 24, or 48 hours post-treatment using antibodies against hMLH1 or hMSH2. (A) Inputs representing 2.5% of the total ChIP reaction were assessed by Western blotting. hMSH2 and hMLH1 accumulate on chromatin in response to DNA damage; however, hMLH1 accumulation is dependent on hMSH2. PCNA chromatin levels are unchanged in response to hMSH2 siRNA. However, the levels of a higher-migrating PCNA species, believed to be monoubuiquitinated PCNA, increase in response to damage in an hMSH2-dependent manner. Histone H3 was used as the loading control. (B) The ChIP assay was performed using an antibody against either hMLH1 or hMSH2 from MNNG-treated cells revealing a complex formation on chromatin including hMSH2, hMLH1 and PCNA. Additional bands in the PCNA immunoblot appeared consistently during this assay that were demonstrated to be non-specifically detected by the secondary antibody (NS). (C-F) Chromatin-enriched extracts were prepared and ChIP was performed against hMLH1 at 4 (C), 8 (D), 24 (E), or 48 (F) hours post-release from the DTB. A chromatin-localized complex exists in cells harvested at 4 and 8 hours post-release including hMSH2, hMLH1 and PCNA. The complex is enhanced by DNA damage and requires hMSH2. MNNG-treated cells harvested 24 hours post-release are mostly in the G2 phase. These cells maintain a chromatin bound complex containing hMSH2 and hMLH1; however, PCNA is no longer in the complex. This complex persists until at least 48 hours post-release. (G and H) Co-immunoprecipitations were performed from the soluble fractions of the cells treated as described above, but in the absence of formaldehyde cross-linking using antibodies against hMLH1, hMSH2 or IgG as a negative control. Inputs (G) and coimmunoprecipitations (H) are shown. No co-immunoprecipitation is detected in these soluble fractions suggesting these complexes only form on chromatin. A weak PCNA band is detected in every co-immunoprecipitation including the co-IP against hMSH2 from hMSH2 siRNA treated cells indicating that this band is non-specific (H, right panels).
As hMSH2 is required for hMLH1 localization to damaged chromatin during S-phase, we wanted to test if hMLH1 is required for hMSH2 localization and accumulation on the chromatin. siRNA against hMLH1 led to a dramatic reduction in hMLH1 protein levels, yet had no effect on hMSH2 chromatin localization during S-phase in response to MNNG compared to the irrelevant siRNA control (Fig. S3, compare lane 2 of S3A with lane 2 of S3B). Interestingly, the MNNG-induced accumulation of hMSH2 persisted at 8h and 24h post-DTB release even in the absence of hMLH1 (Fig. S3B, lanes 4 and 6).
To determine whether PCNA is part of the MMR complex associated with the chromatin, we performed ChIP from cells in S-phase 4h after treatment with MNNG. Fig. 4B demonstrates the formation of hMSH2/hMLH1/PCNA complexes on the chromatin in response to MNNG. This complex was observed when the ChIP was performed with antibodies against either hMLH1 or hMSH2 supporting the formation of this chromatin-associated complex. In addition, we observed that formation of this protein complex was substantially reduced in the absence of DNA damage (Fig. 4C, compare lanes 2 and 4). In the absence of hMSH2, hMLH1 does not interact with PCNA on the chromatin in either the presence or absence of MNNG (Fig. 4C, lanes 1 and 3). To assess whether interactions between hMSH2, hMLH1, and PCNA persist through S-phase, we released cells from the DTB and harvested them at 8h. hMSH2/hMLH1/PCNA complex formation was similarly observed at 8h after treatment with MNNG in an hMSH2-dependent manner (Fig. 4D).
After treatment with MNNG, both hMSH2 and hMLH1 remained chromatin-associated throughout S-phase and into G2, however, PCNA levels diminished (Fig. 2C). Furthermore, Chk1 remained activated even in the absence of strong chromatin-associated PCNA levels (Fig. S2B). Therefore, to investigate the status of the MMR protein complex during G2 in damaged cells, we performed ChIP against hMLH1 at 24h post-release from the DTB. We observed a strong interaction between hMLH1 and hMSH2; however, PCNA was not part of the complex (Fig. 4E). Interestingly, this complex persisted until at least 48h post-release from the DTB (Fig. 4F).
Although these data demonstrate formation of MMR protein complexes on the chromatin, it does not address whether interactions are taking place away from the chromatin as well during S-phase. It has been suggested that the MMR proteins are imported into the nucleus in a preformed complex [38]. To investigate this, we prepared soluble fractions of cells 4h post-treatment from the DTB. Treatment with MNNG resulted in a reduction of both hMLH1 and hMSH2 proteins from the soluble fraction (Fig. 4G and Fig. S1E), which is concomitant with the accumulation of these proteins on the chromatin. Although we were able to IP hMLH1 from the soluble fraction, we did not detect an interaction with hMSH2 in either the absence or presence of MNNG (Fig. 4H, middle panel lanes 1 and 4). The reciprocal IP of hMSH2 yielded a similar result (Fig. 4H, middle panel lanes 2 and 5). This was consistent with our results in asynchronous cells (Fig. 3C). PCNA has been proposed to play additional roles in MMR prior to the resynthesis step [25]. Therefore, we also investigated the interaction between hMLH1/hMSH2 and PCNA. A weak interaction was observed between hMSH2 and PCNA (Fig. 4H, lanes 2 and 5) as well as between hMLH1 and PCNA (Fig. 4H, lanes 1 and 4) that was independent of MNNG treatment. However, immunoprecipitation of IgG alone also demonstrated a similar interaction (Fig. 4H, lanes 3 and 6), suggesting that the interaction was nonspecific. To further test this, we first depleted hMSH2 levels by siRNA then immunoprecipitated the soluble fraction using an antibody against hMSH2. A similar level of PCNA was detected even in the absence of hMSH2 (Fig. 4H, lane 8), suggesting that the low-level of PCNA detected on our Western blot was non-specific. Therefore, we conclude that formation of the hMSH2/hMLH1/PCNA complex occurs only once these proteins have accumulated on the chromatin.
3.4 Formation of MMR protein complexes on chromatin in response to MNNG is dose-dependent
The cellular response to MNNG differs depending on the concentration of alkylating agent [39]. We observed that cells treated with 10 μM MNNG appear to arrest in G2 at 24h post-release from the DTB (Fig. 2A). However, previous reports have demonstrated that cells treated with low doses of alkylating agents do not arrest in G2 until two rounds of DNA replication have been completed [29,30]. To investigate this further, we monitored the cell cycle status of cells treated with 10 μM MNNG at 48h and 72h and found that the cells maintained the G2 arrest. Therefore, at this concentration of MNNG in HeLa cells, the G2 arrest requires only a single round of DNA replication (Fig. 5A). Treatment of cells with high doses of MNNG have been shown to induce damage signaling that is MMR-independent [39]. Therefore, we wished to examine the function of the MMR proteins in cells treated with lower doses of drug. We first monitored the progression of cells treated with either 0.2 or 2 μM MNNG. At 2 μM MNNG, the G2 arrest was not observed until the second cell cycle (Fig. 5A). Treatment with 0.2 μM MNNG also resulted in a significant G2 arrest after the second round of replication, although some cells continued to traverse through the cell cycle (Fig. 5A).
Fig. 5.
Formation of chromatin-associated MMR protein complexes in response to different concentrations of MNNG. HeLa cells were synchronized by a DTB then treated with O6-BG and either 0.2, 2, or 10 μM MNNG or a DMSO control and then harvested at the indicated time points. (A) Cell cycle progression was monitored by flow cytometry. The majority of cells treated with either 0.2 or 2 μM MNNG do not arrest in G2 until two rounds of DNA replication have been completed. In contrast, cells treated with 10 μM MNNG arrest in G2 after the first round of replication and remain until at least 72h post-release from the DTB. (B-E) ChIP assays were performed on cells harvested 4 (B), 8 (C), 24 (D), or 48 (E) hours post-release from the DTB using an antibody against hMLH1. Cells treated with 2 μM MNNG form hMLH1/hMSH2/PCNA complexes as early as 4h post-release from the DTB. However, as cells traverse through the cell cycle PCNA is no longer detectable in the hMSH2/hMLH1 complex.
We next investigated the status of MMR protein complexes in response to lower concentrations of MNNG. Treatment with 0.2 μM MNNG did not result in the formation of significant MSH2/MLH1/PCNA complexes at 4, 8, 24, or 48h post-release from the DTB compared to mock-treated cells (Fig. 5B-E). The sensitivity of this assay may be such that a slight increase in complex formation would not be detected. Complex formation was enhanced by treatment with 2 μM MNNG with significant complexes forming at 4h and to a lesser extent at 8h post-DTB (Fig. 5B and 5C). Interestingly, we also observed the complex at 24h post-release from the DTB when the majority of the population is undergoing a second round of DNA replication. However, PCNA was no longer associated with the complex (Fig. 5D). Furthermore, this hMSH2/hMLH1 complex persisted without detectable PCNA in cells that progressed into G2, where they ultimately arrested (Fig. 5E).
In addition to differences in MMR complex formation, we also investigated the activation of the DNA damage signaling molecules Chk1 and Chk2 after treatment with high and low concentrations of MNNG. 10 μM MNNG treatment led to the activation of Chk1 in early S-phase that declined as cells progressed into G2 (Fig. S2B, compare lanes 2 and 9, and Fig. S2E, compare lanes 1 and 3). A slight Chk2 activation, as determined by measuring the levels of Chk2 phosphorylated at threonine 68, was also observed in cells treated with 10 μM MNNG during early S-phase, but declined as cells entered later S-phase (Fig. S2E, lanes 1 and 2). This activated Chk2 re-emerged as cells arrest in G2 at 24 and 48-hours post release (Fig. S2E, lanes 3 and 4). At 2 μM MNNG, Chk1 was similarly activated as early as 4h post-DTB (Fig. S2D). However, Chk1 activation was also observed at 24h and even more strongly at 48h post-DTB (Fig. S2D). This result is strikingly different from cells treated with 10 μM MNNG. Furthermore, Chk2 was not activated during the first S-phase in cells treated with 2 μM MNNG, and was only slightly activated during the second S-phase (Fig. S2D, compare lanes 1 and 3). Chk2 phosphorylation increased even more as the cells arrested in G2 at 48h (Fig. S2D, lane 4). These differences in Chk1 and Chk2 activation suggest that different modes of damage signaling occur in response to high and low doses of MNNG.
3.5 Analysis of MMR protein localization by confocal immunofluroescence
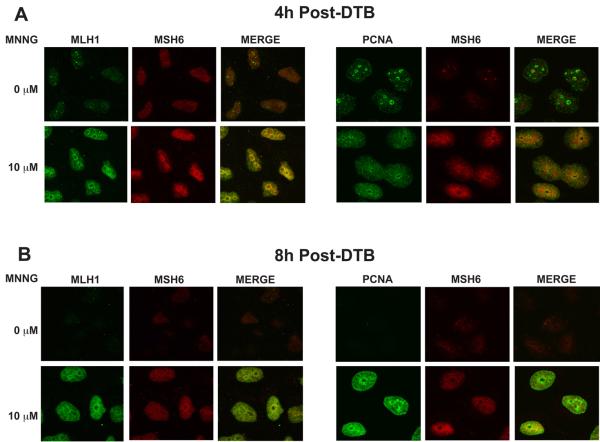
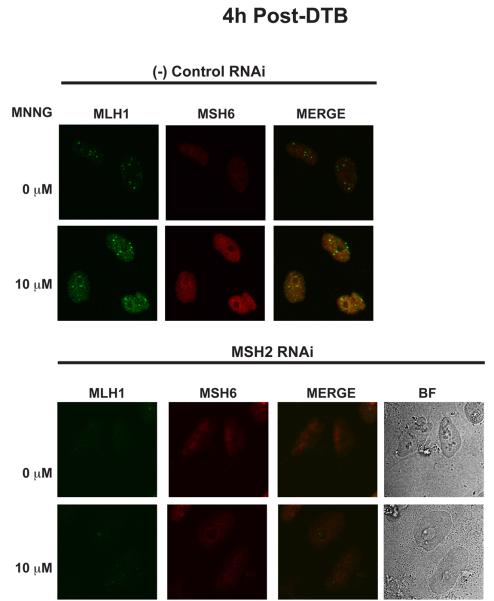
In order to further support the formation of chromatin-associated MMR protein complexes, HeLa cells were grown on cover slips, synchronized by DTB, treated with MNNG or DMSO and released into S-phase as described above. At 4 and 8 hours post-treatment, the cover slips were removed. To identify chromatin-bound complexes, all soluble proteins were removed by detergent extraction prior to fixation. Cells were then stained with antibodies against hMSH6, hMLH1 or PCNA. Given the instability of hMSH6 in the absence of its partner hMSH2 [40], we used hMSH6 as a marker for hMSH2 localization. In mock-treated cells, hMSH6 and hMLH1 staining was barely detectable on the chromatin at 4h post-release (Fig 6A, left panels). However, due to its role in DNA replication, we observed significant PCNA foci formation (Fig. 6A, right panels). Treatment with MNNG resulted in the accumulation of insoluble hMSH6 and hMLH1 with some, though not complete, colocalization between the two proteins (Fig. 6A, left panels). In addition, PCNA staining appeared more diffuse throughout the nucleus with moderate colocalization with hMSH6 (Fig. 6A, right panels). 8h after treatment with MNNG and release from the DTB, some colocalization of hMSH6 and hMLH1 was still evident (Fig. 6B, left panels). Insoluble PCNA levels remained intense and there was overlap in staining with hMSH6 (Fig. 6B, right panels). Due to the only modest colocalization between hMSH2 and hMLH1, we wished to test the specificity of the observed hMLH1 staining. Having demonstrated an hMSH2-dependence for hMLH1 localization to the chromatin in our earlier experiments, we treated cells with either siRNA against hMSH2 or a negative control siRNA prior to immunofluorescence analysis. Cells treated with the negative control siRNA demonstrated a similar increase in both hMSH6 and hMLH1 after treatment with MNNG as expected (Fig. 7, top panel). In contrast, depletion of hMSH2 abrogated the accumulation of both detergent resistant hMSH6 and hMLH1, supporting the specificity of the staining and reaffirming an hMSH2-dependence for hMLH1 chromatin localization. (Fig. 7, bottom panel).
Fig. 6.
DNA damage-dependent colocalization of hMLH1, hMSH6, and PCNA on the chromatin as determined by immunofluorescence. HeLa cells were plated onto glass coverslips, synchronized by a DTB and treated with 10 μM MNNG and O6-BG or DMSO. The coverslips were removed at 4h (A) or 8h (B) following release from the DTB. The cells were extracted with CSK buffer to remove all soluble proteins and examined by confocal immunofluorescence with antibodies against hMLH1, hMSH6, and PCNA. Detergent resistant hMLH1 and hMSH6 are visible in MNNG-treated cells and co-localize to a modest extent. PCNA is detergent resistant in mock and MNNG-treated cells at 4 hours (A), when cells are in S-phase, however, only MNNG-treated cells contain detergent resistant PCNA at 8 hours (B). A sub-population of the chromatin-bound PCNA co-localizes with hMSH6.
Fig. 7.
hMLH1 localization to the chromatin in response to MNNG is abrogated in cells depleted of hMSH2. HeLa cells were plated onto glass coverslips, treated with siRNA against hMSH2, synchronized by a DTB and treated with 10 μM MNNG and O6-BG or DMSO. The coverslips were removed at 4h following release from the DTB. The cells were extracted with CSK buffer as in Figure 5 and examined by confocal immunofluorescence with antibodies against hMLH1and hMSH6. hMSH6 and hMLH1 are no longer resistant to detergent extraction in the absence of hMSH2. (BF) Bright-field.
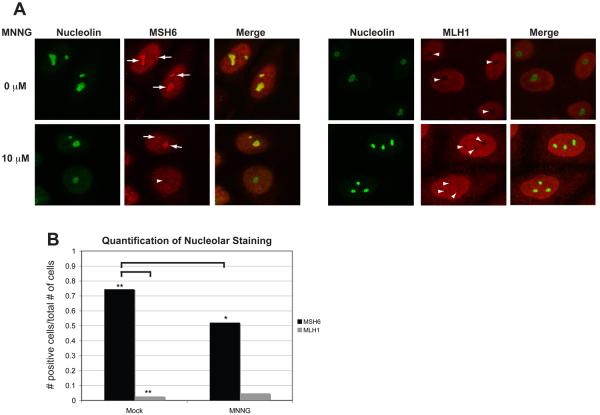
To visualize soluble MMR proteins, cells were synchronized and treated as above, but were not extracted with detergent prior to fixation. During S-phase, both hMSH6 and hMLH1 are diffuse throughout the nucleus (Fig. 8A). However, during an unperturbed S-phase, we observed a population of hMSH6 that was clearly segregated from the rest of the nucleus (Fig. 8A, left panel, arrows). To test whether this population was contained within the nucleolus, we labeled the nucleoli with an antibody against the nucleolar protein nucleolin. hMSH6 appeared to localize within the nucleolus, however, treatment with MNNG resulted in a significant redistribution of this hMSH6 population in some cells (Fig. 8A, left panel, arrowhead and 8B). Interestingly, hMLH1 was excluded from within the nucleolus regardless of treatment with MNNG (Fig. 8A, right panel and 8B). These results suggest a novel mechanism for the sequestration of hMSH2-hMSH6 from hMLH1 in the absence of DNA damage.
Fig. 8.
A population of hMSH6, but not hMLH1, localizes to the nucleolus in the absence of DNA damage. (A) HeLa cells were plated onto glass coverslips, synchronized by a DTB and treated with 10 μM MNNG and O6-BG or DMSO. Coverslips were removed at 4 hours post-release and immediately processed for confocal immunofluorescence with antibodies against hMSH6, hMLH1, and Nucleolin. A population of hMSH6 can be detected within nucleoli as marked by positive nucleolin staining in mock-treated cells. The nucleolar localization is dramatically reduced in MNNG-treated cells. hMLH1 rarely appears to localize within the nucleoli. (B) Quantification of nucleolar staining. >150 cells for each condition were counted. p<0.0001 when comparing hMSH6 mock versus hMSH6 MNNG by a paired T-test. p< 0.0001 when comparing hMSH6 mock versus hMLH1 mock by a paired T-test.
4. Discussion
The DNA MMR pathway is essential for maintaining the fidelity of DNA replication and genomic stability, which is emphasized by the fact that loss of MMR underlies the hereditary cancer syndrome HNPCC. Many studies have investigated the biochemical details of the DNA MMR pathway using a variety of in vitro methods. From these data, three current models have been proposed for how MMR recognition is coupled to the strand discrimination signal and excision. These include the static transactivation model [41], the translocation model [42], and the molecular switch model [11,12]. All three models are consistent in proposing the initial recruitment of hMSH2-hMSH6 and hMLH1-hPMS2 heterodimers to the chromatin and the formation of a ternary protein complex composed of both heterodimers. This study examines the initial steps in the MMR pathway including the localization of MMR proteins and the formation of MMR protein complexes in response to DNA damage inside the cell. Our goal was to determine whether molecular models based on in vitro observations were consistent with the dynamics of the MMR proteins in the nucleus of a replicating cell.
The prevailing models for DNA MMR suggest that hMSH2-hMSH6 recognizes and binds to the DNA lesion and then recruits the hMLH1-PMS2 heterodimer to the DNA to form a ternary protein complex. In support of this, we found that hMSH2 was required for the increased localization of hMLH1 to the chromatin in response to DNA damage. In the absence of DNA damage we observed a subtle accumulation of both hMSH2 and hMLH1 on the chromatin during S-phase. This population was reduced as cells cycled into G2/M demonstrating that the increase in chromatin-associated hMSH2 and hMLH1 occurs specifically during DNA replication and is not maintained throughout the duration of the cell cycle. Furthermore, we demonstrated that a robust increase in chromatin-bound hMSH2 and hMLH1 occurs during S-phase in cells synchronized by a nocodazole block providing evidence that the accumulation cannot be attributed solely to DTB-induced damage. Our ChIP assay indicates that hMSH2 and hMLH1 appear to form a complex during S-phase even in the absence of exogenous damage, though at reduced levels compared to in damaged cells. Previously, hMSH2 and PCNA have been demonstrated to interact on the chromatin in asynchronously growing cells [43]. In our study using synchronized populations, we observed an interaction between hMSH2 and PCNA during an unperturbed round of DNA replication. We observed hMLH1 in this complex as well and could co-immunoprecipitate the complex using hMLH1 as the target antigen. However, hMLH1 interaction with PCNA on the chromatin is dependent upon functional hMSH2, as this interaction is abolished in cells with depleted hMSH2. These chromatin-associated MMR complexes in the absence of exogenous damage may represent active repair complexes responding to low-level DNA replication errors or damaged caused by the DTB, though our results using a nocodazole block argue against the latter. Alternatively, these complexes may be part of a DNA scanning mechanism to ensure for immediate activation and execution of MMR. Consistent with this idea, PCNA has been proposed to play a possible role in recruiting hMSH2-hMSH6 to actively replicating regions of DNA where it may more readily find a mismatch [44].
Previous studies have demonstrated the localization of MMR proteins to the chromatin following alkylation damage with either MNNG or N-Methyl-N-nitrosourea (MNU) [24,43]. In asynchronous cells exposed to MNU, both localization and formation of hMSH2/hMLH1/PCNA protein complexes was maximal 12h after treatment [43]. In cells synchronized to the G1/S boundary, treatment with MNNG resulted in the rapid (<1h post-treatment) accumulation of MMR proteins on the chromatin and formation of MMR protein complexes [24]. Consistent with these data, we also observed the rapid (1h post-treatment) association of hMSH2 and hMLH1 on the chromatin in cells treated with MNNG. In addition, we observed an increase in chromatin-associated PCNA levels after treatment with MNNG. However, these levels diminished as the cells cycled into G2, which contrasts with the persistent association of hMSH2 and hMLH1 throughout G2 in MNNG-treated cells.
We next demonstrated the interaction of hMSH2 and hMLH1 on the chromatin after treatment with MNNG. PCNA interacts with the hMSH2/hMLH1 complex in MNNG-treated cells at 4h post-release from the DTB. This is consistent with previous ChIP experiments that observed the formation of this complex in synchronized cells [24]. We also observed this complex at 8h after release from the DTB in damaged cells, demonstrating that these complexes persist throughout S-phase. However, ChIP against hMLH1 from cells in the G2-phase of the cell cycle at 24h post release from the DTB revealed that while hMLH1 and hMSH2 remain strongly associated with each other on the chromatin, the interaction of this complex with PCNA is almost completely abrogated. The drop in chromatin-bound PCNA levels and its absence from the MMR protein complex is inconsistent with the required role for PCNA in MMR in vitro [9], suggesting either low levels of PCNA remain below detection in our assays or that the MMR complexes harbor PCNA-independent functions, perhaps involved in damage signaling.
The experiments described above were performed with a high dose of MNNG (10 μM). We found that HeLa cells treated with this dose of MNNG, in combination with O6-BG to inhibit MGMT activity, arrest after only one round of replication. However, treatment with high doses of MNNG have been reported to generate MMR-independent damage signaling events [39]. Consistent with this idea, we observed Chk2 activation in the first round of S-phase in cells treated with high doses of MNNG. MMR-independent damage signaling and cell cycle arrest in cells treated with high doses of MNNG may occur as a result of base excision repair (BER) enzymes processing N-methylguanines. The abundance of BER processing of lesions in close proximity might lead to the generation of double-strand breaks that would trigger Chk2 activation [39]. However, the MMR proteins still likely recognize a high proportion of O6-meG/T lesions. This is evident by the increase in chromatin-associated hMSH2 and hMLH1 levels and suggests that the pathway is activated, though the predominant cellular response is MMR-independent. As multiple models of MMR [11,12,42], including the sliding clamp model, predict that the MMR proteins move away from the lesion after recognition, it is possible that the same O6-meG/T could initiate both MMR and BER responses.
When cells are treated with a lower dose of MNNG (2 μM), the cells complete one cell cycle and progress through a second S-phase before arresting in G2, consistent with previously published results [29,30]. Chk2 activation is not observed during the first cell cycle in these cells. This result is consistent with a reduction in double-strand breaks caused by a MMR-independent response. Chk2 activation is observed during the second S-phase and increases as cells arrest in the second G2-phase. The mechanism by which Chk2 is activated during the second cell cycle is not clear. Two models have been proposed to explain the MMR-dependent cell cycle arrest and subsequent cell death in response to alkylating agents. First, the “futile cycle” model proposes that O6-meG/T mismatches are generated during the first S-phase that trigger repeated rounds of excision and resynthesis by the MMR machinery eventually resulting in the generation of persistent single-strand breaks. During the next round of DNA replication, these breaks are converted into cytotoxic double-strand breaks [45-47]. The production of these double-strand breaks during the second cell cycle would explain the activation of Chk2 observed.
Alternatively, a “direct signaling” model proposes that recognition of O6-meG/T mismatches by MMR proteins leads directly to the recruitment of DNA damage signaling molecules that initiate the signaling cascade [35,48-50]. How the MMR proteins would directly signal the presence of an O6-meG/T is not clear. Yoshioka et al., suggest that the MSH2-MSH6 complex may recognize these lesions in a manner that distinguishes them from a normal G/T mismatch [35]. The solved crystal structures of hMSH2-hMSH6 interacting with a G/T mismatch or an O6-meG/T do not reveal any significant differences in lesion recognition [51], though structural data that capture the conformation of hMSH2-hMSH6 binding to the mismatch in the presence of ATP is lacking. In vitro data suggest that the O6-meG/T mismatch is capable of inducing the ATPase activity of the hMSH2-hMSH6 heterodimer as well as provoking ADP→ATP exchange to only a slightly reduced extent compared to G/T [52]. Interestingly, mouse models with knocked-in missense alterations in either Msh2 or Msh6 generate MMR proteins that fail to repair DNA mismatches, yet retain the ability to induce damage-dependent cell death [53,54]. The resultant mutant Msh2/Msh6 heterodimers appear to become trapped on DNA mismatches even in the presence of exogenous ATP. The inability of Msh2/Msh6 to bind ATP and release from a mismatch yet still activate a damage response appears inconsistent with a futile cycling model. These results are consistent, however, with a model in which increased resident time of the MMR proteins on the DNA can recruit downstream signaling molecules to activate a damage response.
A direct signaling model does not immediately explain the activation of Chk2 during the second cell cycle, however, there are reports suggesting that hMSH2 can interact with Chk2 [55,56]. In addition, MMR proteins have been shown to interact with Chk1, ATM and ATR [50,55,56]. Thus, the MMR proteins may recruit these signaling kinases to the sites of damage where they become activated. One can not exclude the possibility, however, that excess MMR proteins bound to DNA indirectly result in a damage response by interfering with the DNA replication machinery, generating secondary DNA damage that may activate both Chk1 and Chk2.
In both high and low dose MNNG treated cells, Chk1 becomes phosphorylated early in the first S-phase. However, we also observed the activation of Chk1 in cells undergoing an unperturbed round of DNA replication, with the phosphorylation levels decreasing as the cells cycle through. This result suggests that the early activation of Chk1 is coupled to DNA replication and not necessarily an MNNG-induced damage response. In support of this, both human Chk1 and its activating kinase, ATR, fractionate with the chromatin during normal DNA replication [57,58]. Furthermore, Chk1 phosphorylation on Serine 317 has been shown to be important for efficient DNA replication fork progression [59]. Taken together, these data suggest that Chk1 activation is a normal response to initiation and progression through S-phase.
MNNG treatment appears to induce a persistent activation of Chk1 that continues as cells exit S-phase and into G2. In cells treated with 10 μM MNNG, this persistent activation occurs during the first cell cycle. In 2 μM MNNG treated cells, however, this persistent activation does not occur until the second cell cycle. In both cases, this persistent activation of Chk1 coincides with the formation of MMR protein complexes on chromatin that lack PCNA. As the protein half-life of Chk1 has been reported to be 3.4h [60] and there is clear evidence that the persistent activation of Chk1 in response to alkylation damage is MMR-dependent [30,61], it is possible that continued activation of Chk1 during G2 is mediated by this population of chromatin-bound hMSH2/hMLH1 that signals a DNA damage response independently of DNA repair. It will be interesting to determine whether these chromatin-bound MMR protein complexes can directly recruit Chk1 to damaged DNA resulting in its activation. If such a model is correct, though, it is not clear why this would occur during the first cell cycle in cells treated with high doses of MNNG and during the second cell cycle with low doses. A simplistic explanation is that cells treated with low doses require two rounds of replication to generate enough O6-meG/T lesions to evoke a response.
Another important question regarding the regulation of MMR proteins in the cell is whether these proteins form a complex prior to interacting with DNA. There has been a suggestion that MMR protein complexes form off the chromatin, and that these interactions may be important for proper nuclear localization [38]. We did not find any evidence that hMSH2 and hMLH1 interact away from the chromatin regardless of treatment with MNNG, suggesting that these complexes only form on the DNA. Additionally, neither hMSH2 nor hMLH1 interact with PCNA off the chromatin suggesting that if PCNA does play a role in loading the MMR proteins on to damaged DNA, it is a population of PCNA already associated with the chromatin.
In addition, we observed differences in the localization of the MMR proteins when not bound to chromatin. In the absence of DNA damage, we observed the nucleolar localization of a subpopulation of hMSH6. The nucleolus is a nuclear organelle that is primarily involved in ribosome biogenesis [62]. Consequently, nucleolar formation appears to be coupled to rDNA transcriptional activity. In addition to its role in ribosome biogenesis, there is evidence that the nucleolus functions to segregate proteins in response to stress. In response to DNA damage, PML sequesters MDM2 in the nucleolus allowing for p53 stabilization [63]. While hMSH6 was identified in the nucleolus in undamaged cells, hMLH1 was clearly excluded from the nucleolus. Upon treatment with MNNG, the nucleolar population of hMSH6 was dramatically reduced, likely relocalized to the chromatin. These data may suggest that a subpopulation of hMSH2-hMSH6 is sequestered in the nucleolus away from hMLH1 to prevent non-specific activation of the MMR pathway in the absence of significant damage. Recently, both hMSH6 and hMSH2 were identified in a large-scale screen of proteins enriched in nucleolar localization [64]. However, hMLH1 was not identified in this screen, consistent with our data. Several pertinent questions remain including the mechanism by which hMSH6 is localized to the nucleolus and how the heterodimer is shuttled from the nucleolus to the nucleoplasm after DNA damage.
Supplementary Material
Supplementary Fig. 1. Chromatin localization of MMR proteins synchronized by a nocodazole block. (A) Mock or MNNG-treated HeLa cells were synchronized in mitosis by a nocodazole block and released until the indicated time points. Cell cycle analysis was measured by flow cytometry. Cells began entering S-phase by 10 hours post-release and were mainly in S-phase at 14 hours post-release. Chromatin enriched (B and C) and soluble (D and E) extracts were prepared from cells mock (B and D) or MNNG (C and E) treated as described under A. The membranes were then probed with antibodies against hMSH6, hMSH2, hMLH1, PCNA and Orc2 or Actin as loading controls. The mismatch repair proteins localize to the chromatin during S-phase in mock-treated cells and this localization is enhanced by DNA damage. The chromatin localization of hMSH2 and hMLH1 persists into G2 similar to cells synchronized by the DTB. The soluble fractions were also examined for phospho-Chk1 Serine 317 and total Chk1. Phospho-Chk1 levels increase in cells as they enter S-phase and are further enhanced by DNA damage.
Supplementary Fig. 2. Differential activation of Chk kinases in response to varied concentrations of MNNG. HeLa cells were synchronized by a DTB and treated with O6-BG and either 2 or 10 μM MNNG or a DMSO control. Soluble fractions were prepared at the various time points post-DTB and examined by Western blotting against PhosphoChk1 Serine 317, Chk1-Total, Phospho-Chk2 Threonine 68, Chk2-Total, and Actin. (A and B) Chk1-P S317 levels are increased during S-phase in soluble extracts from DTB synchronized cells treated with 10 μM MNNG (B). The levels decrease in mock-treated cells (A) as they exit S-phase, however, the levels persist throughout the extended S-phase and into G2 in MNNG-treated cells. (C-E) Treatment with 2 μM MNNG (D) results in a delayed activation of both Chk1-P S317 and Chk2-P Thr68 at 24 and 48h. In contrast, treatment with 10 μM MNNG (E) results in the early activation of both Chk2-P Thr 68 and Chk1-P S317; however, Chk1-P S317 begin to diminish at 24h and is strongly reduced by 48h post-release from the DTB.
Supplementary Fig. 3. Chromatin localization of hMSH2 in response to MNNG is not dependent on hMLH1. HeLa cells were transfected with a non-targeting control siRNA (A) or siRNA targeting hMLH1 (B), synchronized by a DTB then treated with 10 μM MNNG and O6-BG or DMSO. Chromatin-enriched fractions were prepared from mock and MNNG-treated cells at the various time points post-DTB and examined by Western blotting against hMSH2, hMLH1, and TBP.
Acknowledgements
We would like to thank Drs. Thomas Kunkel and Alan Clark for providing us with the Hec59 and Hec59+Chrm2 cell lines. This work was supported by NIH grant CA115783 (to C.D.H.) and American Cancer Society grant RSG-07-145-01-CCG (to C.D.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 2.Cannavo E, Marra G, Sabates-Bellver J, Menigatti M, Lipkin SM, Fischer F, Cejka P, Jiricny J. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 3.Bielas JH, Loeb LA. Mutator phenotype in cancer: timing and perspectives. Environ Mol Mutagen. 2005;45:206–213. doi: 10.1002/em.20111. [DOI] [PubMed] [Google Scholar]
- 4.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 5.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 7.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 10.Fishel R. Mismatch repair, molecular switches, and signal transduction. Genes Dev. 1998;12:2096–2101. doi: 10.1101/gad.12.14.2096. [DOI] [PubMed] [Google Scholar]
- 11.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 12.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 13.Heinen CD, Wilson T, Mazurek A, Berardini M, Butz C, Fishel R. HNPCC mutations in hMSH2 result in reduced hMSH2-hMSH6 molecular switch functions. Cancer Cell. 2002;1:469–478. doi: 10.1016/s1535-6108(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 14.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 15.Gu L, Hong Y, McCulloch S, Watanabe H, Li GM. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plotz G, Raedle J, Brieger A, Trojan J, Zeuzem S. hMutSalpha forms an ATP-dependent complex with hMutLalpha and hMutLbeta on DNA. Nucleic Acids Res. 2002;30:711–718. doi: 10.1093/nar/30.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plotz G, Raedle J, Brieger A, Trojan J, Zeuzem S. N-terminus of hMLH1 confers interaction of hMutLalpha and hMutLbeta with hMutSalpha. Nucleic Acids Res. 2003;31:3217–3226. doi: 10.1093/nar/gkg420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matton N, Simonetti J, Williams K. Identification of mismatch repair protein complexes in HeLa nuclear extracts and their interaction with heteroduplex DNA. J Biol Chem. 2000;275:17808–17813. doi: 10.1074/jbc.M909794199. [DOI] [PubMed] [Google Scholar]
- 19.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. PNAS. 2007;104:10956–10961. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional Interaction of Proliferating Cell Nuclear Antigen with MSH2-MSH6 and MSH2-MSH3 Complexes. J. Biol. Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 22.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 23.Lee SD, Alani E. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J Mol Biol. 2006;355:175–184. doi: 10.1016/j.jmb.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 24.Schroering AG, Williams KJ. Rapid induction of chromatin-associated DNA mismatch repair proteins after MNNG treatment. DNA Repair (Amst) 2008;7:951–969. doi: 10.1016/j.dnarep.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 26.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair. 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Duckett D, Drummond JT, Murchie AI, Reardon JT, Sancar A, Lilley DM, Modrich P. Human MutSalpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc Natl Acad Sci U S A. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stojic L, Mojas N, Cejka P, di Pietro M, Ferrari S, Marra G, Jiricny J. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 2004;18:1331–1344. doi: 10.1101/gad.294404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- 32.Schroering AG, Kothandapani A, Patrick SM, Kaliyaperumal S, Sharma VP, Williams KJ. Prolonged Cell Cycle Response of HeLa Cells to Low-Level Alkylation Exposure. Cancer Res. 2009;69:6307–6314. doi: 10.1158/0008-5472.CAN-09-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bostock CJ, Prescott DM, Kirkpatrick JB. An evaluation of the double thymidine block for synchronizing mammalian cells at the G1-S border. Exp Cell Res. 1971;68:163–168. doi: 10.1016/0014-4827(71)90599-4. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe Y, Haugen-Strano A, Umar A, Yamada K, Hemmi H, Kikuchi Y, Takano S, Shibata Y, Barrett JC, Kunkel TA, Koi M. Complementation of an hMSH2 defect in human colorectal carcinoma cells by human chromosome 2 transfer. Mol Carcinog. 2000;29:37–49. [PubMed] [Google Scholar]
- 35.Yoshioka K, Yoshioka Y, Hsieh P. ATR Kinase Activation Mediated by MutSa and MutLa in Response to Cytotoxic O6-Methylguanine Adducts. Mol Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- 37.Moldovan G-L, Pfander B, Jentsch S. PCNA, the Maestro of the Replication Fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Knudsen NO, Nielsen FC, Vinther L, Bertelsen R, Holten-Andersen S, Liberti SE, Hofstra R, Kooi K, Rasmussen LJ. Nuclear localization of human DNA mismatch repair protein exonuclease 1 (hEXO1) Nucl. Acids Res. 2007;35:2609–2619. doi: 10.1093/nar/gkl1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stojic L, P C, Jiricny J. High doses of SN1 type methylating agents activate DNA damage signaling cascades that are largely independent of mismatch repair. Cell Cycle. 2005;4:473–477. doi: 10.4161/cc.4.3.1528. [DOI] [PubMed] [Google Scholar]
- 40.Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci U S A. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 42.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. Embo J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hidaka M, Takagi Y, Takano TY, Sekiguchi M. PCNA-MutS{alpha}-mediated binding of MutL{alpha} to replicative DNA with mismatched bases to induce apoptosis in human cells. Nucl Acids Res. 2005;33:5703–5712. doi: 10.1093/nar/gki878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau PJ, Kolodner RD. Transfer of the MSH2-MSH6 Complex from Proliferating Cell Nuclear Antigen to Mispaired Bases in DNA. J. Biol. Chem. 2003;278:14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- 45.Goldmacher VS, Cuzick RA, Jr., Thilly WG. Isolation and partial characterization of human cell mutants differing in sensitivity to killing and mutation by methylnitrosourea and N-methyl-N′-nitro-N-nitrosoguanidine. J Biol Chem. 1986;261:12462–12471. [PubMed] [Google Scholar]
- 46.York SJ, Modrich P. Mismatch repair-dependent iterative excision at irreparable O6 -methylguanine lesions in human nuclear extracts. J. Biol. Chem. 2006;281:22674–22683. doi: 10.1074/jbc.M603667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cejka P, Mojas N, Gillet L, Schär P, Jiricny J. Homologous Recombination Rescues Mismatch-Repair-Dependent Cytotoxicity of SN1-Type Methylating Agents in S. cerevisiae. Curr Biol. 2005;15:1395–1400. doi: 10.1016/j.cub.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 48.Fishel R. Signaling mismatch repair in cancer. Nat Med. 1999;5:1239–1241. doi: 10.1038/15191. [DOI] [PubMed] [Google Scholar]
- 49.Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci U S A. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc Natl Acad Sci U S A. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the Human MutSalpha DNA Lesion Recognition Complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 52.Berardini M, Mazurek A, Fishel R. The Effect of O6-Methylguanine DNA Adducts on the Adenosine Nucleotide Switch Functions of hMSH2-hMSH6 and hMSH2-hMSH3. J. Biol. Chem. 2000;275:27851–27857. doi: 10.1074/jbc.M003589200. [DOI] [PubMed] [Google Scholar]
- 53.Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, Jin B, Werling U, Parris T, Kurihara N, Umar A, Kucherlapati R, Lipkin M, Kunkel TA, Edelmann W. An Msh2 Point Mutation Uncouples DNA Mismatch Repair and Apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 54.Yang G, Scherer SJ, Shell SS, Yang K, Kim M, Lipkin M, Kucherlapati R, Kolodner RD, Edelmann W. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 55.Adamson A, Beardsley D, Kim W, Gao Y, Baskaran R, Brown K. Methylator-induced, Mismatch Repair-dependent G2 Arrest Is Activated through Chk1 and Chk2. Mol Biol Cell. 2005;16:1513–1526. doi: 10.1091/mbc.E04-02-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown KD, Rathi A, Kamath R, Beardsley DI, Zhan Q, Mannino JL, Baskaran R. The mismatch repair system is required for S-phase checkpoint activation. Nat Genet. 2002;33:80–84. doi: 10.1038/ng1052. [DOI] [PubMed] [Google Scholar]
- 57.Dart DA, Adams KE, Akerman I, Lakin ND. Recruitment of the Cell Cycle Checkpoint Kinase ATR to Chromatin during S-phase. J Biol Chem. 2004;279:16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- 58.Jiang K, Pereira E, Maxfield M, Russell B, Goudelock DM, Sanchez Y. Regulation of Chk1 Includes Chromatin Association and 14-3-3 Binding following Phosphorylation on Ser-345. J Biol Chem. 2003;278:25207–25217. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- 59.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc Natl Acad Sci U S A. 2008;105:20752–20757. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y-W, Otterness D, Chiang G, Xie W, Liu Y-C, Mercurio F, Abraham R. Genotoxic Stress Targets Human Chk1 for Degradation by the Ubiquitin-Proteasome Pathway. Mol Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 61.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech Ageing Devel. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 64.Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. NOPdb: Nucleolar Proteome Database--2008 update. Nucl Acids Res. 2009;37:D181–184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Chromatin localization of MMR proteins synchronized by a nocodazole block. (A) Mock or MNNG-treated HeLa cells were synchronized in mitosis by a nocodazole block and released until the indicated time points. Cell cycle analysis was measured by flow cytometry. Cells began entering S-phase by 10 hours post-release and were mainly in S-phase at 14 hours post-release. Chromatin enriched (B and C) and soluble (D and E) extracts were prepared from cells mock (B and D) or MNNG (C and E) treated as described under A. The membranes were then probed with antibodies against hMSH6, hMSH2, hMLH1, PCNA and Orc2 or Actin as loading controls. The mismatch repair proteins localize to the chromatin during S-phase in mock-treated cells and this localization is enhanced by DNA damage. The chromatin localization of hMSH2 and hMLH1 persists into G2 similar to cells synchronized by the DTB. The soluble fractions were also examined for phospho-Chk1 Serine 317 and total Chk1. Phospho-Chk1 levels increase in cells as they enter S-phase and are further enhanced by DNA damage.
Supplementary Fig. 2. Differential activation of Chk kinases in response to varied concentrations of MNNG. HeLa cells were synchronized by a DTB and treated with O6-BG and either 2 or 10 μM MNNG or a DMSO control. Soluble fractions were prepared at the various time points post-DTB and examined by Western blotting against PhosphoChk1 Serine 317, Chk1-Total, Phospho-Chk2 Threonine 68, Chk2-Total, and Actin. (A and B) Chk1-P S317 levels are increased during S-phase in soluble extracts from DTB synchronized cells treated with 10 μM MNNG (B). The levels decrease in mock-treated cells (A) as they exit S-phase, however, the levels persist throughout the extended S-phase and into G2 in MNNG-treated cells. (C-E) Treatment with 2 μM MNNG (D) results in a delayed activation of both Chk1-P S317 and Chk2-P Thr68 at 24 and 48h. In contrast, treatment with 10 μM MNNG (E) results in the early activation of both Chk2-P Thr 68 and Chk1-P S317; however, Chk1-P S317 begin to diminish at 24h and is strongly reduced by 48h post-release from the DTB.
Supplementary Fig. 3. Chromatin localization of hMSH2 in response to MNNG is not dependent on hMLH1. HeLa cells were transfected with a non-targeting control siRNA (A) or siRNA targeting hMLH1 (B), synchronized by a DTB then treated with 10 μM MNNG and O6-BG or DMSO. Chromatin-enriched fractions were prepared from mock and MNNG-treated cells at the various time points post-DTB and examined by Western blotting against hMSH2, hMLH1, and TBP.